Abstract
Mitochondria are a power organelles generating biochemical energy, ATP, in the cell. Mitochondria play a variety of roles that include integrating extracellular signals and executing critical intracellular events, such as neuronal cell survival and death. Increasing evidence suggests that a cross-talk mechanism between mitochondria and the nucleus is closely related to neuronal function and activity. Nuclear receptors (estrogen receptors, thyroid (T3) hormone receptor, peroxisome proliferators-activated receptor gamma2) and transcription factors (cAMP response binding protein, p53) have been found to target mitochondria and exert pro-survival and pro-death pathways. In this context, the regulation of mitochondrial function via the translocation of nuclear receptors and transcription factors may underlie some of the mechanisms involved in neuronal survival and death. Understanding the function of nuclear receptors and transcription factors in the mitochondria may provide important pharmacological utility in the treatment of neurodegenerative conditions. Thus, the modulation of signaling pathways via mitochondria-targeting nuclear receptors and transcription factors is rapidly emerging as a novel therapeutic target.
1. Introduction
Mitochondria provide energy synthesis in the form of ATP by passing electrons derived from the oxidation of nutrients down the respiratory chain to react with oxygen, using redox energy to translocate protons across the mitochondrial inner membrane (Saraste, M., 1999). A proton electrochemical potential gradient is generated across the inner membrane consisting of a membrane potential (negative inside) and a pH gradient (basic inside) that drives ATP synthesis through F0F1-ATP synthase (Nicholls and Ferguson, 1992). The established roles of mitochondria and its dysfunction in terms of energy deficiency and oxidative stress are of importance in characterizing the pathogenesis of neurodegenerative disorders (Albers and Beal, 2000; Beal, 1993; Zamzami and Kroemer, 2001). Mitochondrial DNA defects or mutations are also closely linked to neurological disorders. Excitotoxicity is a well-established mechanism of neuronal cell death in neurodegenerative disorders. N-Methyl D-Aspartate (NMDA) stimulation induces the decrease of the mitochondrial membrane potential associated with neuronal excitotoxicity. Mitochondria play an indirect role as executioner in the excitotoxic pathway. Mitochondria are the source of 80% or more of the reactive oxygen species generated in neurons. Neuronal toxins and stress blocking mitochondrial functions cause excessive neuronal damage and cell death by the dysregulation of oxyradicals. The mitochondrial toxins malonate and 3-nitropropionic acid produce striatal lesions which mimic Huntington’s disease (HD) and are mediated by excitotoxic mechanisms. Consistent with this finding, mitochondrial electron transport enzymes are altered in HD. Mitochondria in HD lymphoblasts and fibroblasts show an increased susceptibility to depolarization which directly correlates with CAG repeat length (Sawa et al.,1999). The maximal rate of mitochondrial ATP generation in muscle is significantly reduced in both symptomatic HD patients and in presymptomatic HD gene carriers. There has been some debate regarding the vulnerability of transgenic HD mice to neurotoxins. It has been reported that both R6/1 and R6/2 mice are resistant to excitotoxic lesions (Hansson et al. 1999; Hansson et al. 2001). In contrast, we have shown that R6/2 mice are more susceptible to the mitochondrial toxin, 3 nitropropionic acid (Bogdanov et al. 1998). As such, the discrepancy in the findings may be methodological with regards to periodicity and dosage of injections. It is of interest, however, that YAC mice containing full-length huntingtin are more susceptible to excitotoxicity (Zeron et al., 2001). In addition, the fact that NMDA antagonists prolong survival in R6/2 mice clearly implicates excitotoxicity (Schiefer et al. 2002; Ferrante et al. 2002). Rotenone and MPTP/MPP+ also induce mitochondrial dysfunction that is relevant to Parkinson’s disease (PD). Thus, while the role of mitochondrial dysfunction has been proposed in neurodegenerative diseases, the exact mechanism of mitochondrial pathogenesis is unclear. Identification of specific molecules and signaling cascades, which may ultimately lead to neuronal dysfunction and/or cell death, may provide important clues in understanding the pathogenesis of neurological disorders. Recent findings have suggested a paradigm shift in that nuclear receptors and transcription factors target mitochondria and modulate molecular mechanisms to mediate mitochondria-dependent cellular events (Table 1). In this review, we address what nuclear receptors and factors target mitochondria and how they may trigger mitochondrial signaling pathways.
Table 1.
Nuclear receptors and transcription factors that are translocated and found in mitochondria.
| Mitochondria Targeting Proteins | Reference |
|---|---|
| Nuclear receptors | |
| Estrogen receptors (α and β) | Yang et al., 2004; Chen et al., 2004a and b |
| c-ErbAα1 (Thyroid hormone receptor) | Wrutniak et. al., 1995 and 2001 |
| Glucocorticoid receptor (GR) | Ioannou et al., 1988; Demonacos et al, 1996 |
| Nur 77 | Liu et al., 1994; Li et al., 2000; Lin et al., 2004 |
| PPARγ2 | Casas et al., 2000; Smith and Muscat, 2005 |
|
| |
| Transcription Factors | |
| AP1 (c-fos/c-jun) | Ogita et al., 2002 and 2003 |
| CREB | Cammarota et al., 1999; Lee et al., 2005; Ryu et al., 2005 |
| NF-κB | Cogswell et al., 2003 |
| p53 | Marchenko et al., 2000 |
| TFAM (mt-TFA) | Parisi et al., 1991 |
| TFB1M | Falkenberg et al., 2002 |
| TFB2M | Falkenberg et al., 2002 |
2. Nuclear receptors and transcription factors are present in mitochondria Mitochondrial CREB
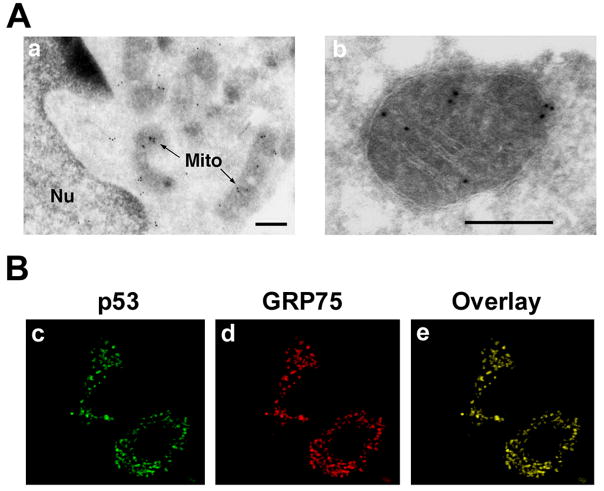
The cAMP response element binding protein (CREB) is a transcription factor known to activate genes that are critical for plasticity, memory, and survival of neurons (Lonze and Ginty, 2002; Mayr and Montminy, 2001). CREB proteins are ubiquitously expressed 38 kDa and 43 kDa proteins, respectively. CREB activates transcription by binding to cAMP response elements upstream of the target gene, CRE; 5′-TGACGTCA-3′. CREB proteins belong to the leucine zipper class of proteins, which form heterodimers in specific combinations. The carboxyl terminus of CREB contains a leucine zipper that is required for dimerization and DNA binding (Meyer and Habener, 1993). The transactivation domain contains several motifs, including a kinase inducible domain containing phosphorylation sites for several kinases including protein kinase A. The involvement of CREB in cell death has been demonstrated in several, non-neuronal paradigms. For example, overexpression of a dominant negative form of CREB, KCREB, in melanoma cells, enhanced apoptosis induced by thapsigargin (Jean et al., 1998). Furthermore, transgenic mice overexpressing KCREB, show increased thymoctye apoptosis. One mechanism by which CREB proteins may inhibit apoptosis is through the upregulation of Bcl-2 expression (Wilson et al., 1996). This has been shown to be the result of CREB binding to a CRE element in the Bcl-2 promoter. Despite these intriguing observations, CREB’s role in regulating oxidative stress-induced apoptosis in HD is not well understood. A number of stimuli, including growth factors, neuropeptides, and neurotransmitters, alter intracellular second messengers in neurons, such as cAMP and calcium, and activate CREB by phosphorylation at serine133. While wild type CREB prevents cell death induced by growth factor deprivation, the over-expression of a dominant negative form of CREB (A-CREB) in sympathetic neurons leads to three changes: decreased Bcl-2 expression, a loss of cytochorome c from the mitochondria, and activation of apoptotic pathways. Recent reports by our group and others show that phosphorylated CREB (pCREB) is present and active in mitochondria and raises the possibility that pCREB may mediate neuronal survival in response to various stimuli by regulating mitochondrial gene expression (Bevilaqua et al., 1999; Lee et al., 2005; Ryu et al., 2005) (Table 1 and Figure 1). CREB may protect striatal neurons by up-regulating mitochondrial genes and interacting with anti-apoptotic molecules that enhances neuronal survival.
Figure 1. Mitochondrial localization of nuclear transcription factors.
A. CREB is presented in the mitochondrial matrix of primary cortical neuron (a and b) (Lee et al, J Biol Chem (2005)). Scale bars: 200nm. B, p53 (c) colocalizes with mitochondrial heat shock protein 70 (mtHsp70/GRP75) (d) in human striatal neurons. Panel e is an overlay image of c and d. Scale bar: 10 μm.
Mitochondrial p53
p53 is a nuclear transcription factor and a tumor suppressor protein. Exposure of cells to DNA damaging agents induces accumulation of p53 protein along with a half-life lengthened by four to five fold without significant changes in its mRNA level (Halaby and Yang, 2007). As p53 has a short half-life in normal cells, it has been found that an efficient steady-state ubiquitination system for p53 is constitutively operating. Interestingly, p53 localizes to the mitochondria under conditions that provoke apoptosis, and that mitochondria-localized p53 is sufficient to launch cell death via the release of cytochrome c (Marchenko et al., 2000; Sansome et al., 2001) (Table 1 and Figure 1). Mitochondrially localized p53 achieves this through direct molecular interaction via its DNA-binding domain with the anti-apoptotic Bcl-2 and Bcl-xL proteins (Mihara et al., 2003). We also found that p53 is continually localized in mitochondria of neuronal cells (Figure 1B), even in the absence of a death stimulus, which coincides with a recent report (Mahyar-Roemer et al., 2004). In spite of the abundance of p53 in the mitochondria in neurons and its importance in neurodegenerative disorders, the exact functions of p53 remain to be determined. It has recently been found that p53 plays a specific role in the mitochondria-associated cellular dysfunction and behavioral abnormalities of HD mice (Bae et al., 2005). Mutant huntingtin (MtHtt) binds to p53 and upreguates p53 transcriptional activity (Bae et al., 2005). This study suggests that p53, in part, links nuclear and mitochondrial pathological features of HD. Furthermore, it is proposed that mitochondrial translocalization of p53 by mtHtt and other cellular stresses, may trigger or accelerate the apoptotic cascade in striatal neurons. In spite of the abundance of p53 in neuronal mitochondria and its importance in neurodegenerative disorders, the exact functions of p53 remain to be determined.
Mitochondrial ER
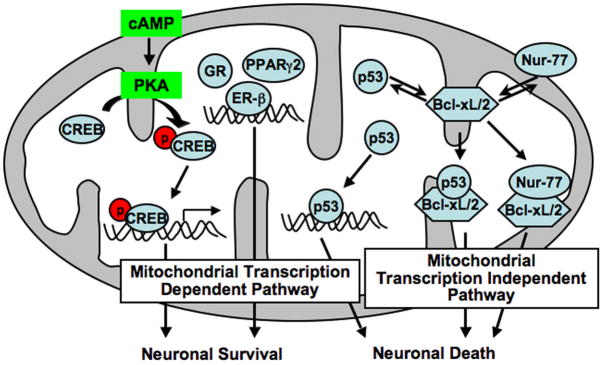
There are two distinct estrogen receptor (ER) subtypes that mediate physiological responses to estrogen. ER alpha and ER beta are encoded by a unique gene, differing in the C-terminal ligand-binding domain and the N-terminal trans-activation domain. Both ERs are expressed in neurons in various stages of development and mediate direct effects of estrogen on brain. Recent findings on the mitochondrial localization of ERs raise question as to how these mitochondrial hormone receptors regulate the transcription of the mitochondrial genome (Yang et al., 2004; Chen et al., 2004a) (Table 1). Mitochondrial ERs bind to estrogen response elements (EREs) or ERE-like sites in the non-coding region of the mitochondrial genome (Chen et al., 2004b; Chen and Yager 2004). Based on previous findings, it is proposed that mitochondrial ERs may act through several different scenarios of mitochondrial transcription pathways (Figure 2). First, mitochondrial ERs can activate the transcription of mitochondrial genes directly through binding to ERE-like sites, which is the mitochondrial ERE-dependent transcription pathway. The classical mode of ERs that target estrogen receptor-responsive areas in the promoter regions of nuclear genes could be applied to mitochondrial gene activation. Second, mitochondrial ERs may trigger other mitochondrial transcription factors such as CREB, and cross talk with other intracellular signaling pathways, which is transcription-dependent but not through binding to ERE-like sites.
Figure 2. The proposed scheme depicts the mitochondrial transcription dependent and transcription independent pathways for neuronal survival and death.
The transcription regulation of mitochondrial gene expression by CREB, ER, GR, and PPARγ2 have been reported. PKA mediated phosphorylation of CREB leads to CREB binding to cyclic AMP response elements (CREs) within the mitochondrial genome that promotes neuronal survival. Alongside p53-dependent transcription from mitochondrial genome may also result in neuronal cell death. It is well established that p53 and Nur-77 complexes with Bcl-xL and Bcl-2 present on mitochondrial membrane which subsequently triggers the downstream cascade of apoptosis via mitochondrial transcription independent pathway.
Mitochondrial PPARγ2
The peroxisome proliferator activated receptors (PPARs) are members of the nuclear hormone receptor superfamily and are involved in the regulation of lipid metabolism. Three distinct subtypes have been identified: PPARα, PPAR γ and PPAR δ. While PPAR δ is ubiquitously expressed, PPAR α is predominantly expressed in the liver and PPAR γ in the adipose tissue. PPAR α regulates fatty acid catabolism, PPAR γ regulates adipocyte differentiation and PPAR δ regulates cholesterol efflux, lipid catabolism and energy expenditure (Smith and Muscat 2005). Casas et al (2000) have found that a 45 kDa protein (mitochondrial PPAR) related to PPAR γ2 isoform is located in the mitochondrial matrix and binds specifically in the D-loop region of mitochondrial DNA which suggests that mitochondrial PPAR may also serve to regulate mitochondrial gene expression (Figure 2).
Mitochondrial Nur 77
The nuclear-orphan receptor Nur-77 (also known as TR3) has been identified as an immediate early gene induced by serum and growth factors (Hazel et al., 1988). The most fascinating aspect regarding Nur-77 is that it can exhibit differential functions of cell survival or cell death. While overexpression of Nur-77 in cancer cells is linked to survival, suppression of the expression is associated with inhibition of the transformation phenotype (Ke al., 2004; Moll et al., 2006). Studies on apoptotic thymocytes and T-cell hybridomas demonstrate that high levels of Nur-77 in these cells are correlated to apoptotic function (Liu et al, 1994; Woronicz et al., 1994). Transactivation of nuclear gene expression by Nur-77 is thought to play a part in its apoptotic function. It is also emerging from recent studies that Nur-77 mediates apoptosis by translocating from the nucleus to the mitochondria where it interacts with Bcl-2 and induces cytochrome c release (Jeong et al, 2003; Lin et al., 2004) (Figure 2).
Other mitochondrial nuclear receptors and transcription factors
Glucocorticoid receptor (GR) and c-Erb Aα1 (triiodothyronine (T3) receptor) have been found in the mitochondria (Table 1). On the basis of these findings, glucocorticoid and thyroid hormones effect directly on the mitochondrial genes concomitant with the effects on nuclear genes involving similar molecular mechanisms to those mediating glucocorticoid and thyroid hormone actions on nuclear gene transcription (Ioannou et al., 1988; Demonacos et al., 1996). c-Erb Aα1 specifically binds to a direct repeat 2 sequence located in the D-loop of the mitochondrial genome. Moreover, expression of a truncated form of the c-Erb Aα1 nuclear receptor is associated with a mitochondrial localization and a stimulation of mitochondrial activity. These results supply evidence of the localization of a member of the nuclear receptor superfamily in the mitochondrial matrix involved in the regulation of mitochondrial activity that could act as a mitochondrial T3-dependent transcription factor. However, the T-binding 43-kDa protein related to c-Erb A 1 is not detectable in adult rat brain mitochondria, in agreement with the previously reported lack of T receptors in these mitochondria (Sterling et al., 1977; Wrutniak et al., 1995). Thus, the mitochondrial presence of other c-Erb subunits in neurons remains to be investigated.
Ogita and colleagues have found that kainate injection increases AP-1 complex (c-fos and c-jun) translocation into mitochondria followed by enhanced DNA-binding activity to mitochondrial AP-1-like sites in mouse hippocampus (Ogita et al., 2002 and 2003) (Table 1). They have demonstrate that AP-1-like sites, but not the genuine AP-1 site (5,-TGAGTCA-3′), are found in the non-coding region (D-loop) of the mitochondrial genome.
It has emerged from recent studies that the nuclear factor- kappa B (NF-kB) and IkB localize in the mitochondria and NF-kB also regulates the mitochondrial gene expression (Cogswell et al., 2003) (Table 1). The NF-kB family comprises 5 members: NF-kB1 (p500), RelA(p65), NF-kB2(p52) RelB and c-Rel. The NF-kB predominantly resides in the cytoplasm complexed with inhibitor proteins (members of the IkB family). Activation of NF-kB occurs upon release from the inhibitor complex by phosphorylation and subsequent degradation of IkB proteins, mostly by IkB kinase (IKK). NF-kB exerts its antiapoptotic effects by inducing antiapoptotic genes thereby promoting cell survival and proliferation. Thus, it antagonizes the proapoptotic functions of p53. NF-kB has been shown to negatively regulate p53 stability by modulating the p53 E3 ubiquitin ligase Mdm2 levels (Tergaonkar et al.,2002). NF-kB was shown to negatively regulate the mitochondrially encoded cytochrome c oxidase III and cytochrome b in response to TNFα stimulation (Cogswell et al., 2003).
3. Traveling mechanisms of nuclear transcription factors to mitochondria
Mitochondrial DNA (mt DNA) encodes only a small fraction of the mitochondrial proteins with the vast majority being nuclear encoded and thus have to be imported into mitochondria. Mitochondria are bound by two membranes: the outer mitochondrial membrane (OMM) and the inner mitochondrial membrane (IMM). While, the outer membrane is highly permeable and non selective, the inner membrane has a limited permeability. The voltage-dependent anion channel (VDAC) in the OMM, a large water filled pore, allows polar molecules up to 5 kDa to pass freely (Lee and Thevenod, 2006). In contrast, the inner membrane allows only a few molecules to pass freely with the majority of the small molecules requiring specific transporters.
The nuclear encoded mitochondrial proteins contain signal sequences either at the N-terminus or within their sequences that target them to the mitochondria. The proteins are synthesized on ribosomes in the cytosol and they associate with chaperones that help in their mitochondrial translocation. One of the major chaperones in this category is the mitochondrial heat shock protein 70 (mtHsp70)/GRP75. Interestingly, p53 has been shown to localize to the mitochondria by associating with mtHsp70 at the onset of p53 dependent apoptosis. Based on the signal information in the precursor protein, it could be targeted to any of the four locations: the mitochondrial outer membrane, inner membrane, the intermembrane space or the mitochondrial matrix. This signal also determines the translocation pathway and the energy requirement (Hood and Joseph, 2004). Further, it is now being increasingly suggested in literature that the translocation process happens co-translationally (Mukhopadhyay et al., 2004; Ni et al., 1999). This process is mediated by translocases present in the mitochondrial outer and inner membranes. These comprise the translocase of the outer membrane (TOM) complex, which is the entrance gate for almost all mitochondrial proteins from the cytosol, and the TIM complex of the inner membrane for transporting proteins into/across the inner membrane. These translocases are multi-subunit protein assemblies utilizing ATP and the membrane potential as energy sources (Neupert and Herrmann, 2007). After the initial entry through the TOM complex, distinct pathways sort proteins into different mitochondrial compartments, such as, sorting and assembly machinery complex (SAM) for the β-barrel proteins of the outer membrane, the mitochondrial intermembrane space import and assembly (MIA) pathway for intermembrane space proteins bearing cysteine motifs, and the TIM 22/TIM 23 complexes for targeting to inner membrane or mitochondrial matrix (Bohnert et al., 2007). Further study is necessary to elucidate whether TOM/TIM complexes are involved in importation of nuclear receptors and transcription factors into mitochondria.
4. Novel roles of nuclear receptors and transcription factors in mitochondria
4-1. Mitochondrial Transcription-Dependent Pathway
Mitochondrial DNA translates 37 genes; 22 tRNA genes, 2 rRNA genes, and 13 polypeptide encoding genes. All 13 mitochondrially-encoded proteins are mainly components of the respiratory chain or the F0F1-ATP synthase (Wallace, 1999). Many subunits of mitochondrial oxidative phosphorylation enzymes are encoded by nuclear DNA, translated on cytoplasmic ribosomes, imported into mitochondria and assembled there into functional complexes along with mitochondrially-encoded proteins (Neupert, 1997; Schatz, 1996). The two strands of the mammalian mtDNA, named on the basis of their buoyant density heavy- (H) and light- (L) strand, are replicated asynchronously and asymmetrically (Clayton, 1982, 1987). The mammalian mtDNA contains three promoters (L, H1 and H3) located in the non-coding D-loop region. The transcription of the H-strand starts at two sites, ITH1 and ITH2 whereas L-strand transcription starts at a single site, ITL1 (Montoya et al., 1983). Transcripts synthesized from ITL1 and ITH2, which is located within the tRNAphe gene, are polycistronic and their corresponding transcription units encompass the complete L and H DNA strands, respectively (Taanman, 1999). ITH1 is placed just before the tRNAphe gene, and the transcription unit ends in a strong termination signal located in the downstream of the 16S rRNA gene. The termination signal is evolutionarily conserved (Valverde et al., 1994) and binds a trans-acting factor called mTERF (Fernandez-Silva et al., 1997, 2003). Therefore, the main function of the H1 promoter is to regulate synthesis of the two mitochondrial rRNAs. The primary polycistronic transcripts are further processed to monocistronic or bicistronic mRNAs with the tRNAs acting as punctuation signals (Ojala et al., 1981). The enzymatic activities responsible for the 5′ and 3′ processing of tRNAs have been characterized (Rossmanith et al., 1995), but the genes encoding the mitochondrial RNase P and precursor tRNA 3′ endonuclease remain to be identified (Garesse and Vallejo, 2001). Despite the fact that mitochondrial mRNAs are polyadenylated, the mitochondrial poly (A) polymerase and potential associated factors also remain to be further characterized. The mitochondrial transcriptional machinery is relatively simple compared to the nuclear machinery and consists of a single unspecific RNA polymerase that is evolutionarily conserved to bacteriophage T7, T3 and SP6 RNA polymerases and at least a specificity factor, mitochondrial transcription factor A (mtTFA/TFAM), a small protein that belongs to the family of high mobility group (HMG) DNA binding factors (Dairaghi et al., 1995). Not only does mtTFA play a role as a transcription factor, but also is essential for mtDNA maintenance in yeast (Diffley and Stillman, 1991) and most likely in mammals (Larsson et al., 1998), due to its binding capacity to non-specific DNA. Thus, mtTFA shows a dual role as a mitochondrial specific factor/transcriptional activator and in mitochondrial genome packaging. While it has been proposed that mtTFA may interact with the mitochondrial transcription factor B (mtTFB), the mitochondrial transcription machinery and mechanisms are poorly characterized (Shadel and Clayton, 1997).
CREB and ERs are nuclear transcription factors that have been shown to act in the nucleus to regulate gene expression. Other’s and we provide evidence supporting the hypothesis that CREB and ERs also target mitochondrial DNA (Yang et al., 2004; Chen et al., 2004a and b; Lee et al, 2005; Ryu et al., 2005). These studies suggest that some nuclear transcription factors may participate in regulating mitochondrial function through transcriptional regulation of mitochondrial DNA (Figure 2). For example, CREB is localized in the mitochondrial matrix as well as nuclei in rat hippocampus (Cammarota et al., 1999; Lee et al., 2005) (Figure 1). Mitochondrial CREB is phosphorylated on its serine-133 after one trial of inhibitory avoidance training procedures in rat hippocampus (Bevilaqua et al., 1999). While direct evidence that CREB regulates mitochondrial gene expression has been lacking, we have demonstrated that CREB is present in the inner mitochondrial matrix of neurons, and directly binds to cAMP response elements (CREs) in the promoter (D-loop) regions of rodent mitochondrial DNA (mtDNA). Putative mitochondrial CRE-like sites have been found by chromatin immunoprecipitation (ChIP), EMSA and DNAse footprint analysis (Lee et al., 2005; Ryu et al., 2005). It seems likely that the secondary structure of mitochondrial D-loop DNA is important for its occupancy by mitochondrial CREB when long D-loop template DNA is used for baiting CREB. Our group has shown that disruption of CREB activity in the mitochondria decreases the expression of a subset of mitochondrial genes, including the ND5 subunit of complex 1, reduces mitochondrial respiration, and increases susceptibility to 3-nitropropionic acid, a mitochondrial toxin known to induce a clinical and pathological phenotype similar to HD (Lee et al., 2005). In addition, transgenic HD mice contain decreased levels of mitochondrial CREB, correlating with lowered mitochondrial ND5 gene expression. These results show that CREB is transcriptionally active in mitochondria (Figure 2). It further suggests that regulation of mitochondrial gene expression by mitochondrial CREB may underlie some of the established protective effects of CREB, and raises the possibility that decreased mitochondrial CREB activity contributes to the mitochondrial dysfunction and neuronal loss in HD as well as in other neurodegenerative conditions.
4-2. Mitochondrial Transcription-Independent Pathway
The transcription independent pathway of p53 mediated apoptosis has been proposed since studies showed that p53 dependent apoptosis proceeded in the absence of transcription or protein synthesis (Caelles et al., 1994; Marchenko et. al, 2000) (Figure 2). Studies with p53 mutants that are inactive in transcription but capable of inducing apoptosis also support a mechanism that is independent of transcription (Haupt et al., 1995). Marchenko and coworkers have shown that a fraction of stress-induced p53 protein rapidly localizes to mitochondria at the onset of p53 dependent apoptosis that precedes the release of cytochrome c and procaspase-3 activation (Marchenko et. al, 2000).
The anti-apoptotic proteins Bcl-xL and Bcl2 present at the outer mitochondrial membrane (OMM) stabilize the mitochondrial membrane. Interestingly, p53 destabilizes the membrane by complexing with these proteins. Furthermore, p53 directly activates the cytoplasmic proapoptotic protein Bax, inducing Bax oligomerization and permeabilization of the outer membrane (Mihara et al., 2003). Permeabilization of the mitochondrial outer membrane leads to release of cytochrome c along with other apoptogenic factors which activate the caspase leading to apoptosis. These studies have also demonstrated that p53 interacts with Bcl-xL and Bcl2 through its DNA binding domain. Studies with tumor-derived mutants of p53 showed that these mutant proteins lose or are severely compromised in their abilities to form inhibitory complexes with Bcl-xL or Bcl-2 (Mihara et al., 2003; Tomita et al., 2006) (Figure 2). Chipuk et al. (2005) have shown that the p53/Bcl-xL complex requires an additional ‘p53 upregulated modifier of apoptosis’ (PUMA) to displace p53 from Bcl-xL and engage cell death, thus coupling the nuclear and cytoplasmic proapoptotic functions of p53.
5. Therapeutic regulation of mitochondrial function
5-1. Mitochondria as a Therapeutic Target
Mitochondria are thread shape organelles consisting of several compartments each with different compositions and functions (Murphy and Smith, 2000). Therefore, mitochondria have been considered as intracellular targets for small compounds (Murphy, 1997). The porous mitochondrial outer membrane is permeable to molecules which are smaller than ~5kDa. The mitochondrial intermembrane space contains many specific proteins, but is continuous with the cytoplasm for small molecules. The mitochondrial inner membrane with a convoluted and invaginated structure, contains oxidative phosphorylation enzymes and a series of metabolic pathways. Mitochondrial membrane damage contributes to the pathogenesis of many neurodegenerative diseases. Neuronal cell fate is dependent on mitochondria that play key roles in apoptotic and necrotic cell death. Necrotic neuronal cell death occurs in response to acute damage and insults and results in rapid, uncontrolled death with subsequent cell lysis and an inflammatory response. Necrotic cell death follows ATP depletion and cellular calcium overloading, with extensive mitochondrial damage leading to necrotic cell death (Nicotera et al., 1998). Otherwise, apoptotic neuronal cell death accompanies the activation of cell death program that causes the ordered self-destruction of the cell, ending with phagocytosis without leakage of damaging contents and thus no inflammatory response. The difference between apoptotic and necrotic neuronal death is rather arbitrary as completion of the apoptotic program requires ATP, and if ATP levels drop lower than a critical threshold after initiation of apoptosis, apoptosis is aborted and neurons die by necrosis (Leist and Nicotera, 1998).
5-2. Mitochondrial CREB regulation by antioxidants
The catalytic subunit of protein kinase A (PKA) is found in the mitochondrial matrix to phosphorylate mitochondrial CREB in neurons (Ryu et al., 2005). The therapeutic approaches increasing mitochondrial PKA and mitochondrial CREB activity may provide a novel direction in both preclinical and human trials. Deferoxamine (DFO), an antioxidant and iron chelator known to inhibit oxidative stress-induced cell death, activates mitochondrial PKA and increased mitochondrial CREB phosphorylation (Ser 133) (Ryu et al, 2005). DFO increases CREB binding to CRE in the mitochondrial D-loop DNA and D-loop CRE-driven luciferase activity. In contrast, KT5720, a specific inhibitor of PKA, reduces DFO-mediated neuronal survival against oxidative stress induced by glutathione depletion. Neuronal survival by DFO may be, in part, mediated by the mitochondrial PKA-dependent pathway. These results suggest that the regulation of mitochondrial function via the mitochondrial PKA and CREB pathways may underlie some of the salutary effects of DFO in neurons. Taken together, the idea of targeting biologically active molecules to the mitochondria is to modulate selective mitochondrial functions in a specific manner. Therapeutic strategies will allow mitochondria to better cope with oxidative stress, mitochondrial damage by excitotoxicity, and maintain efficient oxidative phosphorylation and respiratory function. This study provides a novel mechanism for preventing mitochondrial transcriptional dysfunction in neurodegenerative conditions and in the design of applicable therapeutic treatments to modulate mitochondrial hormone receptors and transcription factors.
5-3. Specific Estrogen Receptor Modulators (SERMs) to Target Mitochondria
Estrogen attenuates NMDA receptor-mediated excitotoxic neuronal death and oxidative neuronal death (Weaver et al., 1997; Montal, 1998; Kajta et al., 2002; Linford et al., 2002; Dribben et al., 2003; Xue and Hay 2003). Estrogen has a number of neurotrophic effects mediated via different signaling pathways, including activation of PKA, ERK and phosphatidylinositol 3-kinase (PI3K) cascades and inactivation of glycogen synthase kinase 3beta (GSK3 beta) (Mendez et al., 2003; Yu et al., 2004). PKA localizes to the matrix of mitochondria in neurons. Thus, it has been proposed that estrogen and agonist-dependent mitochondrial transcription is, in part, mediated via the mitochondrial PKA signaling pathways.
The important structural motif that elicits estrogenic effects is a phenol ring that is relatively unhindered and attached to a bulky hydrophobic structure (Schultz et al., 2002). The phenolic A ring is related to its neuroprotective function (Green et al., 1997). Steroids with a hydroxyl group in the C3 position of the A ring provides an antioxidant property. E2 can suppress intracellular ROS and prevent neurons from oxidative stress-induced cell death. However, the antioxidant property requires a higher concentration of E2 (10–100 microM). Furthermore, antiapoptotic neuroprotection may be blocked by ICI 182,720, which has hydroxyl group in C3 (Murashov et al., 2004; Stroppolo et al., 2004). Therefore, it is unlikely that the anti-cell death effect is solely due to the antioxidant property of E2. As neuroprotective molecules, SERMs may act through both the mitochondria-dependent and independent signaling pathway. First, they may activate the transcription of mitochondrial genes directly through binding to mitochondrial ERs and subsequently to estrogen response element (ERE) in mitochondrial genome, which is the mitochondrial transcription-dependent pathway (Yager and Chen, 2007). This classical mode of estrogen action works through the activation of estrogen receptors that target estrogen receptor-responsive areas in the promoter regions of mitochondrial genomes. Therefore, mitochondrial estrogen receptors that are activated by estrogen directly act as mitochondrial transcription factors (Chen et al., 2004a and b; Yang et al., 2004). Second, SERMs directly regulate gene expression through ERs and ERE, as well as indirectly activating gene transcription by performing a crosstalk with various intracellular signaling pathways (Leong et al., 2004). In this case, it is predicted that estrogenic compounds bind to estrogen receptors that do not directly bind to the mitochondrial ERE, but rather interact with other signaling cascades in the mitochondrial matrix. Such signaling partners of interaction may include the mitochondrial PKA and the CREB-signaling processes. Third, SERMs may directly effect mitochondrial membranes by modulating Ca2+ fluxes and protect neurons through their antioxidant effects, which promote the transcription-independent pathway (Farhat et al., 1996).
6. Conclusion
The extraordinary dependence of neurons on the energy provided by mitochondrial oxidative metabolism is directly linked to neurodegenerative conditions. The central role of mitochondria in neurodegeneration has become apparent over the last decade as the molecular mechanisms causing neuronal cell death have been underscored. Previous findings on the mitochondrial localization and function of nuclear receptors and transcription factors have unraveled interesting mechanisms in the mitochondria. The effectiveness of drug treatment may depend upon targeting bioactive molecules to the appropriate organ, cell type, and subcellular organelle. Therefore, future studies using small compounds to target the mitochondria directly or to modulate nuclear receptors and transcription factors that subsequently convey the signal to the mitochondria, may contribute to improve mitochondrial functions in a specific manner. We expect that novel therapeutic strategies will enable nuclear receptors and transcription factors via mitochondria to better cope with oxidative stress, excitotoxicity, and transcriptional dysfunction, as well as maintain efficient respiratory function in neurons.
References
- Albers DS, Beal MF. Mitochondrial dysfunction and oxidative stress in aging and neurodegenerative disease. J Neural Transm Suppl. 2000;59:133–154. doi: 10.1007/978-3-7091-6781-6_16. [DOI] [PubMed] [Google Scholar]
- Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH, Montell C, Ross CA, Snyder SH, Sawa A. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47(1):29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Beal MF, Hyman BT, Koroshetz W. Do defects in mitochondrial energy metabolism underlie the pathology of neurodegenerative diseases? Trends Neurosci. 1993;16(4):125–131. doi: 10.1016/0166-2236(93)90117-5. [DOI] [PubMed] [Google Scholar]
- Bevilaqua LR, Cammarota M, Paratcha G, de Stein ML, Izquierdo I, Medina JH. Experience-dependent increase in cAMP-responsive element binding protein in synaptic and nonsynaptic mitochondria of the rat hippocampus. Eur J Neurosci. 1999;11(10):3753–3756. doi: 10.1046/j.1460-9568.1999.00830.x. [DOI] [PubMed] [Google Scholar]
- Bogdanov MB, Ferrante RJ, Kuemmerle S, Klivenyi P, Beal MF. Increased vulnerability to 3-nitropropionic acid in an animal model of Huntington’s disease. J Neurochem. 1998;71(6):2642–2644. doi: 10.1046/j.1471-4159.1998.71062642.x. [DOI] [PubMed] [Google Scholar]
- Bohnert M, Pfanner N, van der Laan M. A dynamic machinery for import of mitochondrial precursor proteins. FEBS Lett. 2007;581(15):2802–2810. doi: 10.1016/j.febslet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370(6486):220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- Cammarota M, Paratcha G, Bevilaqua LR, Levi de Stein M, Lopez M, Pellegrino de Iraldi A, Izquierdo I, Medina JH. Cyclic AMP-responsive element binding protein in brain mitochondria. J Neurochem. 1999;72(6):2272–2277. doi: 10.1046/j.1471-4159.1999.0722272.x. [DOI] [PubMed] [Google Scholar]
- Casas F, Domenjoud L, Rochard P, Hatier R, Rodier A, Daury L, Bianchi A, Kremarik-Bouillaud P, Becuwe P, Keller J, Schohn H, Wrutniak-Cabello C, Cabello G, Dauca M. A 45 kDa protein related to PPARgamma2, induced by peroxisome proliferators, is located in the mitochondrial matrix. FEBS Lett. 2000;478(1–2):4–8. doi: 10.1016/s0014-5793(00)01814-7. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am J Physiol Endocrinol Metab. 2004a;286(6):E1011–1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Eshete M, Alworth WL, Yager JD. Binding of MCF-7 cell mitochondrial proteins and recombinant human estrogen receptors alpha and beta to human mitochondrial DNA estrogen response elements. J Cell Biochem. 2004b;93(2):358–373. doi: 10.1002/jcb.20178. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Yager JD. Estrogen’s effects on mitochondrial gene expression: mechanisms and potential contributions to estrogen carcinogenesis. Ann N Y Acad Sci. 2004;1028:258–272. doi: 10.1196/annals.1322.030. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309(5741):1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Nuclear gene products that function in mitochondrial DNA replication. Philos Trans R Soc Lond B Biol Sci. 1987;317(1187):473–482. doi: 10.1098/rstb.1987.0074. [DOI] [PubMed] [Google Scholar]
- Cogswell PC, Kashatus DF, Keifer JA, Guttridge DC, Reuther JY, Bristow C, Roy S, Nicholson DW, Baldwin AS., Jr NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J Biol Chem. 2003;278(5):2963–2968. doi: 10.1074/jbc.M209995200. [DOI] [PubMed] [Google Scholar]
- Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim Biophys Acta. 1995;1271(1):127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- Demonacos CV, Karayanni N, Hatzoglou E, Tsiriyiotis C, Spandidos DA, Sekeris CE. Mitochondrial genes as sites of primary action of steroid hormones. Steroids. 1996;61(4):226–232. doi: 10.1016/0039-128x(96)00019-0. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci U S A. 1991;88(17):7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dribben W, Nemmers B, Nardi A, Taylor G, Olney J, Farber N. Chronic but not acute estradiol treatment protects against the neurodegenerative effects of N-methyl-D-aspartate receptor antagonists. Endocrine. 2003;21(1):53–58. doi: 10.1385/endo:21:1:53. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31(3):289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- Farhat MY, Abi-Younes S, Ramwell PW. Non-genomic effects of estrogen and the vessel wall. Biochem Pharmacol. 1996;51(5):571–576. doi: 10.1016/s0006-2952(95)02159-0. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88(1):41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G. The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. Embo J. 1997;16(5):1066–1079. doi: 10.1093/emboj/16.5.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF. Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington’s disease. J Neurosci. 2002;22(5):1592–1599. doi: 10.1523/JNEUROSCI.22-05-01592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garesse R, Vallejo CG. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263(1–2):1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- Green PS, Gordon K, Simpkins JW. Phenolic A ring requirement for the neuroprotective effects of steroids. J Steroid Biochem Mol Biol. 1997;63(4–6):229–235. doi: 10.1016/s0960-0760(97)00124-6. [DOI] [PubMed] [Google Scholar]
- Halaby MJ, Yang DQ. p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene. 2007;395(1–2):1–7. doi: 10.1016/j.gene.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Hansson O, Castilho RF, Korhonen L, Lindholm D, Bates GP, Brundin P. Partial resistance to malonate-induced striatal cell death in transgenic mouse models of Huntington’s disease is dependent on age and CAG repeat length. J Neurochem. 2001;78(4):694–703. doi: 10.1046/j.1471-4159.2001.00482.x. [DOI] [PubMed] [Google Scholar]
- Hansson O, Petersen A, Leist M, Nicotera P, Castilho RF, Brundin P. Transgenic mice expressing a Huntington’s disease mutation are resistant to quinolinic acid-induced striatal excitotoxicity. Proc Natl Acad Sci U S A. 1999;96(15):8727–8732. doi: 10.1073/pnas.96.15.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9(17):2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- Hazel TG, Nathans D, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci U S A. 1988;85(22):8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood DA, Joseph AM. Mitochondrial assembly: protein import. Proc Nutr Soc. 2004;63(2):293–300. doi: 10.1079/PNS2004342. [DOI] [PubMed] [Google Scholar]
- Ioannou IM, Tsawdaroglou N, Sekeris CE. Presence of glucocorticoid responsive elements in the mitochondrial genome. Anticancer Res. 1988;8(6):1405–1409. [PubMed] [Google Scholar]
- Jean D, Harbison M, McConkey DJ, Ronai Z, Bar-Eli M. CREB and its associated proteins act as survival factors for human melanoma cells. J Biol Chem. 1998;273(38):24884–24890. doi: 10.1074/jbc.273.38.24884. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Park JS, Moon B, Kim MC, Kim JK, Lee S, Suh H, Kim ND, Kim JM, Park YC, Yoo YH. Orphan nuclear receptor Nur77 translocates to mitochondria in the early phase of apoptosis induced by synthetic chenodeoxycholic acid derivatives in human stomach cancer cell line SNU-1. Ann N Y Acad Sci. 2003;1010:171–177. doi: 10.1196/annals.1299.029. [DOI] [PubMed] [Google Scholar]
- Kajta M, Lason W, Bien E, Marszal M. Neuroprotective effects of estrone on NMDA-induced toxicity in primary cultures of rat cortical neurons are independent of estrogen receptors. Pol J Pharmacol. 2002;54(6):727–729. [PubMed] [Google Scholar]
- Ke N, Claassen G, Yu DH, Albers A, Fan W, Tan P, Grifman M, Hu X, Defife K, Nguy V, Meyhack B, Brachat A, Wong-Staal F, Li QX. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64(22):8208–8212. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18(3):231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim CH, Simon DK, Aminova LR, Andreyev AY, Kushnareva YE, Murphy AN, Lonze BE, Kim KS, Ginty DD, Ferrante RJ, Ryu H, Ratan RR. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J Biol Chem. 2005;280(49):40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WK, Thevenod F. A role for mitochondrial aquaporins in cellular life-and-death decisions? Am J Physiol Cell Physiol. 2006;291(2):C195–202. doi: 10.1152/ajpcell.00641.2005. [DOI] [PubMed] [Google Scholar]
- Leist M, Nicotera P. Apoptosis versus necrosis: the shape of neuronal cell death. Results Probl Cell Differ. 1998;24:105–135. doi: 10.1007/978-3-540-69185-3_6. [DOI] [PubMed] [Google Scholar]
- Leong H, Riby JE, Firestone GL, Bjeldanes LF. Potent ligand-independent estrogen receptor activation by 3,3′-diindolylmethane is mediated by cross talk between the protein kinase A and mitogen-activated protein kinase signaling pathways. Mol Endocrinol. 2004;18(2):291–302. doi: 10.1210/me.2003-0196. [DOI] [PubMed] [Google Scholar]
- Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD, Lin B, Chen G, Lu J, Lin F, Xie Z, Fontana JA, Reed JC, Zhang X. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289(5482):1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, Dawson MI, Reed JC, Zhang XK. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116(4):527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- Linford NJ, Dorsa DM. 17beta-Estradiol and the phytoestrogen genistein attenuate neuronal apoptosis induced by the endoplasmic reticulum calcium-ATPase inhibitor thapsigargin. Steroids. 2002;67(13–14):1029–1040. doi: 10.1016/s0039-128x(02)00062-4. [DOI] [PubMed] [Google Scholar]
- Liu ZG, Smith SW, McLaughlin KA, Schwartz LM, Osborne BA. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature. 1994;367(6460):281–284. doi: 10.1038/367281a0. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Mahyar-Roemer M, Fritzsche C, Wagner S, Laue M, Roemer K. Mitochondrial p53 levels parallel total p53 levels independent of stress response in human colorectal carcinoma and glioblastoma cells. Oncogene. 2004;23(37):6226–6236. doi: 10.1038/sj.onc.1207637. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275(21):16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Mendez P, Azcoitia I, Garcia-Segura LM. Estrogen receptor alpha forms estrogen-dependent multimolecular complexes with insulin-like growth factor receptor and phosphatidylinositol 3-kinase in the adult rat brain. Brain Res Mol Brain Res. 2003;112(1–2):170–176. doi: 10.1016/s0169-328x(03)00088-3. [DOI] [PubMed] [Google Scholar]
- Meyer TE, Habener JF. Cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr Rev. 1993;14(3):269–290. doi: 10.1210/edrv-14-3-269. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11(3):577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 - transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25(34):4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- Montal M. Mitochondria, glutamate neurotoxicity and the death cascade. Biochim Biophys Acta. 1998;1366(1–2):113–126. doi: 10.1016/s0005-2728(98)00124-8. [DOI] [PubMed] [Google Scholar]
- Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34(1):151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Ni L, Weiner H. A co-translational model to explain the in vivo import of proteins into HeLa cell mitochondria. Biochem J. 2004;382(Pt 1):385–392. doi: 10.1042/BJ20040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashov AK, Islamov RR, McMurray RJ, Pak ES, Weidner DA. Estrogen increases retrograde labeling of motoneurons: evidence of a nongenomic mechanism. Am J Physiol Cell Physiol. 2004;287(2):C320–326. doi: 10.1152/ajpcell.00542.2003. [DOI] [PubMed] [Google Scholar]
- Murphy MP. Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol. 1997;15(8):326–330. doi: 10.1016/S0167-7799(97)01068-8. [DOI] [PubMed] [Google Scholar]
- Murphy MP, Smith RA. Drug delivery to mitochondria: the key to mitochondrial medicine. Adv Drug Deliv Rev. 2000;41(2):235–250. doi: 10.1016/s0169-409x(99)00069-1. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Ni L, Heard TS, Weiner H. In vivo mitochondrial import. A comparison of leader sequence charge and structural relationships with the in vitro model resulting in evidence for co-translational import. J Biol Chem. 1999;274(18):12685–12691. doi: 10.1074/jbc.274.18.12685. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Ferguson SJ. Bioenergetics. Vol. 2. London, UK: Academic Press; 1992. [Google Scholar]
- Nicotera P, Leist M, Ferrando-May E. Intracellular ATP, a switch in the decision between apoptosis and necrosis. Toxicol Lett. 1998;102–103:139–142. doi: 10.1016/s0378-4274(98)00298-7. [DOI] [PubMed] [Google Scholar]
- Ogita K, Fujinami Y, Kitano M, Yoneda Y. Transcription factor activator protein-1 expressed by kainate treatment can bind to the non-coding region of mitochondrial genome in murine hippocampus. J Neurosci Res. 2003;73(6):794–802. doi: 10.1002/jnr.10720. [DOI] [PubMed] [Google Scholar]
- Ogita K, Okuda H, Kitano M, Fujinami Y, Ozaki K, Yoneda Y. Localization of activator protein-1 complex with DNA binding activity in mitochondria of murine brain after in vivo treatment with kainate. J Neurosci. 2002;22(7):2561–2570. doi: 10.1523/JNEUROSCI.22-07-02561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290(5806):470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252(5008):965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisa E. Human mitochondrial tRNA processing. J Biol Chem. 1995;270(21):12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ. Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci U S A. 2005;102(39):13915–13920. doi: 10.1073/pnas.0502878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488(3):110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283(5407):1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF, Jr, Greenamyre JT, Snyder SH, Ross CA. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999;5(10):1194–1198. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271(50):31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Schiefer J, Landwehrmeyer GB, Luesse HG, Sprunken A, Puls C, Milkereit A, Milkereit E, Kosinski CM. Riluzole prolongs survival time and alters nuclear inclusion formation in a transgenic mouse model of Huntington’s disease. Mov Disord. 2002;17(4):748–757. doi: 10.1002/mds.10229. [DOI] [PubMed] [Google Scholar]
- Schultz TW, Sinks GD, Cronin MT. Structure-activity relationships for gene activation oestrogenicity: evaluation of a diverse set of aromatic chemicals. Environ Toxicol. 2002;17(1):14–23. doi: 10.1002/tox.10027. [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Smith AG, Muscat GE. Skeletal muscle and nuclear hormone receptors: implications for cardiovascular and metabolic disease. Int J Biochem Cell Biol. 2005;37(10):2047–2063. doi: 10.1016/j.biocel.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Sterling K, Milch PO, Brenner MA, Lazarus JH. Thyroid hormone action: the mitochondrial pathway. Science. 1977;197(4307):996–999. doi: 10.1126/science.196334. [DOI] [PubMed] [Google Scholar]
- Stroppolo A, Tian C, Guinea B, Olm V, Sheffield R, Sommer J, Ehrlich ME. 17beta-Estradiol promotes striatal medium size spiny neuronal maturation in vitro. Neuroendocrinology. 2004;79(5):259–267. doi: 10.1159/000079320. [DOI] [PubMed] [Google Scholar]
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410(2):103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1(5):493–503. doi: 10.1016/s1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, Pan H, Kessler H, Pancoska P, Moll UM. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem. 2006;281(13):8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- Valverde JR, Marco R, Garesse R. A conserved heptamer motif for ribosomal RNA transcription termination in animal mitochondria. Proc Natl Acad Sci U S A. 1994;91(12):5368–5371. doi: 10.1073/pnas.91.12.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Weaver CE, Jr, Park-Chung M, Gibbs TT, Farb DH. 17beta-Estradiol protects against NMDA-induced excitotoxicity by direct inhibition of NMDA receptors. Brain Res. 1997;761(2):338–341. doi: 10.1016/s0006-8993(97)00449-6. [DOI] [PubMed] [Google Scholar]
- Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16(10):5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367(6460):277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- Wrutniak C, Cassar-Malek I, Marchal S, Rascle A, Heusser S, Keller JM, Flechon J, Dauca M, Samarut J, Ghysdael J, et al. A 43-kDa protein related to c-Erb A alpha 1 is located in the mitochondrial matrix of rat liver. J Biol Chem. 1995;270(27):16347–16354. doi: 10.1074/jbc.270.27.16347. [DOI] [PubMed] [Google Scholar]
- Wrutniak-Cabello C, Casas F, Cabello G. Thyroid hormone action in mitochondria. J Mol Endocrinol. 2001;26(1):67–77. doi: 10.1677/jme.0.0260067. [DOI] [PubMed] [Google Scholar]
- Xue B, Hay M. 17beta-estradiol inhibits excitatory amino acid-induced activity of neurons of the nucleus tractus solitarius. Brain Res. 2003;976(1):41–52. doi: 10.1016/s0006-8993(03)02629-5. [DOI] [PubMed] [Google Scholar]
- Yager JD, Chen JQ. Mitochondrial estrogen receptors--new insights into specific functions. Trends Endocrinol Metab. 2007;18(3):89–91. doi: 10.1016/j.tem.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Yang SH, Liu R, Perez EJ, Wen Y, Stevens SM, Jr, Valencia T, Brun-Zinkernagel AM, Prokai L, Will Y, Dykens J, Koulen P, Simpkins JW. Mitochondrial localization of estrogen receptor beta. Proc Natl Acad Sci U S A. 2004;101(12):4130–4135. doi: 10.1073/pnas.0306948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Rajala RV, McGinnis JF, Li F, Anderson RE, Yan X, Li S, Elias RV, Knapp RR, Zhou X, Cao W. Involvement of insulin/phosphoinositide 3-kinase/Akt signal pathway in 17 beta-estradiol-mediated neuroprotection. J Biol Chem. 2004;279(13):13086–13094. doi: 10.1074/jbc.M313283200. [DOI] [PubMed] [Google Scholar]
- Zamzami N, Kroemer G. The mitochondrion in apoptosis: how Pandora’s box opens. Nat Rev Mol Cell Biol. 2001;2(1):67–71. doi: 10.1038/35048073. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Chen N, Moshaver A, Lee AT, Wellington CL, Hayden MR, Raymond LA. Mutant huntingtin enhances excitotoxic cell death. Mol Cell Neurosci. 2001;17(1):41–53. doi: 10.1006/mcne.2000.0909. [DOI] [PubMed] [Google Scholar]