Abstract
5-Lipoxygenase (5LO) plays a pivotal role in cellular leukotriene synthesis. To identify proteins interacting with human 5LO, we used a two-hybrid approach to screen a human lung cDNA library. From a total of 1.5 × 107 yeast transformants, nine independent clones representing three different proteins were isolated and found to specifically interact with 5LO. Four 1.7- to 1.8-kb clones represented a 16-kDa protein named coactosin-like protein for its significant homology with coactosin, a protein found to be associated with actin in Dictyostelium discoideum. Coactosin-like protein thus may provide a link between 5LO and the cytoskeleton. Two other yeast clones of 1.5 kb encoded transforming growth factor (TGF) type β receptor-I-associated protein 1 partial cDNA. TGF type β receptor-I-associated protein 1 recently has been reported to associate with the activated form of the TGF β receptor I and may be involved in the TGF β-induced up-regulation of 5LO expression and activity observed in HL-60 and Mono Mac 6 cells. Finally, three identical 2.1-kb clones contained the partial cDNA of a human protein with high homology to a hypothetical helicase K12H4.8 from Caenorhabditis elegans and consequently was named ΔK12H4.8 homologue. Analysis of the predicted amino acid sequence revealed the presence of a RNase III motif and a double-stranded RNA binding domain, indicative of a protein of nuclear origin. The identification of these 5LO-interacting proteins provides additional approaches to studies of the cellular functions of 5LO.
Keywords: yeast, two-hybrid system, leukotriene, protein interaction, coactosin-like protein
5-Lipoxygenase (5LO; arachidonate:oxygen 5-oxidoreductase, EC 1.13.11.34) is found primarily in polymorphonuclear leukocytes, macrophages, and mast cells, where it plays a central role in cellular leukotriene synthesis. 5LO converts arachidonic acid, released from the membranes by phospholipase A2, into 5(S)-hydroperoxy-6,8,11,14-eicosatetraenoic acid (5-HPETE), and subsequently into the epoxide intermediate leukotriene A4 (LTA4) (1). Hydrolysis of LTA4 by LTA4 hydrolase leads to the formation of the potent neutrophil chemoattractant LTB4, whereas conjugation of LTA4 with glutathione through the action of LTC4 synthase yields LTC4, which then is sequentially degraded into LTD4 and LTE4. The cysteinyl-leukotrienes, which constitute slow-reacting substance of anaphylaxis, are known to contract airway smooth muscle, increase vascular permeability, and promote mucus secretion (2).
Import of 5LO into the nucleus and association with the perinuclear membrane have been observed for several cell types (see for example refs. 3–5). Different patterns were found, particularly for peripheral blood leukocytes as compared with alveolar macrophages. In blood polymorphonuclear leukocytes, 5LO translocates from the cytosol to nuclear membrane when the cells are activated to produce leukotrienes. In alveolar macrophages, a large part of 5LO is found inside the nucleus (associated to euchromatin) already when cells are isolated, and upon activation to produce leukotrienes, the intranuclear 5LO binds to nuclear membrane. Recently, an N-terminal fragment of 5LO was found to direct nuclear localization, whereas three classical nuclear localization sequences in 5LO appeared less important (6).
Translocation and activation of 5LO may involve interactions with other proteins. A study by Lepley and Fitzpatrick (7) first suggested that 5LO may contain an Src homology 3 binding motif that could enable its interaction with growth factor receptor-bound protein 2 and cytoskeletal proteins in vitro. In fact, such an association of 5LO with cytoskeletal structures could have important implications for translocation and modulation of 5LO activity.
In our aim at determining the protein partners of 5LO by using the two-hybrid system, we identified three distinct proteins probably acting at different levels of the cellular machinery, including a protein named ΔK12H4.8 homologue for its high homology with the hypothetical helicase K12H4.8 in Caenorhabditis elegans. The identification of these 5LO-interacting proteins may help to improve our understanding of the cellular roles of 5LO.
MATERIALS AND METHODS
Two-Hybrid Constructs.
The Gal4 DNA binding domain (BD) vector pGBT9 (CLONTECH), carrying the TRP1 gene, and the Gal4 activating domain (AD) vector pACT2 (CLONTECH), carrying the LEU2 gene, were used for two-hybrid experiments. The human 5LO cDNA was obtained from the pT3–5LO plasmid (8) as an EcoRI/SalI fragment and was cloned in-frame into the EcoRI and SalI restriction sites of pGBT9 to get pGBT9–5LO. The construct pGBT9-SNF1 was prepared by ligating the 2.3-kb SNF1 BamHI/SalI insert from pSE1112 (American Type Culture Collection) in-frame into the BamHI/SalI sites of pGBT9. The 0.9-kb SNF4 insert was excised from pSE1111 (American Type Culture Collection) as a BglII fragment and cloned in-frame into pACT2 to obtain pACT2-SNF4. Microsomal glutathione S-transferase-I (MGST-I) cDNA (kindly provided by R. Morgenstern, Karolinska Institute, Stockholm) was amplified by PCR and cloned in-frame into the BamHI/SalI restriction sites of pGBT9. The vectors pGBT9-MGST-I and pTD1–1 [simian virus 40 (SV40) large T-antigen in pACT2] (CLONTECH) were used as control plasmids. All of the constructs were verified by restriction analysis and DNA sequencing on an Applied Biosystems PRISM 377 sequencer with the Applied Biosystems PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin–Elmer).
Yeast Two-Hybrid Screening.
The yeast strain PJ69–4A (MATa trp1–901 leu2–3,112 ura3–52 his3–200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ) was maintained in yeast extract/peptone/dextrose plus adenine medium and was used for the two-hybrid cDNA library screening. PJ69–4A harboring the Gal4 DNA-BD vector pGBT9–5LO was transformed with 70 μg of a human lung cDNA library (HL4044AH, CLONTECH), constructed in the Gal4 AD vector pACT2, by the lithium acetate/single-stranded DNA/polyethylene glycol method, as described by Gietz and Schiestl (9). Freshly transformed yeast cells were plated on synthetic dropout (SD)/-Leu/-Trp/-Ade plates, and growers were streaked on SD/-Leu/-Trp/-Ade, on SD/-Leu/-Trp/-His + 4 mM 3-amino-1,2,4-triazole (Sigma), and on SD/-Leu/-Trp + 5-bromo-4-chloro-3-indolyl β-d-galactoside (Sigma) plates to test for the adenine, histidine, and lacZ reporter genes, respectively. Positive clones were grown in selective medium, and plasmid DNA was prepared (10). The interacting pACT2 plasmid was rescued by complementation of the leuB6 deficiency in the bacterial strain HB101 (Invitrogen), transformed by electroporation, and plated on minimal medium lacking leucine. Plasmid DNA was prepared from the HB101 growers and retransformed back into PJ69–4A with the DNA-BD vector pGBT9–5LO to confirm the interaction, and both strands of the interacting cDNA inserts were sequenced.
Liquid β-Galactosidase Assay.
One OD600 of yeast cell transformants, grown in selective medium at 30°C, was centrifuged and resuspended in 750 μl of Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/50 mM β-mercaptoethanol, pH 7.0). After addition of 10 μl of 0.1% SDS and 20 μl of chloroform, the cell suspensions were vortexed and equilibrated at 28°C for 15 min. The reaction was initiated by adding 150 μl of o-nitrophenyl β-d-galactopyranoside (4 mg/ml in Z buffer; Sigma). When a pale yellow color had developed, the reaction was stopped by the addition of 375 μl of 1 M Na2CO3. The tubes were centrifuged, and the supernatant was transferred to a microcentrifuge tube. The ODs at 420 nm and 550 nm then were determined, and the β-galactosidase units were calculated.
Immunoblot Analysis.
Yeast transformants were grown in 12 ml of selective medium at 30°C overnight. The cell suspensions were centrifuged, and the cells were washed in ice-cold buffer A (20 mM Tris-Cl, pH 8.0/10% glycerol/10 mM EDTA/1 mM DTT) and resuspended in 1 vol of ice-cold buffer A containing Complete Protease Inhibitor Mix (Boehringer Mannheim). Acid-washed glass beads (425–600 μm; Sigma) were added, and the tubes were vortexed at high speed for 5 min. The homogenates were centrifuged, and the pellets were resuspended in 1 vol of buffer A containing Complete Protease Inhibitor Mix and 1% SDS. One volume of 2× SDS sample buffer (62.5 mM Tris-Cl, pH 6.8/10% glycerol/2% SDS/1% β-mercaptoethanol/5 μg/ml bromophenol blue, final concentrations) was added, and the samples were boiled for 5 min. The yeast protein extracts were analyzed by SDS/PAGE by using the Mini Protean system (Bio-Rad) and immunoblotted to nitrocellulose membranes (Amersham Pharmacia). The membranes were blocked in 20 mM Tris⋅HCl/137 mM NaCl/0.1% Tween 20, pH 7.6 (TBST) containing 5% nonfat dry milk for 1 h, washed, and incubated with the 5LO antibody 1551 or anti-AD (CLONTECH) primary antibody for 1 h at room temperature. The blots then were washed with TBST and incubated with 1:1,000 dilutions of alkaline phosphatase (AP)-conjugated anti-rabbit IgG (anti-5LO) or anti-mouse IgG (anti-AD) (Sigma) for 1 h at room temperature. After washing with TBST, the proteins were visualized by using AP substrates (nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate, Sigma) in AP buffer (100 mM Tris⋅HCl/100 mM NaCl/5 mM MgCl2, pH 9.5).
Sequence Analysis and Alignments.
The blast searches of the SWALL NonRedundant Protein Sequence Database, which included Swissprot, TREMBL, and TREMBLNEW databases, and amino acid sequence alignments by using the clustal w program were performed with the Wisconsin Package Version 9.1, Genetics Computer Group, or via the European Bioinformatics Institute server, Cambridge, U.K. (http://www.ebi.ac.uk). Search of the PROSITE database was performed by using the scanprosite program via the Swiss Institute of Bioinformatics server, Geneva, Switzerland (http://www.expasy.ch).
RESULTS
Experimental Approach.
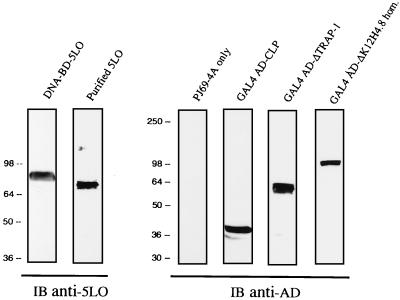
The aim of the present study was to identify cellular proteins interacting with 5LO by using the yeast two-hybrid approach. However, in establishing the appropriate conditions to perform a two-hybrid screen with the 5LO as a bait, we faced the problem of autoactivation of the reporter genes by using the high-expression vector pAS2–1 in the yeast strains Y190, Y187, or CG-1945. To circumvent this problem, we opted for a low-expression vector (pGBT9) with the use of the yeast strain PJ69–4A (11). The suitability of this approach first needed to be ascertained by cotransforming PJ69–4A with pGBT9–5LO and pACT2, pACT2-SNF4, or pTD1–1 and testing for the reporter genes. Under these conditions, no activation of the reporter genes was observed. We then verified that the DNA-BD-5LO hybrid protein was stably expressed in PJ69–4A. Immunoblot analysis using the anti-5LO antibody showed expression of the DNA-BD-5LO fusion as a single band of approximately 85 kDa (Fig. 1, lane 1), with a decreased electrophoretic mobility as compared with purified 5LO (Fig. 1, lane 2), as expected. Thus, the absence of autoactivation of the reporter genes by 5LO, together with the stable expression of the DNA-BD-5LO fusion protein, allowed us to move forward in our aim at finding 5LO-interacting proteins.
Figure 1.
Protein immunoblots showing expression of the Gal4 DNA-BD-5LO (Left) and the Gal4 AD-5LO-interacting protein fusions (Right) in the yeast strain PJ69–4A. Yeast protein extracts were fractionated by 7.5% or 10% SDS/PAGE and subjected to immunoblot (IB) analysis with anti-5LO (IB Anti-5LO) or anti-activating domain (IB Anti-AD) antibody, respectively. Purified 5LO (100 ng) was included as a control. The proteins were visualized by using alkaline phosphatase substrates (nitro blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate).
Two-Hybrid cDNA Library Screening.
For the two-hybrid cDNA library screening, PJ69–4A was sequentially transformed with pGBT9–5LO, and then with 70 μg of a human lung cDNA library. From a total of 1.5 × 107 transformants, nine independent clones were found to interact with 5LO and were isolated. Four of the clones contained coactosin-like protein (CLP) cDNA, two identical clones encoded the partial transforming growth factor (TGF) β receptor-I-associated protein 1 (TRAP-1) cDNA (ΔTRAP-1), and three identical clones represented a unique K12H4.8 homologue partial cDNA (ΔK12H4.8 homologue). Because several identical clones were isolated, it appeared that our screen was saturated. Yeast cells cotransformed with 5LO and CLP, ΔTRAP-1, or the ΔK12H4.8 homologue were able to grow in the absence of adenine or histidine (in the presence of 3-amino-1,2,4-triazole), and turned blue when incubated in the presence of the substrate 5-bromo-4-chloro-3-indolyl β-d-galactoside (Table 1). No activation of the reporter genes was observed when 5LO was coexpressed with SNF4, SV40 large T-antigen, or the empty vector, as well as for the 5LO-interacting proteins coexpressed with SNF1, microsomal glutathione S-transferase I, or the empty vector.
Table 1.
5LO interacts specifically with CLP, partial TRAP-1 (ΔTRAP-1), and the partial K12H4.8 (ΔK12H4.8) homologue in the yeast two-hybrid system
| Gal4 DNA-BD constructs | Gal4 DNA-AD constructs | Reporter genes
|
||
|---|---|---|---|---|
| Adenine | Histidine | lacZ | ||
| Empty | Empty | − | − | White |
| SNF1 | Empty | − | − | White |
| Empty | SNF4 | − | − | White |
| SNF1 | SNF4 | + | + | Blue |
| 5LO | Empty | − | − | White |
| 5LO | SNF4 | − | − | White |
| 5LO | SV40 large T-antigen | − | − | White |
| Empty | CLP | − | − | White |
| SNF1 | CLP | − | − | White |
| MGST-I | CLP | − | − | White |
| 5LO | CLP | + | + | Blue |
| Empty | ΔTRAP-1 | − | − | White |
| SNF1 | ΔTRAP-1 | − | − | White |
| MGST-I | ΔTRAP-1 | − | − | White |
| 5LO | ΔTRAP-1 | + | + | Blue |
| Empty | ΔK12H4.8 homologue | − | − | White |
| SNF1 | ΔK12H4.8 homologue | − | − | White |
| MGST-I | ΔK12H4.8 homologue | − | − | White |
| 5LO | ΔK12H4.8 homologue | + | + | Blue |
+, growth; −, no growth. The plasmids expressing the Gal4 fusion proteins were cotransformed into the yeast strain PJ69-4A, and transformants were selected on SD/−Leu/−Trp. Three or four independent colonies were streaked on SD/−Leu/−Trp/−Ade, on SD/−Leu/−Trp/−His + 4 mM 3-amino-1,2,4-triazole, and on SD/−Leu/−Trp + 5-bromo-4-chloro-3-indolyl β-d-galactoside plates to test for the adenine, histidine, and lacZ reporter genes respectively, and incubated at 30°C for 3–5 days. Growth and color of the colonies then were determined.
The relative affinity between 5LO and CLP, ΔTRAP-1, or the ΔK12H4.8 homologue was evaluated by liquid β-galactosidase assays (Table 2). In these experiments, the β-galactosidase activity induced by the coexpression of SNF1 and SNF4, two proteins known to interact in the two-hybrid system and used as a positive control, averaged 6.6 units. These levels were approximately 15- to 20-fold higher than those obtained by coexpressing 5LO with an unrelated protein (SV40 large T-antigen) or with the empty vector. When assaying the interaction of 5LO with its interacting proteins, the β-galactosidase activity levels induced by CLP were comparable to that of the SNF1 + SNF4 combination, whereas ΔTRAP-1 more weakly and the ΔK12H4.8 homologue more vigorously stimulated the lacZ reporter gene. Taken together, these results show that CLP, ΔTRAP-1, and ΔK12H4.8 homologue interact specifically with 5LO, with the ΔK12H4.8 homologue being the strongest interactor.
Table 2.
Evaluation of the relative affinity between 5LO and CLP, ΔTRAP-1, or the ΔK12H4.8 homologue by liquid β-galactosidase assays
| Gal4 DNA-BD constructs | Gal4 AD constructs | β-Galactosidase activity, units |
|---|---|---|
| SNF1 | SNF4 | 6.6 ± 0.4 |
| 5LO | Empty | 0.3 ± 0.1 |
| 5LO | SV40 large T-antigen | 0.4 ± 0.1 |
| 5LO | CLP | 6.5 ± 0.9 |
| 5LO | ΔTRAP-1 | 3.2 ± 0.3 |
| 5LO | ΔK12H4.8 homologue | 11.7 ± 0.6 |
In these experiments, pSE1112 (SNF1) and pSE1111 (SNF4) were used as a positive control. The plasmids expressing the Gal4 fusion proteins were cotransformed into the yeast strain PJ69-4A, and transformants were selected on SD/−Leu/−Trp. Four to eight independent colonies were grown in selective medium and assayed for β-galactosidase activity, as described in Materials and Methods. Mean ± SEM.
5LO Interacts with CLP.
Of the nine positive yeast clones, four encoded the complete coding region of CLP cDNA (12) with various lengths of 5′ untranslated sequence fused in-frame to the Gal4 AD. Analysis of the CLP nucleotide sequence revealed an ORF of 426 bp with the presence of an in-frame stop codon (TAG) located 119 bp upstream from the starting ATG. This ATG start codon is surrounded by the sequence GCGGCG in positions −6 to −1, and a G in position +4, characteristic of a consensus sequence for initiation of translation (13). Interestingly, the nucleotide sequence of two of the clones found to interact with 5LO in the two-hybrid system started immediately after the in-frame stop codon located upstream (at position −119) from the starting ATG, suggesting that this stop codon is functional.
Immunoblot analysis with the anti-AD antibody showed expression of the Gal4 AD-CLP fusion protein as a single band of approximately 40 kDa (Fig. 1, lane 4). This finding is in agreement with the predicted protein sequence consisting of 142 aa and a molecular mass of 15,945 Da. The CLP shows a high degree of homology with coactosin, an actin-binding protein from Dictyostelium discoideum (14), sharing 33.3% identity and 74.9% homology, as reported (12). A blast search of the SWALL database revealed no other homologous proteins. Although the structure of the CLP gene, present in two copies on chromosome 17 band p11.2, is not fully characterized, the 3′ untranslated region shows more than 95% sequence identity over 210 bp to a human cDNA containing a sequence with autonomously replicating activity (12).
5LO Interacts with the C-Terminal Part of ΔTRAP-1.
Two of the nine positive yeast clones were identical and contained nucleotides 1426–2859 of the reported TRAP-1 cDNA sequence (15) (accession no. AF022795, GenBank), covering more than half of the ORF to the stop codon (TGA), plus an additional 54 noncoding nt at the 3′ end. This construct resulted in an in-frame fusion of TRAP-1 amino acids 462–860 with the Gal4 AD, which was confirmed by immunoblot analysis with the anti-AD showing expression of the fusion protein as a single band of approximately 60 kDa (Fig. 1, lane 5). As shown in Table 1, ΔTRAP-1, representing the C-terminal half of the protein, specifically interacts with 5LO. A blast search of the SWALL database revealed no other homologous proteins.
5LO Interacts with ΔK12H4.8 Homologue.
Finally, three identical clones represented a unique cDNA that we named ΔK12H4.8 homologue, based on its high degree of similarity to the hypothetical helicase K12H4.8 in C. elegans (16) (accession no. L14331, GenBank). The two proteins share 58% identity and 74% similarity over 275 aa (P = 1.1e-114). The nucleotide as well as the predicted amino acid sequences of the ΔK12H4.8 homologue are shown in Fig. 2. The partial cDNA is composed of 2,083 nt, with a continuous ORF of 2,025 nt, interrupted by a TGA stop codon, and a 3′ untranslated region of 55 nt. No potential starting ATG in a favorable translation initiation context was found. In our two-hybrid experiments, the encoded Gal4 AD-ΔK12H4.8 homologue fusion was expressed as a 105-kDa protein (Fig. 1, lane 6), as expected. Interestingly, the ΔK12H4.8 homologue was found to be the strongest interactor with 5LO.
Figure 2.
cDNA and deduced amino acid sequence of the ΔK12H4.8 homologue, a protein homologous to the hypothetical helicase K12H4.8 in C. elegans. The predicted amino acid sequence is indicated in single-letter code. The stop codon TGA is underlined.
A blast search of the SWALL database with the predicted 675 aa of the ΔK12H4.8 homologue revealed high homology with a hypothetical helicase in C. elegans, known as K12H4.8, and with RNases III. Search of the PROSITE database with the partial amino acid sequence of the ΔK12H4.8 homologue demonstrated the presence of the RNase III signature [QE] - RLEFLGD - [AS]. This motif appears in ΔK12H4.8 homologue, K12H4.8, E. coli RNase III (17, 18), and a putative Bacillus subtilis RNase III (19) (see Fig. 3). The K12H4.8 gene, situated in the central gene cluster of chromosome III in C. elegans, first was found to have sequence similarities to a putative ATP-dependent RNA helicase in E. coli (16); however, we obtained a better score in comparison to the putative RNase III from B. subtilis. In addition to the RNase III signature, a double-stranded (ds) RNA-binding domain has been found in C. elegans K12H4.8 (20). Interestingly, all 13 conserved amino acid residues in the K12H4.8 dsRNA binding domain are also present in the ΔK12H4.8 homologue. As in the RNases III from bacteria and yeast, the dsRNA binding domain appears subsequent to (C terminal of) the RNase III signature, in ΔK12H4.8 homologue between residues 610–665.
Figure 3.
The ΔK12H4.8 homologue contains an RNase III motif. Amino acids 422–511 of the ΔK12H4.8 homologue were aligned with the hypothetical helicase K12H4.8 in C. elegans (P34529) and with two bacterial RNases III by using the clustal w program (European Bioinformatics Institute). The RNase III signature [DEQ] - [RQ] - [LM] - E - [FYW] - [LV] - G - D - [SAR], present in the four proteins, is shown in bold. The four proteins share 24% (22/93) identity and 57% (53/93) similarity over 93 aa centered around the RNase III motif. Amino acid identity and conservative differences between the proteins are indicated by ∗ and . or :, respectively.
DISCUSSION
To study the regulation and cellular roles of 5LO, we deemed it of particular interest to identify proteins that are able to bind 5LO. With this aim in view, a yeast two-hybrid system that enabled us to perform a genetic screen for 5LO-interacting proteins was developed. The use of the yeast strain PJ69–4A, together with the low-expression vector pGBT9, solved the problems of autoactivation of the reporter genes induced by 5LO that we observed in preliminary experiments (11). The yeast strain PJ69–4A has the advantage of combining three reporter genes under the control of three different Gal4-responsive promoters (GAL1-HIS3, GAL2-ADE2, and GAL7-lacZ), thereby reducing the number of false positives (11).
One of the proteins interacting with 5LO was CLP. Human CLP initially was found as a sequence flanking a deletion on chromosome 17 characterizing the Smith-Magenis syndrome (12). The CLP amino acid sequence shares 33.3% identity and 74.9% homology with coactosin, an actin-binding protein isolated from D. discoideum (14). Although it is not known whether CLP shares the ability of coactosin to bind actin, our findings may provide a link between 5LO and the actin cytoskeleton. Such a link has been proposed previously (7). CLP might act as an anchor that would retain 5LO in the cytosol of resting cells and/or silence 5LO activity by steric hinderence of its active site or cofactor binding sites. Association of 5LO with the cytoskeleton, which provides a ramified, complex, and dynamic network along which 5LO could be vehicled within the cell, also has been put forward as a potential initiator step in the translocation process. Interestingly, it recently was found that monohydroxyacids, including 5(S)-hydroxy-6,8,11,14-eicosatetraenoic acid, could bind to cytosolic actin (21).
A second 5LO-interacting protein was identified as TRAP-1. The cDNA of TRAP-1 was cloned recently (15). The C-terminal half of TRAP-1, which also interacts with 5LO, has been shown to specifically interact only with the activated form of the type I TGF β receptor, and thus could distinguish it from its quiescent state. In addition, partial TRAP-1 was shown to inhibit TGF β receptor-I-mediated signaling (15). Previous work in our laboratory has shown a significant up-regulation of 5LO protein expression and activity in myeloid cells (HL-60 and Mono Mac 6 cells) differentiated with TGF β and vitamin D3 (22–24). It is tempting to speculate that TRAP-1 could physically associate 5LO to the TGF β receptor, thereby providing a functional link between the activated TGF β receptor and 5LO.
Among the three 5LO-interacting proteins, the ΔK12H4.8 homologue represented the strongest 5LO interactor. The absence of a potential start codon in a favorable translation initiation context, together with immunoblot analyses, suggest that the ΔK12H4.8 homologue cDNA is still incomplete. We nevertheless were able to identify two putative functional domains: an RNase III motif and a double-stranded RNA binding domain. These functional domains related to RNA processing suggest a nuclear function for this human protein.
RNase III is a double-stranded endoribonuclease that plays important roles in several aspects of RNA metabolism, including processing of mRNA and rRNA precursors (see ref. 25 for review). These functions have been described in studies of E. coli RNase III, a relatively small protein (226 amino acid residues). In Saccharomyces cerevisiae, the RNT1 gene encodes a 473-aa RNase III essential for ribosome synthesis (26). The proteins from C. elegans K12H4.8 (1,822 amino acid residues) and the human ΔK12H4.8 homologue are larger than the RNases III in bacteria and yeast. This finding might suggest additional roles complementing the putative RNase III activity in K12H4.8 and its human homologue.
The interactions of 5LO with CLP, ΔTRAP-1, and ΔK12H4.8 homologue are of interest in connection with the regulation and nuclear translocation of the enzyme. Moreover, the binding to the human ΔK12H4.8 homologue might suggest a noncatalytic role of the 5LO.
Acknowledgments
We thank Philip James for kindly providing the yeast strain PJ69–4A and Liz Muller for fruitful discussions. P.P. is a recipient of a Fellowship from the Heart and Stroke Foundation of Canada and the Medical Research Council of Canada. This study was supported by grants from the Swedish Medical Research Council (03X-217), the European Union (BMH4-CT96–0229), and the Verum Foundation.
ABBREVIATIONS
- CLP
coactosin-like protein
- 5LO
5-lipoxygenase
- TGF
transforming growth factor
- TRAP-1
TGF type β receptor I-associated protein 1
- BD
binding domain
- AD
activating domain
- SD
synthetic dropout
- SV40
simian virus 40
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ132261).
References
- 1.Samuelsson B. Science. 1983;220:568–575. doi: 10.1126/science.6301011. [DOI] [PubMed] [Google Scholar]
- 2.Samuelsson B, Dahlén S-E, Lindgren J A, Rouzer C A, Serhan C N. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 3.Woods J W, Coffey M J, Brock T G, Singer I I, Peters-Golden M. J Clin Invest. 1995;95:2035–2046. doi: 10.1172/JCI117889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brock T G, McNish R W, Peters-Golden M. J Biol Chem. 1995;270:21652–21658. doi: 10.1074/jbc.270.37.21652. [DOI] [PubMed] [Google Scholar]
- 5.Pouliot M, McDonald P P, Krump E, Mancini J A, McColl S R, Weech P K, Borgeat P. Eur J Biochem. 1996;238:250–258. doi: 10.1111/j.1432-1033.1996.0250q.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen X S, Zhang Y Y, Funk C D. J Biol Chem. 1998;273:31237–31244. doi: 10.1074/jbc.273.47.31237. [DOI] [PubMed] [Google Scholar]
- 7.Lepley R A, Fitzpatrick F A. J Biol Chem. 1994;269:24163–24168. [PubMed] [Google Scholar]
- 8.Zhang Y Y, Rådmark O, Samuelsson B. Proc Natl Acad Sci USA. 1992;89:485–489. doi: 10.1073/pnas.89.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz R D, Schiestl R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 10.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 11.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault A C, Lee C C, Lupski J R. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 14.de Hostos E L, Bradtke B, Lottspeich F, Gerisch G. Cell Motil Cytoskeleton. 1993;26:181–191. doi: 10.1002/cm.970260302. [DOI] [PubMed] [Google Scholar]
- 15.Charng M-J, Zhang D, Kinnunen P, Schneider M D. J Biol Chem. 1998;273:9365–9368. doi: 10.1074/jbc.273.16.9365. [DOI] [PubMed] [Google Scholar]
- 16.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 17.Nashimoto H, Ushida H. Mol Gen Genet. 1985;201:25–29. doi: 10.1007/BF00397981. [DOI] [PubMed] [Google Scholar]
- 18.March P E, Ahnn J, Inouye M. Nucleic Acids Res. 1985;13:4677–4685. doi: 10.1093/nar/13.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oguro A, Kakeshita H, Takamatsu H, Nakamura K, Yamane K. Gene. 1996;172:17–24. doi: 10.1016/0378-1119(96)00181-3. [DOI] [PubMed] [Google Scholar]
- 20.Mian I S. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang L-T, Vanderhoek J Y. J Lipid Res. 1998;39:1476–1482. [PubMed] [Google Scholar]
- 22.Steinhilber D, Rådmark O, Samuelsson B. Proc Natl Acad Sci USA. 1993;90:5984–5988. doi: 10.1073/pnas.90.13.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brungs M, Rådmark O, Samuelsson B, Steinhilber D. Proc Natl Acad Sci USA. 1995;92:107–111. doi: 10.1073/pnas.92.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Härle D, Rådmark O, Samuelsson B, Steinhilber D. Eur J Biochem. 1998;254:275–281. doi: 10.1046/j.1432-1327.1998.2540275.x. [DOI] [PubMed] [Google Scholar]
- 25.Deutscher M P. J Biol Chem. 1993;268:13011–13014. [PubMed] [Google Scholar]
- 26.Elela S A, Igel H, Ares M., Jr Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]