Abstract
Previous studies have indicated a paucity of SINEs within the genomes of the guinea pig and nutria, representatives of the Hystricognathi suborder of rodents. More recent work has shown that the guinea pig genome contains a large number of B1 elements, expanding to various levels among different rodents. In this work we utilized A–B PCR and screened GenBank with sequences from isolated clones to identify potentially uncharacterized SINEs within the guinea pig genome, and identified numerous sequences with a high degree of similarity (>92%) specific to the guinea pig. The presence of A-tails and flanking direct repeats associated with these sequences supported the identification of a full-length SINE, with a consensus sequence notably distinct from other rodent SINEs. Although most similar to the ID SINE, it clearly was not derived from the known ID master gene (BC1), hence we refer to this element as guinea pig ID-like (GPIDL). Using the consensus to screen the guinea pig genomic database (Assembly CavPor2) with Ensembl BlastView, we estimated at least 100,000 copies, which contrasts markedly to just over 100 copies of ID elements. Additionally we provided evidence of recent integrations of GPIDL as two of seven analyzed conserved GPIDL-containing loci demonstrated presence/absence variants in Cavia porcellus and C. aperea. Using intra-IDL PCR and sequence analyses we also provide evidence that GPIDL is derived from a hystricognath-specific SINE family. These results demonstrate that this SINE family continues to contribute to the dynamics of genomes of hystricognath rodents.
Keywords: Genome Evolution, ID elements, Retrotransposons, Rodent Genomes, SINEs
1. Introduction
Short interspersed DNA elements (SINEs) are non-autonomous retrotransposons found throughout eukaryotic species. Based on DNA sequence analyses, SINEs are presumptively derived from either the 7SL RNA gene, tRNA genes, or 5S rRNA genes (Deininger et al., 2003; Kapitov and Jurka, 2003), and believed to rely on the proteins encoded by long interspersed DNA elements (LINEs) for amplification in the genome. This hypothesis has been supported using a cell culture retrotransposition assay (Dewannieux et al., 2003; Dewannieux and Heidmann, 2005a). Genomes from a variety of mammalian species have shown a high abundance of retrotransposons including roughly 42% of genome mass in humans (Lander et al., 2001) and 37% in the mouse (Waterston et al., 2002). Of that, there are 1.5 million SINEs in the mouse that comprise 8.2% of its genome, as well as roughly 1.5 million SINEs in the human, predominantly the Alu element, that comprise about 13% of its genome. Therefore, retrotranspositions have made a significant contribution to the organization of mammalian genomes.
Certain families of SINEs are specific to different taxonomic levels. For example Alu elements are found in primates (Deininger, 1989); whereas the B1, B2, and ID are among elements that have been found in rodents (Deininger, 1989). However, two analyzed rodents in the suborder Hystricognathi, the guinea pig and nutria, were shown to either have very low copies or no copies of ID and B2 elements, respectively (Kass et al., 1996; 1997). Two potential explanations for this find are (i) this group of rodents has avoided a “take-over” by these genomic parasites due to either containing specific repressors or lacking the necessary retrotransposition machinery (perhaps LINEs), or (ii) these rodents contain a unique group of SINEs not found in the Sciurognathi suborder which includes mice, rats, and squirrels among other rodents. There has been controversy as to whether the guinea pig is a rodent (Graur et al., 1991; Frye and Hedges, 1995; D’Erchia et al., 1996; Philippe, 1997; Konno et al., 1999), but even if guinea pigs are considered rodents, the time of divergence between suborders may be sufficient for different SINE families to have been derived and amplified. While we were investigating the paucity of SINEs in the guinea pig, Vassetzky et al. (2003) determined that the B1 family does exist in high copy number (150,000) in the guinea pig and therefore demonstrated SINEs can amplify among hystricognaths. This study therefore investigates the potential finding of undefined SINEs that may be in the guinea pig and other hystricognaths to assess their contributions to the dynamics of the genomes of this understudied group of rodents.
2. Materials and Methods
2.1. Tissue and DNA samples
Tissue samples of various hystricognath rodents were kindly provided by the Museum of Vertebrate Zoology of the University of California at Berkely. These include the paca (Agouti paca; MVZ 153575), rock rat (Aconaemys porteri; MVZ 184959), Mesomys hispidus (MVZ 155158), prehensile-tail porcupine (Coendou bicolor; MVZ 155200), and the pacarana (Dinomys branikii; MVZ 153574). Nutria (Myocaster coypus), grey squirrel (Sciurus carolinensis), European house mouse (Mus musculus), Norway rat (Rattus norvegicus), white-footed mouse (Peromyscus leucopus), guinea pig (Cavia porcellus), tree shrew (Tupaia glis) and human (Homo sapiens) DNA were isolated in previous studies (Kass et al., 1996; Kass et al., 1997). Degu tissue was kindly provided by Dr. Theresa Lee of the University of Michigan, and rabbit tissue by Dr. Michael Angell of Eastern Michigan University. Brazilian guinea pig tissue (Cavia aperea), one from Peru (USNM 579693) and two from Bolivia (USNM 584588 and USNM 584589), was kindly provided by the Smithsonian Institute.
2.2. A–B PCR
In order to identify undiscovered SINEs in the guinea pig, we incorporated the technique of A–B PCR, which involves using primers based on conserved sequences of A and B RNA polymerase III promoters to obtain unique sequence between those sites. This technique was developed and successfully used to identify SINEs in various species including bats (Borodulina and Kramerov, 1999). The primers used were: 5′-trgctcagtgg-3′ and 5′-ggratygaacy-3′. Conditions for AB PCR were as follows in a 25 μl volume: 200 ng genomic DNA, 1 × GoTaq buffer (Promega Inc., Madison, WI), 2.5 mM MgCl2, 200μM dNTPs, 300 nM each primer, and 1 u GoTaq cycled for 95°C for 1 min 1x; 30 cycles of 95°C for 1 min, 30°C for 1 min, 72°C for 1 min; concluding with a final extension of 72°C for 3 min. PCR products were analyzed using 2% agarose gels stained with ethidium bromide.
2.3. Cloning and sequencing A–B PCR amplicons
The A–B PCR products were shotgun cloned by ligation into the pGEMTEasy TA cloning vector (Promega Inc), and used to transform E. coli JM109 cells according the manufacturer’s protocol. White colonies were selected, plasmid DNA isolated using the Wizard Plus SV Minipreps DNA Purification System (Promega) and analyzed for the correct-size inserts by digestion with EcoRI. Clones were submitted to Functional Biosciences Inc (Madison, WI) for sequencing.
2.4. Use of A–B PCR products to identify guinea pig SINEs
Sequences for A–B PCR products were used to identify potential SINEs by incorporating a BLAST search of the GenBank database. Several of the A–B PCR clones, including clone GP7 and GP5 matched well to an apparent ID-like element within the guinea pig locus of accession number AC175606.3, hence we refer to this locus as GP7; whereas the top hit for a BLAST search for the GP5 A–B PCR clone was guinea pig locus AC166752.4 and we refer to this guinea pig ID-like element as GP5. “Hits” were analyzed for hallmarks of SINEs such as polyA tail, and flanking direct repeats to distinguish full-length SINEs. We then used the top hits of a rescreened GenBank BLAST search queried with GP5 and GP7 for identifying additional full-length elements. These were contrasted to other known rodent SINEs using a MacVector ClustalW alignment to assess the relative identity to other elements. Additionally, a full-length element was used to screen the guinea pig genomic library (Assembly CavPor2) in Ensembl, using Ensembl Multi BlastView to estimate copy numbers of this SINE family.
2.5. Contrast newly identified guinea pig SINEs
In order to assess the taxonomic levels to which the newly identified IDL SINEs can be assigned, intra-IDL PCR was performed under the following conditions in 25 μl volumes: 10ng template DNA, 1X GoTaq buffer, 2.5 mM MgCl2, 200 μM dNTPs, 300nM each primer (5′-ggctggggatttagct-3′ and 5′-atcggtaccagggatc-3′), and 1 u GoTaq. Cycling conditions were: 1 cycle of 94°C for 2 min; 27 cycles of 94°C for 15 sec, 50°C for 15 sec, 72° for 15 sec; followed by an extension step of 72°C for 3 min. Products were analyzed on a 2% agarose gel. Additionally, we analyzed hystricognath species for the guinea pig ID (BC1-derived) family of elements using ID-specific primers (5′-ggggttggggatttg-3′ and 5-caggtttgagctgag-3′) with the same reagent concentrations and cycling conditions as GPIDL. Guinea pig ID and intra-IDL derived PCR amplicons from various species were shotgun cloned into the pGEMTEasy cloning vector using the manufacturer’s protocol, with plasmid DNA isolation and sequencing in the same manner as A–B PCR products. The sequenced clones (omitting the primer sequences) were analyzed using the ClustalW (v1.4) phylogenetic analysis with the MacVector DNA Sequence software program using default settings. Additionally, we screened various rodent libraries with Ensembl BlastView using the GPIDL and guinea pig ID sequences to further contrast the evolutionary history of these SINE families.
2.6. Analysis of IDL-containing orthologous loci in guinea pigs
We used the full-length GP7 IDL (see above) to rescreen the GenBank database and we refer to the top hits (all guinea pig loci) as GP7 hit1 (accession number AC 175606.3), GP7 hit2 (accession number AC167437.2), GP7 hit3 (accession number AF 312680.1), GP7 hit4 (AC171739), GP7 hit5 (AC186083), GP7 hit6 (AC183690.3), and GP7 hit7 (AC174609.3). Primers flanking the GPIDL element of interest were generated using the MacVector DNA software primer analysis program and were purchased through Integrated DNA Technologies (Coralville, IA). PCR was performed in 20 μl volumes containing 100ng DNA, 1X GoTaq buffer (Promega), 200 μM dNTP, 250 nM each primer (see below), and 1 unit GoTaq enzyme (Promega), and cycled in an MJ Research thermal cycler under the following conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 54°C for 30 sec, 72°C for 30 sec, with a final extension step of 72°C for 5 min. The primers and expected sizes of PCR products (inclusive of the GPIDL element were as follows): GP7 hit1 333 bp, primers 5′-agtggagtggtgaggactgagttg-3′ and 5′-agaaaagaaaggggaaaaggacc-3′; GP7 hit2 469 bp, primers 5′-atggtgtttccctgtgtccttg-3′ and 5′-tggtgactggagagtagttctgagg-3′; GP7 hit3 403 bp, primers 5′-gaacctctaaaatgtgattctccc-3′ and 5-atttgctgacagaccattctcag-3′; GP7 hit4 539 bp, primers 5′-cagcctgctcaaattcagtattgc-3′ and 5′-gtacagactttctgtggtttagccg-3′; GP7 hit5, 692bp, primers 5′-gcagtaattctcctgtctcagcctc-3′ and 5-cccaaatccctgaactgacaaag-3′; GP7 hit6, 622 bp, primers 5′-gtggaacagaaaatgctgtgttgg-3′ and 5′-caccatcaatttgcagttgtctgc-3′; GP7 hit7, 641 bp, primers 5′-cgctgcttagcattctagtgctg-3′ and 5′-accaagagagctgcacaattctg-3′. PCR products of GP7 hit1 and GP7 hit 4 loci, for the four guinea pig samples, were cloned using the pGEM-TEasy vector as discussed above to verify orthologous loci. Plasmid DNA was isolated using the Wizard Plus SV Minipreps DNA Purification System (Promega) Digest miniprep kit. Insert size was determined by EcoRI digestion of 12 μl of plasmid DNA sample in a 20 μl reaction and analyzed by electrophoretic separation on a 2% agarose gel. Two clones from each species were submitted to Functional Biosciences, Inc. for sequencing.
3. Results
3.1. Isolation of SINEs based on AB-PCR and data mining
The focus of this study was to identify any undiscovered SINE family in the guinea pig and other hystricognath rodents. We utilized the technique of A–B PCR (Borodulina and Kramerov, 1999) since the A and B box regions of SINEs are most conserved. We identified bands of equivalent size in the guinea pig and the rat (Figure 1). Using A–B PCR of the rat genome as a control, we identified a B2 clone demonstrating the effectiveness of this technique (data not shown). Using A–B PCR of the guinea pig genome, two clones demonstrated a high degree of sequence similarity to several guinea pig sequences (many over 92% up to 100%) in the GenBank database. There was no significant sequence identity to other organisms. The two clones (referred to as GP5 and GP7) were used to screen the GenBank database, identifying sequences that demonstrated the hallmarks of SINEs including the A-tail and flanking direct repeats. This allowed us to retrieve sequences that flank the A and the B box and depict full-length SINEs in the guinea pig.
Figure 1.
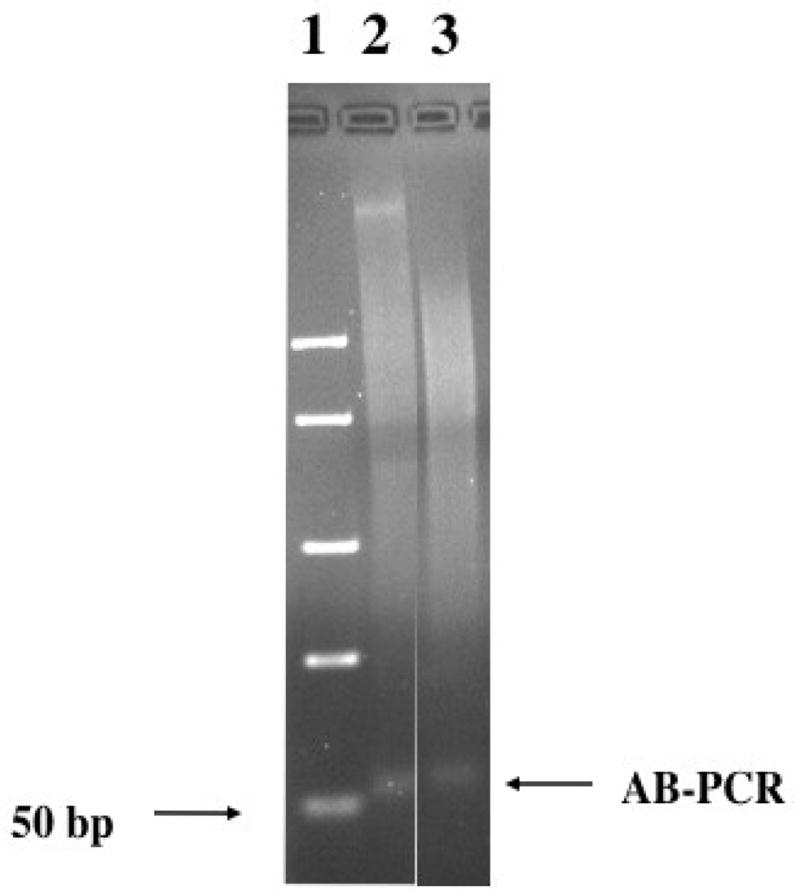
Analysis of AB-PCR. Samples were analyzed on a 2% agarose gel. Lane 1 Low range markers (Fermentas), lane 2 guinea pig, lane 3 rat.
3.2. Determination of a new SINE family in the guinea pig
We used the top “hits” of a rescreened GenBank BLAST search queried with GP5 and GP7 for identifying additional full-length elements. For GP5 BLAST, the first 27 hits were to guinea pig sequences, many with 93–94% sequence identity, and with GP7 BLAST the first 30 hits were to guinea pig with up to 95% match (excluding the query sequence itself). Therefore, we selected some of the top hits (referred to as GP5 BLAST1, GP5 BLAST2, etc. and GP7 BLAST1 GP7 BLAST2, etc.) for further analyses.
Using the MacVector ClustalW alignment, it appeared that the full-length isolates (such as GP5 BLAST 1 and GP7 BLAST 1) were not B1, B2, or ID based on pairwise analyses (note alignment in Figure 2). In contrast, the rat control A–B PCR product alignment appeared to support this as a B2 element (data not shown). Among the known rodent SINEs, this undefined isolated element appeared most closely related to ID, although the guinea pig ID element demonstrated greater sequence similarity to the rat ID than this new element (Figure 2). Hence, this new element is not BC1-derived, BC1 being the is the presumptive master gene of ID elements (Kim et al., 1994), but since it resembles the ID SINE most closely we refer to it as guinea pig ID-like or GPIDL. We then aligned the BLAST hits from GP5 and GP7, which demonstrated a high level of sequence identity among elements from the different loci (Figure 3). The variants at positions 30 and 73 (highlighted in Figure 3) between GP5 and GP7 BLAST hits might suggest the presence of potential diagnostic sites distinguishing distinct GPIDL subfamilies. The sequence GP7 BLAST1 matched the consensus sequence for all the eleven isolated elements and then we used it to screen the guinea pig genomic library (Assembly CavPor2) in Ensembl using Ensembl Multi BlastView (http://www.ensembl.org/Multi/blastview). We determined there are at least 100K copies of this element (using default settings, blocking repeatmasker), as 102K hits satisfied an E value of 0.01 among 773,088 sequence contigs. In stark contrast to GPIDL, when using this tool for the guinea pig ID (BC1-1 based sequence) only 138 hits satisfied an E value of 0.001 and 142 hits for E= 0.01. Therefore, at E = 0.01 GPIDL and the most related SINE, ID, are distinguishable and 100K represents a reasonable estimate, if not an underestimate, for the number of GPIDL elements. Additionally, the ID copy number estimate is consistent with previous findings within the guinea pig genome (Anzai et al., 1987; Kass et al., 1996). To further limit the possibility of false positives in our copy number estimate, we reinvestigated the GPIDL screening at a higher stringency (E = 0.0001), although the guinea pig genome had been upgraded to 3144 sequence contigs (cavPor3), limiting the capability for copy number estimates. However, of the 3144 sequences scanned, the GP7BLAST1 screen generated 1126 hits that satisfy E = 0.0001; 1127 hits at E = 0.001; and 1142 hits that satisfy E = 0.01. Therefore, the modest difference at the highest stringency is consistent with false positives having relatively negligible impact on our copy number assessment. These results demonstrate that although ID elements did not amplify in the guinea pig genome to the several fold greater extent of rat or mouse, an IDL group of elements with a different master gene has been a successful genomic parasite in the guinea pig. This further supports that there is not a paucity of SINEs in the guinea pig and the finding of a previously undiscovered rodent SINE family.
Figure 2.
Alignment of identified guinea pig IDL (GPIDL) sequences resulting from GenBank BLAST searches using cloned AB-PCR products with other known rodent SINEs. The 3′ ends of B1 and B2 were truncated. Dashes were included for maximal alignment, and dots indicate the same nucleotide as in GP7 BLAST1. Positions of the A-box and B-box promoter sequences are provided as well as the location of the polyA tail for GPIDL.
Figure 3.
Alignment of sequences from different loci obtained from GenBank database using GP5 and GP7 (see text) as query sequences. Highlighted boxed nucleotides represent variants that distinguish GP5 and GP7 as potentially distinct subfamilies.
3.3. Extent of Rodent Genomes Containing the GPIDL SINE family
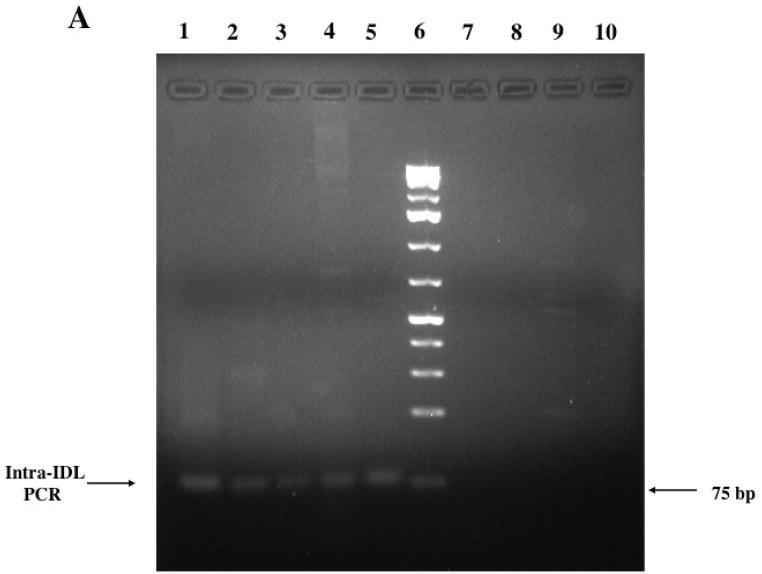
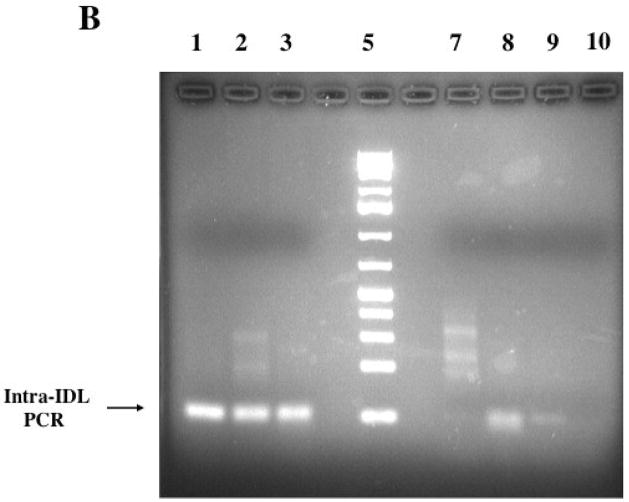
To determine the extent of rodent genomes containing the GPIDL SINE family we performed intra-IDL PCR on a number of hystricognath and non-hystricognath rodents. We observed amplified products of the expected size for all the analyzed hystricognath rodents (Figure 4), which include guinea pig, pacarana, degu, porcupine, paca (not shown), nutria, Mesomys hispidus (not shown) and rock rat. In contrast, when performing intra-ID PCR based on guinea pig ID element sequence among the hystricognath rodents, the amplicon of expected size was only observed within the guinea pig genome (Figure 5). This is consistent with IDL as a more conserved, hence more recently active SINE family within hystricognaths than ID elements. The IDL amplicons of the expected size were not observed in rat, white-footed mouse, rabbit (Figure 4) and hamster (data not shown), although we did observe a diffuse band in the squirrel and relatively faint bands in the tree shrew (Figure 4). Although this could argue against a hystricognath-specific SINE family, a tree shrew Ensembl Blast (release-44) at E = 0.001 revealed no hits, hence this IDL family of elements is apparently not present in the tree shrew, and possibly the tRNA-based Tu-SINE (Nishihara et al., 2002) was amplified. A mouse database search using GPIDL revealed no hits (E = 0.001), hence consistent with the PCR results supporting that these elements are not found in myomorphic rodents. In contrast, screening the mouse database (m37) with the guinea pig ID (BC1-1) sequence twenty two hits of 211 contigs were generated that satisfied E = 0.0001, consistent with the presence of ID elements throughout rodent genomes. However, screening of the rat library (RGSC 3.4 genebuild) with GPIDL generated 14 of 22 sequences that satisfied E = 0.0001 (none beyond 45 of the 76 nt), although 11 of the 14 just barely fit the criteria (all 6.4e−05), whereas screening with ID, 21 of 22 sequences satisfied the E value ranging from 5.2e−22 to 6.6e−24. Therefore, although false positives are possible at the highest stringency used in this analysis, there is an overwhelming demarcation between IDL and ID in analyzing the rat. The squirrel was intriguing as although it is in the Sciurognathi suborder along with rats and mice, our data may support a closer relationship to Hystricognathi. Therefore, we shotgun cloned the squirrel intra-GPIDL PCR products, as well as those from some hystricognaths to further investigate the evolutionary origins of this SINE family.
Figure 4.
Intra-IDL PCR of various rodents. Samples were analyzed on a 2% agarose gel. A. Lane 1 guinea pig, 2 rock rat, 3 porcupine, 4 nutria, 5 pacarana, 6, 1kb+ ladder (Fermentas), 7 rabbit, 8 rat, 9 white-footed mouse, 10 negative control (no DNA). B. Lane 1 pacarana, 2 degu, 3 porcupine, 5 1 kb+ ladder (Fermentas), 7 rat, 8 squirrel, 9 tree shrew, 10 negative control.
Figure 5.
Intra-ID PCR using sequences based on the guinea pig ID. Lane 1 1 kb+ ladder (Fermentas), 2 guinea pig, 3 degu, 4 nutria, 5 porcupine, 6 paca, 7 squirrel, 8 rat, 9 mouse, 10 negative control (no DNA).
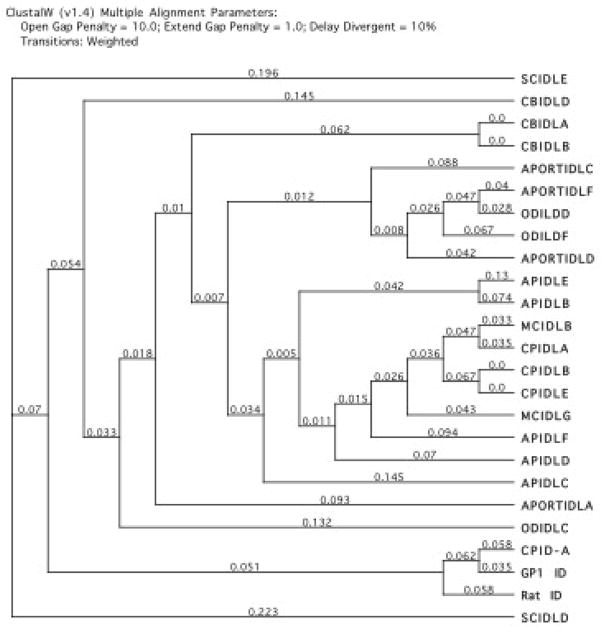
To further assess the nature of the diffuse squirrel PCR intra-IDL product we sequenced five clones. Four clones were identical in sequence to each other, and the fifth having only 61% sequence similarity (data not shown). When comparing these sequences using a ClustalW alignment to that of the hystricognath rodents, they were distinct from the hystricognath IDL clade (Figure 6). UPGMA and neighbor-joining analysis generated comparable results (data not shown). This molecular phylogeny (Figure 6) also suggests the squirrel and hystricognath IDL elements are not derived from a common ancestral master gene based on the closer association of the GPIDL (hystricognath IDL) family to ID elements. Additionally, analyzing the Spermophilus tridecemlineatus genome (thirteen-lined ground squirrel) using Ensembl queried with GP7BLAST1 (E = 0.01) generated just two hits of 41 nt, with no hits satisfying E = 0.0001 among over 147,000 contigs. These results suggest that the guinea pig and squirrel IDL elements are distinct SINE families, and perhaps a distinct IDL family of elements exists in sciuromorph rodents. This is also consistent with our previous findings (Kass et al. 1996), which had involved determination of copy numbers of ID elements among rodents. When using the BC1 gene from the rat as a probe for dot blots, it was found under low stringency that the squirrel contained 50,000 copies of ID elements, but when washed more stringently that number was reduced to 10,000. When screening the Spermophilus library with guinea pig ID, 4235 hits satisfied an E value of 0.01; 2436 hits for E = 0.001; and 1577 hits for E = 0.0001. These results are then consistent with the squirrel having ID elements as well as an ID-like family of elements distinct from myomorphs and hystricognaths.
Figure 6.
Molecular phylogeny, ClustalW (v 1.4) (MacVector) of intra-IDL clones (amplicons only without primer sequences) from analyzed hystricognaths and the squirrel and contrasted to ID elements. IDL clones were isolated from the following species: Sciurus carolinensis (SC), Cavia porcellus (CP), Coendou bicolor (CB), Myocaster coypus (MC), Agouti paca (AP), Aconaemys porteri (APORT), and Octodon degus (OD). Letters following IDL designate specific clones. Rat ID and GP1 ID are previously determined BC1-based ID elements. CPID-A is an intra guinea pig-ID PCR clone.
3.4. GPIDL analysis within the hystricognaths
A ClustalW analysis of intra-IDL PCR products (Figure 6) is indicative of a hystricognath-specific SINE family, as they form a distinct clade from both the BC1-based ID SINE and the squirrel ID-like sequences. This holds true using UPGMA and neighbor joining methods as well (data not shown). The degu and rock rat sequences appear closely related which is consistent with grouping of these two rodents within the same family (Octodontidae). However, the guinea pig and nutria sequences appear to form a clade, but have been shown not to be closely related species (Nedbal et al., 1994). Consensus sequences derived from our clones were also not consistent with known hystricognath phylogeny (data not shown), but potentially the development of consensus sequences from a large number of clones in each species may provide a useful phylogenetic tool. Overall, these results suggest that there is a hystricognath-wide family of IDL elements that apparently successfully proliferated in this suborder, as the ID elements have done within various myomorphic rodents.
3.5. Is the GPIDL family active?
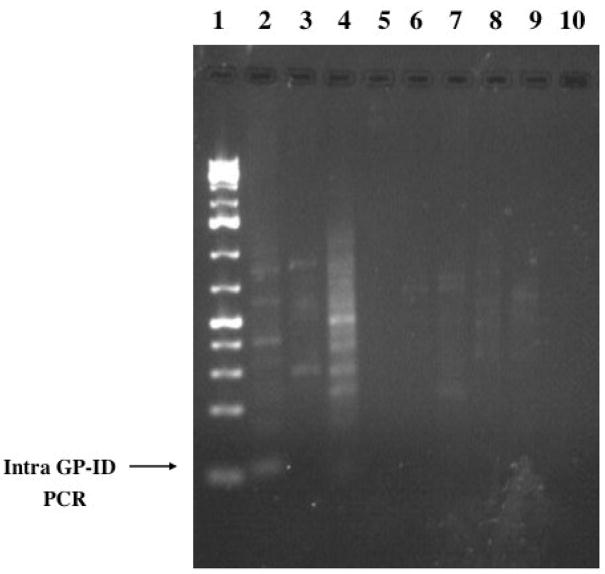
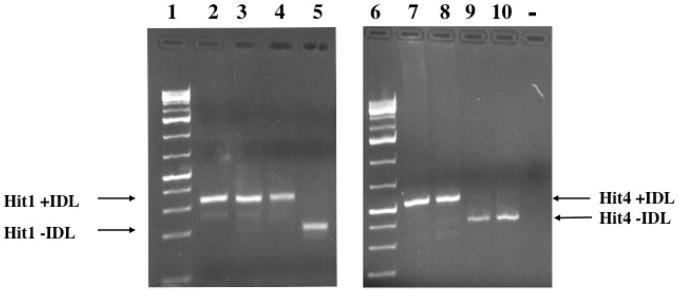
GP7 and GP5 were isolated using A–B PCR, thereby potentially representing younger elements based on the A and B promoter sequences being more conserved. Although database searches were biased using GP7BLAST1 and GP5BLAST1, the finding of numerous elements that have high sequence homology indicates some recent activity of the GPIDL family, even though older elements and the intra-IDL PCR results suggest this SINE family may have existed for millions of years. To further explore recent retrotransposition of GPIDL elements we analyzed seven loci representing top “hits” obtained when screening the GenBank database using BLAST with the GP7 sequence, with results referred to as GP7 hit1, GP7 hit2, etc. (see accession numbers in the methods section). Primers flanking the IDL element were generated and orthologous loci amplified using DNA from Cavia porcellus and three C. aperea individuals (note that these are actually the same species but vary in nomenclature simply as a result of contrasting the domestic and wild forms (Gentry et al., 2004)), to assess for the presence or absence of the GPIDL element. GP7 hit2, hit3, hit5, hit6, and hit7 contain the GPIDL element in the four genomes, (we attempted but could not amplify the orthologous loci from other analyzed hystricognath species), however, GP7 hit1 and hit4 both demonstrated presence/absence variants. The GPIDL element was lacking in the hit1 locus (also referred to as GP7 BLAST1) in one of the C. aperea individuals (Figure 7), whereas the GPIDL element of the hit4 locus was absent in the two C. aperea specimens from Bolivia (Figure 7). Cloning and sequencing the amplicons from each individual verified orthologous loci, noting the flanking sequence similarity, target site duplications, and screening the GenBank and Ensembl databases. The lack of fixation of this element in the species is indicative of an evolutionary recent integration event. Hence GPIDL elements continue to “jump” within the guinea pig genome.
Figure 7.
Presence/absence analysis of orthologous GPIDL-containing loci in guinea pig. Samples were analyzed on a 2% agarose gel. Lane 1 1kb+ ladder (Fermentas), Hit 1 locus – lane 2 Cavia porcellus, 3 C. aperea (Peru), 4 C. aperea (Bolivia-sample 584589), 5 C. aperea (Bolivia sample 584588), 6 1kb+ ladder (Fermentas), Hit 4 locus- 7 C. porcellus, 8 C. aperea (Peru), 9 C. aperea (Bolivia-9), 10 C. aperea (Bolivia-8), - negative control (no DNA).
In a separate ongoing study, we had performed RT-PCR of rodent germ-line tissue to analyze expression of B1 and B2 repeats. Interestingly by performing RT-PCR of guinea pig ovary tissue using a B2 primer and shotgun cloning the products, we found that six of ten clones contained the GPIDL sequence (data not shown), with one clone, which was identical to the consensus of the six clones, containing only a single variant from the genomic consensus sequence (GP7BLAST1). One clone was similar to a BC1-2 generated ID element. Although the RT-PCR study is preliminary, this finding is also consistent with GPIDL as a young active SINE family, as we hypothesize that greater expression in the germline allows for the increased potential to hijack LINE machinery and retrotranspose.
4. Discussion
This study was initiated to investigate the paucity of known rodent SINEs, B2 and ID elements, within the guinea pig and nutria genomes, two members of the Hystricognathi suborder. During this study another group found the rodent B1 SINE to have amplified to various levels in hystricognath rodents, including over 200,000 in the guinea pig (Veniaminova et al., 2007). However, beginning with A–B PCR we unraveled a previously unknown SINE in the guinea pig that is distinct from, but most similar to, the ID element and therefore refer to it as a guinea pig ID-like (GPIDL) family of elements. Additionally, since the master gene of ID elements has been considered the BC1 gene (Kim et al., 1994), and mammalian retrotransposons are derived from a small number of master genes (Deininger et al., 1992), then an alternative master gene is responsible for the amplification of these elements. Therefore, in conjunction with the results of the MultiBlast View, intra-GPIDL and intra-GPID PCR results, it is evident that GPIDL and ID represent distinct SINE families. This newly identified SINE family is found throughout the hystricognath suborder and a related but distinct family, which is not ID, is found in the squirrel. Therefore, although B1 has been relatively successful in both rodent suborders (Hystricognathi and Sciurognathi) ranging from about 10,000 to one million (Veniaminova et al., 2007), we now show that although BC1-based ID elements appear to be unsuccessful in hystricognaths, a previously unidentified ID-like group of elements have amplified prolifically in the guinea pig and are found in all the analyzed hystricognaths.
SINEs have been demonstrated to use LINE machinery for their amplification using a cell culture assay (Dewannieux et al., 2003; Dewannieux and Heidmann, 2005a). The initial findings of low copy numbers of known rodent SINEs in analyzed hystricognaths (Kass et al., 1996;1997) could have potentially been explained by a lack of active LINEs, as it has been shown that LINEs have died out in certain rodent groups (Casavant et al., 2000). Alternatively, the absence of endogenous cellular factors or perhaps the presence of some type of repressor may have been responsible for stalling the analyzed retrotransposons. For example, Bogerd et al. (2006) demonstrated APOBEC3A and APOBEC3B inhibit LINE-1 and Alu retrotransposition. However, the finding of high levels of genomic B1 and IDL integrations suggests the success of the SINE is more a function of the master gene. For example, the BC1 genes of the guinea pig could have served as poor master genes as a result of their highly truncated A-tails (Kass et al., 1996), as the A-tail has been shown to be an important feature of successful proliferation (Dewannieux and Heidmann, 2005b). Additionally, a correlation of A-richness of the BC1 master gene and genomic copy number has been observed among rodents (Roy-Engel et al., 2002). The apparent silencing of B1 activity in two Sigmodontinae species of rodents (Sigmodon hispidus and S. mascotensis), as well as Nyctomys sumichrasti of the Tylominae subfamily prior to LINE extinction (Rinehart et al., 2005) further suggest the master gene itself is the primary factor for successful amplification.
Analysis of retrotransposon subfamilies has provided a useful molecular fossil record to discern phylogenetic relationships (Furano and Usdin, 1995). Additionally, the finding of an expressed BC1 gene in hystricognaths supports their monophyletic relationship with other rodents (Martignetti and Brosius, 1993). Rodents have typically been classified into two suborders (Hystricognathi which include hystricomorphs and caviomorphs, and Sciuorognathi which include the myomorphs, (mice and rats), and sciuromorphs, (squirrels)) (Li et al., 1992), although DeBry (2003) presents data that support Myodonta, Geomyoidea, Castoridae, and Anomaluroidea in one suborder, with Ctenohystrica, Gliridae, and Sciuroidea in the other. This rearranged the positioning of the squirrel-like rodents. However, a common alternative taxonomy includes three suborders, Sciuromorpha, Myomorpha, and Hystricomorpha (Li et al., 1992), which moves the squirrel-like rodents into a separate suborder. A more recent rodent phylogenetic analysis (Huchon et al., 2007) divides rodents into five suborders, although the rodents we list as hystricognaths form a clade of three infraorders grouped in the suborder Ctenohystrica. Despite arguable taxonomic classification of rodents, the squirrel and hystricomorph IDL families appear distinct from each other and from ID elements, suggesting that each was derived from different master genes.
In conclusion, we have provided evidence that the guinea pig IDL SINE family remains active and continues to be involved in the dynamics of the guinea pig genome, and likely IDL continues to contribute to the dynamics of other hystricognath rodent genomes. Further investigations of these elements in conjunction with current research of the known rodent and primate SINEs may offer additional insights into mechanisms and regulation of retrotransposition. Additionally, further analyses of these newly identified SINE families have the potential to further resolve the phylogeny of rodents.
Acknowledgments
This work was supported by a National Institutes of Health AREA grant GM62828-01A1 to DHK, an EMU Biology Department Helwig Research Apprenticeship to BAS, and an EMU Honor’s College Fellowship to NJ. We also thank Sarah Killian for initiating the development of intra-IDL PCR conditions. We greatly appreciate the tissue donations from Drs. Mike Angell and Theresa Lee, as well as the tissue loans from the Museum of Vertebrate Zoology at the University of California at Berkely, and the Smithsonian Institute.
Abbreviations
- GPIDL
guinea pig ID-like
- LINE
long interspersed DNA element
- SINE
short interspersed DNA element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Anzai K, et al. Conservation of the ID sequence and its expression as small RNA in rodent brains: analysis with cDNA for mouse brain-specific small RNA. Brain Res. 1987;388:43–49. doi: 10.1016/0169-328x(87)90019-2. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodulina OR, Kramerov DA. Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett. 1999;457:409–413. doi: 10.1016/s0014-5793(99)01059-5. [DOI] [PubMed] [Google Scholar]
- Casavant NC, et al. The end of the LINE? : lack of recent L1 activity in a group of South American rodents. Genetics. 2000;154:1809–1817. doi: 10.1093/genetics/154.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Erchia AM, Gissi C, Pesole G, Saccone C, Arnason U, et al. The guinea-pig is not a rodent. Nature. 1996;381:597–600. doi: 10.1038/381597a0. [DOI] [PubMed] [Google Scholar]
- Deininger PL. SINEs, short interspersed repeated DNA elements in higher eucaryotes. In: Berg DE, Howe MM, editors. Mobile DNA. American Society for Microbiology; Washington, DC: 1989. pp. 619–636. [Google Scholar]
- Deininger PL, et al. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Deininger PL, et al. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotranspoition. Genomics. 2005a;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol. 2005b;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- Furano AV, Usdin K. DNA “fossils” and phylogenetic analysis. Using L1 (LINE-1, long interspersed repeated) DNA to determine the evolutionary history of mammals. J Biol Chem. 1995;270:25301–25304. doi: 10.1074/jbc.270.43.25301. [DOI] [PubMed] [Google Scholar]
- Gentry A, Clutton-Brock J, Groves CP. The naming of wild animal species and their domestic derivatives. Journal of Archaeological Science. 2004;31:645–651. [Google Scholar]
- Graur D, Hide WA, Li WH. Is the guinea-pig a rodent? Nature. 1991;351:649–652. doi: 10.1038/351649a0. [DOI] [PubMed] [Google Scholar]
- Kapitov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- Kass DH, Kim J, Deininger PL. Sporadic amplification of ID elements in rodents. J Mol Evol. 1996;42:7–14. doi: 10.1007/BF00163205. [DOI] [PubMed] [Google Scholar]
- Kass DH, et al. Evolution of B2 repeats: the muroid explosion. Genetica. 1997;99:1–13. doi: 10.1007/BF02259494. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc Natl Acad Sci USA. 1994;91:3607–3611. doi: 10.1073/pnas.91.9.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno R, et al. Guinea pig D-amino-acid oxidase cDNA and phylogenetic position. DNA Seq. 1999;10:85–91. doi: 10.3109/10425179909008422. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li WH, et al. The molecular taxonomy and evolution of the guinea pig. J Hered. 1992;83:174–181. doi: 10.1093/oxfordjournals.jhered.a111188. [DOI] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. Neural BC1 RNA as an evolutionary marker: guinea pig remains a rodent. Proc Natl Acad Sci USA. 1993;90:9698–9702. doi: 10.1073/pnas.90.20.9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Terai Y, Okada N. Characterization of novel Alu- and tRNA-related SINEs from the tree shrew and evolutionary implications of their origins. Mol Biol Evol. 2002;19:1964–1972. doi: 10.1093/oxfordjournals.molbev.a004020. [DOI] [PubMed] [Google Scholar]
- Philippe H. Rodent monophyly: pitfalls of molecular phylogenies. J Mol Evol. 1997;45:712–715. [PubMed] [Google Scholar]
- Rinehart TA, Grahn RA, Wichman HA. SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet Genome Res. 2005;110:416–425. doi: 10.1159/000084974. [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, et al. Active Alu element “A-tails”: size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky NS, Ten OA, Kramerov DA. B1 and related SINEs in mammalian genomes. Gene. 2003;319:149–160. doi: 10.1016/s0378-1119(03)00805-9. [DOI] [PubMed] [Google Scholar]
- Veniaminova NA, Vassetzky NS, Kramerov DA. B1 SINEs in different rodent families. Genomics. 2007;89:678–86. doi: 10.1016/j.ygeno.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]