Abstract
Although tertiary folding of whole protein domains is prohibited by the cramped dimensions of the ribosomal tunnel, dynamic tertiary interactions may permit folding of small elementary units within the tunnel. To probe this possibility, we used a β-hairpin as well as an α-helical hairpin from the cytosolic N-terminus of a voltage-gated potassium channel and determined a probability of folding for each at defined locations inside and outside the tunnel. Minimalist tertiary structures can form near the exit port of the tunnel, a region that provides an entropic window for initial exploration of local peptide conformations. Tertiary subdomains of the nascent peptide fold sequentially, but not independently, during translation. These studies offer an approach for diagnosing the molecular basis for folding defects that lead to protein malfunction and provide insight into the role of the ribosome during early potassium channel biogenesis.
Keywords: Protein folding, nascent peptides, potassium channel biogenesis, T1 domain
Protein folding begins with the birth of the peptide within the ribosomal tunnel1–8, but the dimensions of this tunnel, 100Å in length and 10–20Å in width9–12, limit the dimensions of a folded peptide that can fit inside the tunnel. For example, the complete folding of a tertiary monomeric functional T1 domain of the voltage-gated potassium channel, Kv1.3, which is approximately 27×27×25Å13,14 (Fig. 1a), is precluded1. However, we may ask whether smaller subdomains can fold inside the ribosomal tunnel. Although compact secondary structures form within the tunnel1–8, including regions of the T1 domain1, subdomain formation inside the tunnel has not been investigated explicitly. The elementary tertiary folding unit has not been established.
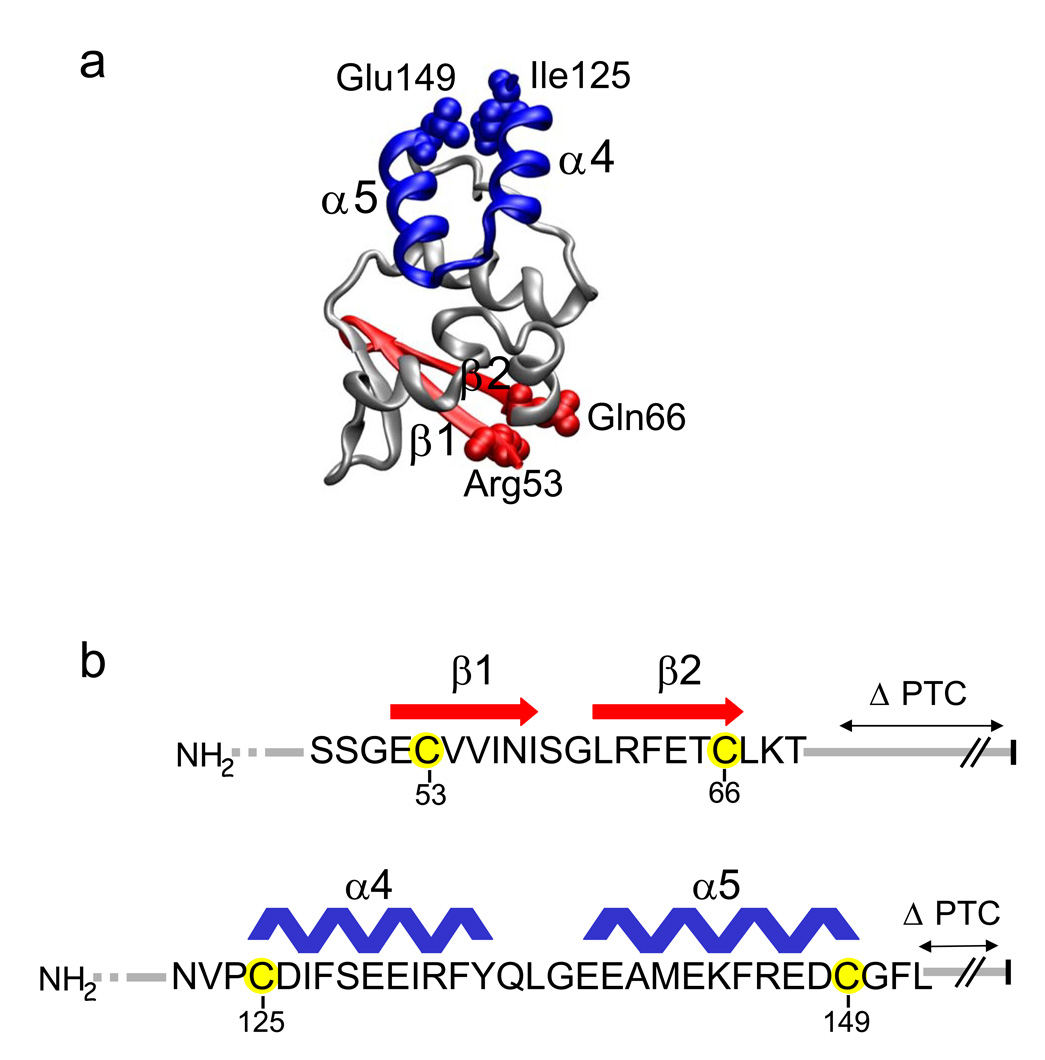
Figure 1. The T1 domain and Experimental Design.
(a) Hairpin structures. The monomeric T1 domain (taken from Minor et al., 200013 for Kv1.2) is shown with the indicated subdomains: a β–hairpin (red) comprised of β1 and β2, and an α-helical hairpin (blue) comprised of α4 and α5. Hairpin terminal residues Arg53, Gln66, Ile125, and Glu149 are shown as spacefilling atoms and are equivalent to residues 34, 47, 106, and 130, respectively, at the homologous hairpin termini in Kv1.2. (b). Amino acid sequence of the T1 Kv1.3 β–hairpin (red) and α-helical hairpin (blue) with secondary structure assignments derived from the highly identical Kv1.2 structure in (a)14. Engineered cysteines 53C and 66C (β–hairpin) and 125C and 149C (α-helical hairpin) in Kv1.3 are highlighted in yellow. The PTC is indicated by the vertical black bar at the right.
The T1 domain is in the cytoplasmic N-terminus of voltage-gated potassium (Kv) channels. It is a recognition domain that plays a crucial role in the folding and oligomerization of Kv channel proteins, enabling formation of the tetrameric channel15–17, its axonal targeting18, and possibly the opening and closing of the permeation pathway for K+ ions13,19–21. We have previously shown that T1 domains tetramerize while still attached to ribosomes22, and that the whole T1 domain folds into its tertiary structure only after the primary sequence emerges from the ribosomal tunnel and the linker between T1 and the first transmembrane segment has been synthesized1.
T1 is comprised of several subdomains, including a β-hairpin, β1/β2, and an α–helical hairpin, α4/α5 (Fig.1). Can these subdomains fold inside the ribosomal tunnel? In this paper, we assess the extent and relative ease of T1 subdomain folding and the compartment in which this folding occurs. We also address a key mechanistic issue in protein folding in general, namely, whether subdomains of nascent peptides fold sequentially in the tunnel or in a concerted fashion upon emergence, and whether the folding of one nascent subdomain depends on the synthesis and/or folding of another subdomain.
Here, we establish that tertiary intrapeptide interactions occur within the tunnel and that stable hairpins form upon emergence from the tunnel. In addition, folding of a C-terminal hairpin depends on the presence and status of the N-terminal β-hairpin. Moreover, the nascent peptide-tunnel complex may be more dynamic than thought previously, and a distal portion of the tunnel (near the exit port) provides an entropic window for exploration of conformational space by the nascent peptide.
RESULTS
Subdomain Folding
To study elementary tertiary folding of T1 subdomains at different stages of biogenesis, we chose two subdomains: a pair of antiparallel β-strands, i.e., a β-hairpin, and an α-helical hairpin (Fig. 1, red and blue, respectively). According to the T1 crystal structure of Kv1.213, which is 95% identical in sequence to the T1 domain of human Kv1.3, these subdomains are located at the N- and C-terminal regions, respectively, of the T1 domain (Fig. 1a). We engineered a pair of cysteines (53Cys and 66Cys, denoted as 53C66C) into the β-hairpin and a different pair (125C149C) into the α-helical hairpin. In each case, the cysteines are exposed in the folded monomer and are thus available to crosslinking reagents under the conditions of our assay1,23. Moreover, these cysteines are predicted to be within 4–6 Å of each other if the subdomain is folded similarly to that modeled in the crystal structure of the mature T1 domain13,14.
To crosslink a cysteine pair, we used ortho-phenyldimaleimide (PDM), a bifunctional crosslinking reagent with an intermaleimide distance of ~6Å. If the engineered cysteines in each of the T1 subdomains shown in Fig. 1b come within 4–7 Å of each other, then they can be crosslinked with PDM. To distinguish crosslinked from non-crosslinked products, we use methoxy-polyethylene glycol maleimide (PEG-MAL) and methoxy-polyethylene glycol thiol (PEG-SH). Addition of the PEG moiety to a protein (pegylation) shifts its molecular mass by ≥10 kDa. A free SH group can be labeled with PEG-MAL24, and a free peptidyl-maleimide can be labeled with PEG-SH23. A crosslinked subdomain will fail to undergo a gel shift with either pegylation reagent, PEG-MAL or PEG-SH. A probability of crosslinking, Pxlink, can be calculated, as described in Kosolapov et al. 20041. We consider that the extent of crosslinking reflects tertiary interactions and, depending on the location of the engineered cysteine pairs, the probability of tertiary folding.
To determine the cellular compartment (e.g., ribosomal tunnel vs cytosol) in which subdomain folding occurs, we generated double-cysteine constructs as nascent peptides (still attached to the ribosome) at various distances from the peptidyl transferase center (PTC; Fig. 1b) using different enzyme restriction sites (Supplementary Table 1) that eliminate the stop codon. This produces different chain lengths between the PTC and the cysteine at the C-terminus of each subdomain. The crosslinking assay was performed on each of these double-cysteine constructs. In addition, similar assays were carried out on constructs containing only a single cysteine of each cysteine pair to calculate the probability of crosslinking, Pxlink (Methods and Kosolapov et al., 20041). Each mRNA for the engineered nascent peptide was translated in a cell-free, membrane-free rabbit reticulocyte system with added 35S-methionine and then subjected to the crosslinking assay1 (see Methods). After labeling with PDM, samples are first treated with SDS and then pegylated to assay for either free available cysteines or free available peptidyl-maleimides.
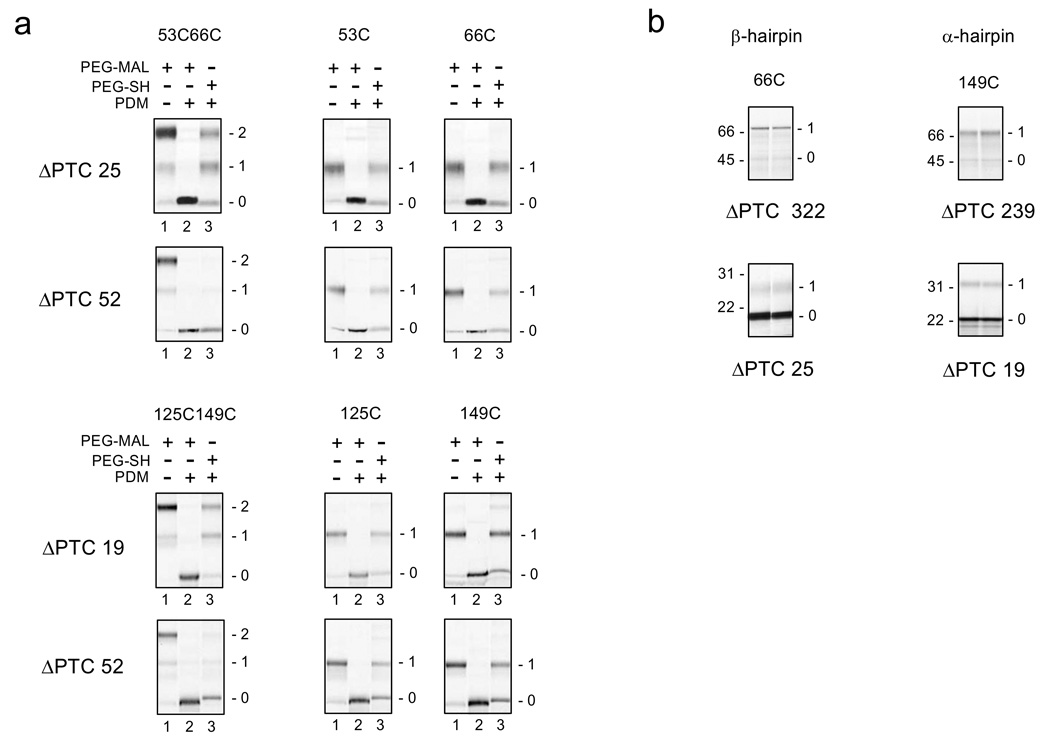
Figure 2a shows data for the crosslinking assays of the β–hairpin (top two rows) and the α-hairpin (bottom two rows) from constructs with different numbers of amino acids between the PTC and the C-terminal cysteine, ΔPTC (calculated as the number of the last residue included by the restriction site minus the residue number of the engineered cysteine, e.g., 91−66 = 25 for the first construct shown in Figure 2a). Regardless of the ΔPTC and number of cysteines in the construct, both N- and C-terminal cysteines were available for pegylation (lanes 1), and were completely labeled by PDM (lanes 2). Moreover, all PDM-modified single-cysteine constructs show a similar efficiency of labeling with PEG-SH, regardless of ΔPTC (lane 3 for all single-cysteine constructs). For a ΔPTC of 52 (outside tunnel), both the β-hairpin and the α-hairpin double-cysteine nascent peptides that were modified with PDM produced negligible gel shifts with PEG-SH (lanes 3, second and fourth rows, leftmost gel). This indicates that no free peptidyl-maleimides were present, i.e., the cysteines are crosslinked and subdomains are mostly folded. In contrast, double-cysteine peptides with ΔPTC of 19 or 25 residues (deeper in the tunnel) were not crosslinked and therefore not folded. In this case, the PDM-treated peptides contained free peptidyl-maleimides that were pegylated with PEG-SH to produce gel shifts (bands 1 and 2 in lanes 3, first and third row, leftmost gels). Single-cysteine constructs were not crosslinked, yet were modified by PDM to contain a free maleimide that is pegylated by PEG-SH (band 1 in lanes 3, two rightmost gels). Crosslinking data for additional constructs with ΔPTC values of 27 and 39 for the β-hairpin and 24 and 29 for the α-hairpin are shown in Supplementary Figure 1.
Figure 2. Crosslinking and Accessibility Assays for β- and α- Hairpins.
(a). Intramolecular crosslinking assay for indicated cysteine pairs (left column of gels) and single cysteines (two right-most columns of gels) for β-hairpin (top two rows) and α-hairpin (bottom two rows). The number of amino acids from the PTC to the C-terminal cysteine of the pair, or to the single cysteine, is indicated to the left of each row of gels as the ΔPTC. Engineered restriction sites, Xba I@91 and Kpn I@118, were used to produce ΔPTCs of 25 and 52, respectively, for the β-hairpin constructs. An engineered restriction site, BstE II@168, and a native site, Xba I@201, were used to produce ΔPTCs of 19 and 52, respectively, for the α-hairpin constructs. Numbers to the right of each gel represent doubly (2) or singly (1) pegylated or unpegylated (0) protein. In all cases, lane 2 background is significantly less than other lanes due to >95% labeling of the peptide with PDM, which leads to unpegylated protein that accumulates entirely in band 0. (b) Accessibility assays for C-terminal cysteines. Nascent peptides were pegylated as described in the Methods for 4 and 6 hours (lanes 1 and 2, respectively) for 66C (left column of gels) or 149C (right column of gels) in constructs with the indicated number of amino acids from the PTC to the C-terminal cysteine (ΔPTC). Long peptides (ΔPTC = 322 and 239), residing outside tunnel are shown in the top row of gels; short peptides (ΔPTC = 25 and 19) residing within the tunnel are shown in the bottom row of gels. Numbers to the left of each gel are molecular weight standards; numbers to the right of each gel represent singly (1) pegylated or unpegylated (0) protein.
Location of the Subdomain in the Tunnel
The interpretation of these crosslinking data requires that we know the location of the subdomain, inside or outside of the tunnel, which depends on both ΔPTC and secondary structure of the peptide between the C-terminal cysteine of the subdomain and the PTC. To determine subdomain location, we used an accessibility assay (Fig. 2b and Supplementary Fig. 2). This assay also relies on a mass-tagging strategy using PEG-MAL2,24, but without the SDS pre-treatment used in the crosslinking assay (see Methods)1. The final extent of PEG-MAL labeling of a cysteine in the last 20Å of the tunnel is monotonically dependent on the distance of the modifiable cysteine from the PTC2. Accessibility is therefore an accurate estimator of distance from the PTC2 (see Discussion). For each subdomain, a single cysteine, corresponding to the C-terminal cysteine of each cysteine pair was engineered into the nascent peptide, which was treated with PEG-MAL and the final extent of labeling determined. As shown in Figure 2b, for β- and α-hairpins located outside of the ribosomal tunnel, i.e., ΔPTC is 322 and 239, respectively (top gels), the fraction of pegylated protein is ≥ 0.88. In contrast, the fraction of pegylated protein is ≤ 0.14 for constructs where C-terminal cysteines of the β- and α-subdomains were placed at 25 and 19 residues, respectively, from the PTC (bottom gels). These cysteines, located inside the tunnel, are 80 Å or less from the PTC2.
Relationship between Subdomain Location and Folding
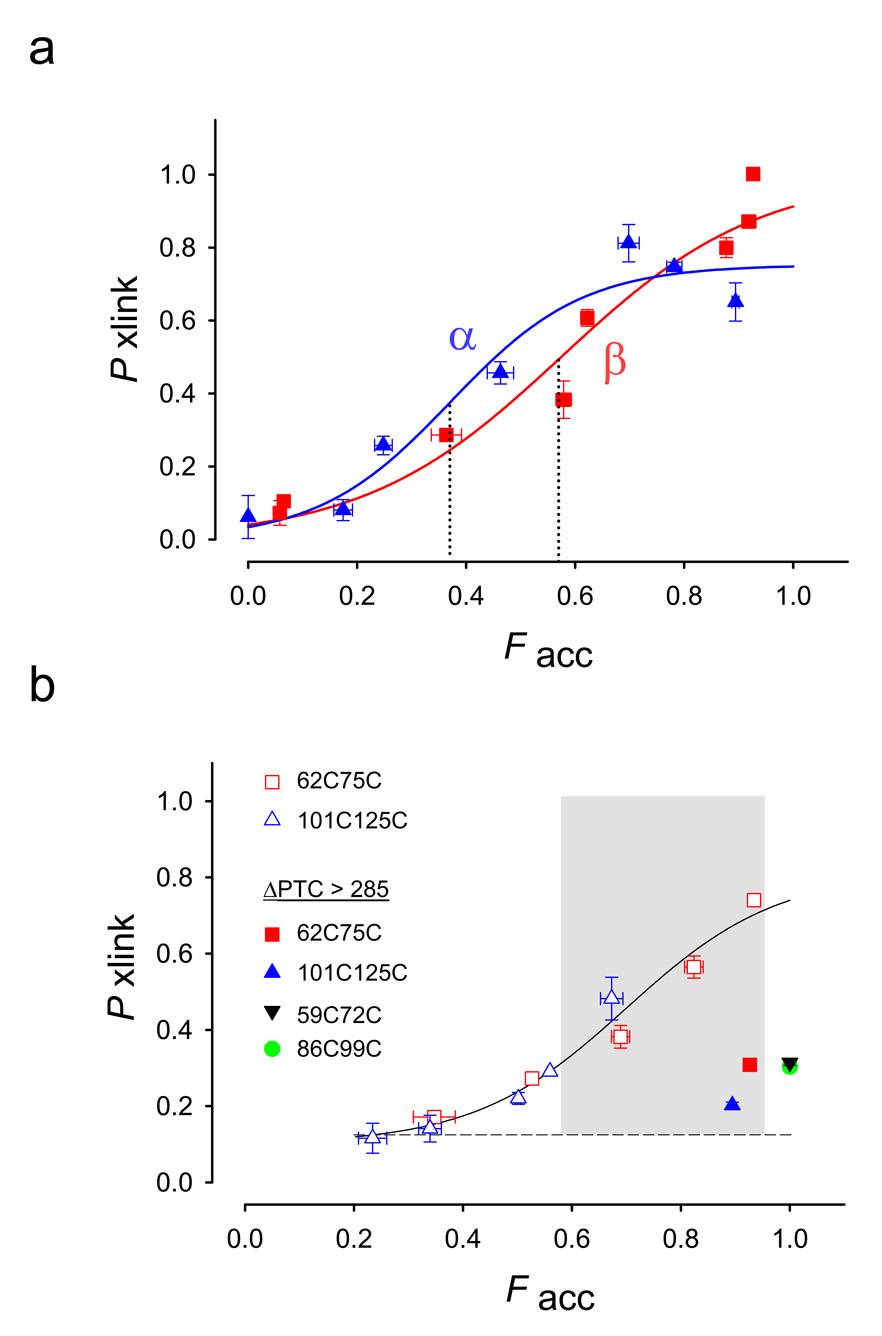
To determine the tunnel location where hairpin folding occurs, we combined the results from the crosslinking and accessibility assays for each hairpin. In Figure 3a, we plot the probability of crosslinking (Pxlink) against the fraction of accessible peptide (Facc). The midpoint of each PF-curve, F50, is the fraction accessible when 50% of the peptide is maximally crosslinked (dotted lines). Thus we can compare Pxlink at different distances from the PTC and the relative ease of folding for each subdomain at the same location. Both subdomains apparently begin to fold before they emerge completely from the ribosomal tunnel, and maximum folding occurs after the subdomains emerge from the tunnel. Pxlink saturates below 100% completion due to rapid binding of a PDM molecule to both cysteines23. The F50 values are 0.57 ±0.10 for the β-hairpin and 0.37 ± 0.06 for the α-hairpin, not significantly different (P = 0.075, Z-test), suggesting that the relative ease of folding of the two different hairpins is similar.
Figure 3. Accessibility-Dependent Probability of Crosslinking.
(a). Probability of crosslinking, Pxlink, as a function of the fraction of accessible peptide, Facc. Data are derived from crosslinking and accessibility assays (exemplified in Fig. 2) for β-hairpins (red squares) α-helical hairpins (blue triangles) and fit to a sigmoidal function with two parameters (SigmaPlot 8.0). Data are means ± SEM for at least triplicate measurements. The dotted lines indicate F50 values as defined in the text. The β-hairpin has F50 = 0.37 ± 0.06, whereas the α-helical hairpin has F50= 0.57 ± 0.10. These midpoints are not significantly different (P= 0.075, Z-test). The ΔPTC values are (increasing order of Facc) 21, 25, 27, 29, 39, 322, 52, and 86 (latter two occur at the same Facc) for the β-hairpin and 3, 19, 24, 29, 40, 52, and 239 for the α-hairpin. (b) Probability of crosslinking of 62C75C (red squares) and 101C125C (blue triangles). A curve was drawn through all the data points, but no particular function is intended. The shaded region indicates a region in which peptides can be crosslinked (an entropic window). All filled symbols represent cysteine pairs far outside the tunnel on a long tether (ΔPTC values > 285). Pxlink for these constructs is 0.2–0.3. Pxlink for both 86C99C (filled green circle) and 59C72C (filled inverted black triangle) is ~0.3, in agreement with 62C75C (filled red square) at this same location. The ΔPTC values are (increasing order of Facc) 30, 35, 37, 43, 67 and 313 for the 62C75C constructs and 27, 38, 41, 48, 43, and 263 for the 101C125C constructs. Data are means ± average error or ± SEM for 2–4 replicate samples, except for points at ΔPTC 48, 67 and the 86C99C construct (green filled circle) at ΔPTC 289. For all other points, errors are either clearly visible or within the symbol.
Is subdomain folding affected by the interaction of the ribosomal tunnel with the subdomain or is it due solely to the intrinsic properties of the peptide? To address this question, we first released the nascent peptides from the ribosome using RNase, isolated the released peptide, and assayed it for crosslinking. A nascent peptide in which the C-terminus of the β-hairpin sequence was located 25 residues from the PTC was released and gave nearly maximal crosslinking for the hairpin (Supplementary Fig. 3), suggesting that it folded completely. In contrast, a released peptide in which the C-terminus of the α-hairpin sequence was 19 residues from the PTC, crosslinked only 55% of the maximum attained while attached to the ribosome. Intriguingly, when this sequence was first positioned more distally in the tunnel, e.g., 24 and 29 residues from the PTC, and then released, the α-hairpin apparently folded more completely, 71% and 76% of the maximum, respectively. This behavior suggests that several factors may determine the probability of folding of the α-hairpin: the location of the α-hairpin sequence in the tunnel prior to release (so that the tunnel promotes a folding-competent state of the peptide), the total length of the nascent peptide, and/or the number and nature of the residues between the hairpin C-terminus and the PTC. In addition, the maximally folded α-hairpin present in the crystal structure may represent only one of several hairpin structures. This α-hairpin behavior contrasts with the folding requirements for the β-hairpin, which is constrained by the tunnel but apparently folds completely once it is released.
Two additional conclusions may be drawn from the PF-curves shown in Figure 3a. First, folding of the upstream β-hairpin does not require the presence and/or folding of the α-hairpin, as evidenced by maximum folding for the ΔPTC 52 peptide, which lacks the α-hairpin. Second, tertiary folding of both hairpins is prohibited in short nascent intermediates with relatively inaccessible C-terminal cysteines (ΔPTC<21). However, the α-hairpin shows a significant amount of folding (Pxlink = 0.26, 34% of its maximum) when ΔPTC = 24, i.e., the C-terminal cysteine is inside the tunnel (Facc = 0.25), which suggests a dynamic equilibrium between folded and unfolded species (Supplementary Results).
Evidence for an Entropic Window
The PF-curves shown in Figure 3a make two predictions. First, at locations far outside the tunnel (ΔPTC>239), the hairpin subdomains, as well as the whole T1 domain1, are folded. We can therefore predict from the crystal structure of the T1 domain which side-chains in the folded T1 are too distant from one another to be intramolecularly crosslinked (>14Å 1) and should therefore exhibit a low Pxlink. Second, at tunnel locations within 80Å of the PTC, no folding occurs. Thus pairs of distant cysteines engineered anywhere along the nascent peptide, including non-hairpin sequences, should also exhibit a low Pxlink within these 80Å. Each prediction derives from a different constraint on tertiary folding. In the first case, the whole stably-folded T1 domain has a network of intramolecular interactions that restricts its mobility, decreasing the degrees of freedom allowed for any given stretch of primary sequence within the folded T1. In the second case, the tunnel itself is too small to permit intramolecular tertiary interactions in the nascent peptide. To test these predictions, we designed two separate double-cysteine constructs, 62C75C and 101C125C, in the region adjacent to the β-hairpin and α-hairpin, respectively. The cysteines in the 62C75C and 101C125C pairs are separated by the same number of amino acids, respectively, as in the 53C66C (β-hairpin) and 125C149C (α-hairpin) pairs. In contrast to intra-pair proximities (6–7Å) for 53C66C and 125C149C, both 62C75C and 101C125C are farther apart in the folded T1 monomer, ~25 and ~17Å, respectively.
As predicted, crosslinking efficiencies for 62C75C (red squares, Fig. 3b) and 101C125C (blue triangles) are low when these constructs were placed either deep inside or completely outside of the tunnel (Facc ≤ 0.5 or ≥ 0.9, respectively), consistent with the lack of crosslinking reported for two cysteines located 14 Å apart in the whole folded T1 domain1.
However, between these two regions of restricted conformational entropy there is a region where some crosslinking is permitted (shaded area, Fig. 3b), even though in each case the introduced cysteines within the pair are distant from one another in the mature T1 structure. We used chain lengths that position 62C75C and 101C125C in the more distal regions of the tunnel, at ΔPTC values of 30, 35, 37, 43, 67, 313 and 27, 38, 41, 43, 48, 263, respectively. Short constructs (open symbols, Fig. 3b) with Facc between 0.6 and 0.95 crosslinked to give a Pxlink of ≥ 0.6, suggesting that this region of the tunnel is dynamic and allows the peptide to explore conformational space, therefore constituting an entropic window (shaded region) for sampling potential folding partners. For these constructs, both accessibility measurements and calculations based on secondary structure between the C-terminal cysteine and the PTC, indicate that these C-terminal cysteines are located either just within the tunnel at the exit port or outside in the immediate vicinity of the tunnel. To confirm findings in the restricted region, we engineered two constructs with ΔPTC > 285. We engineered 86C99C between the β- and α-hairpin sequences. These cysteines are 14Å apart in the mature folded structure. We also engineered 59C72C in the β-hairpin sequence. These cysteines are 25Å apart in the mature folded structure. Pxlink for both 86C99C (filled green circle, Fig. 3b) and 59C72C (filled inverted black triangle) is ~0.3, in agreement with 62C75C (filled red square) at this same location. All filled symbols represent cysteine pairs far outside the tunnel on a long tether, which have Pxlink of 0.2–0.3. To confirm findings in the interaction-permissive region, we engineered a construct containing 59C72C that poises this cysteine pair in the vicinity of the exit port, but outside the tunnel. The Pxlink is >0.6 (data not shown). These findings permit a more precise interpretation of the crosslinking results for hairpin residues 53C66C and 125C149C that are in close proximity in the T1 domain (Fig. 3a). While these residues in each hairpin initially interact as part of the exploration of conformational space allotted by the tunnel’s entropic window, the hairpins fold into stable structures only in the context of the longer, folded T1 domain.
Cooperative Subdomain Folding
The α-hairpin folds when the N-terminal region of the T1 domain, including the β-hairpin has already been synthesized and folded (Fig. 3a and Kosolapov et al., 20041). This folded N-terminus may provide a topological template. This hypothesis predicts folding of the α-hairpin sequence should be sensitive to deletion of its preceding T1 residues. To explore this possibility, we truncated the T1 domain at position 122 (Fig. 4), which yields an N-terminally deleted nascent peptide (ΔN) containing the α-hairpin sequence starting 3 residues after the methionine start site. For a long tether (ΔPTC 239), Pxlink is significantly decreased by 46% (P = 0.004, Fig. 4). It appears therefore that the extent of α-hairpin folding depends on the presence of the N-terminal T1 domain. In addition to these long-tethered ΔN-constructs with a folding domain outside of the tunnel, we investigated shorter ΔN-constructs, ΔN-ΔPTC24 and ΔN-ΔPTC29, in which the folding domains reside near the end of the tunnel. The truncated ΔN-ΔPTC24 folds less efficiently than the non-truncated peptide (Pxlink is 0.06 ± 0.12, n=2 versus 0.26 ± 0.03, n=3, respectively, (Supplementary Fig. 4)). Truncated ΔN-ΔPTC29, which is also near the end of the tunnel and provides a larger dynamic range, has a Pxlink of 0.30 ± 0.02 (n=2) versus 0.46 ± 0.03 (n=5) for the non-truncated construct. For both ΔN-ΔPTC24 and ΔN-ΔPTC29 constructs lacking the N-terminal β-hairpin, folding of the α-hairpin is less efficient, again consistent with folding of a C-terminal hairpin depending on the presence and status of the N-terminal β-hairpin sequence. In contrast, the β-hairpin apparently folds in the absence of the C-terminal region of T1, including the α-hairpin sequence. A 118-residue nascent peptide that includes the β-hairpin sequence (ΔPTC = 86) but not the α-hairpin sequence, folds as much as the longer nascent peptides (ΔPTC = 322 (Fig. 3a)) that includes both β- and α-hairpins.
Figure 4. T1 Domain Mutants.
A schematic of T1 constructs with an N-terminal deletion or a T65D mutation is shown along with the full-length peptide for comparison. The T65D mutation is shown as a black circle. A truncated peptide (ΔN) begins at N122M. All constructs were cut with a BstEII restriction enzyme to give a ΔPTC value of 239 for the 125C149C constructs and 322 for the 53C66C constructs. Nascent peptides were translated, isolated, and assayed for crosslinking as described in the Methods. The Pxlink values for these N-terminal deletion and T65D mutants are depicted as white bars. The gray bars represent data shown in Figure 3a for nascent peptides 125C149C and 53C66C. The results for the α-helical hairpins are indicated by the right-most and left-most sets of bars. The results for the β–hairpin are shown in the middle set of bars. Pxlink data are means ± SEM for triplicate measurements.
The second strategy we used to probe the dependence of α-hairpin formation on the β-hairpin was to use a Kv1.3 mutation, T65D. Residue 65 lies in the β-hairpin at the intersubunit interface of the T1 tetramer. Kv1.3 with a T65D mutation does not tetramerize and only achieves one-half as much tertiary folding as wildtype T123,25. We therefore used the T65D mutation to investigate two issues. Does the T65D mutation impair tertiary folding of the β-hairpin? Does the T65D mutation affect α-hairpin formation? We introduced the T65D mutation into both a 53C66C background (ΔPTC 322) and a 125C149C background (ΔPTC 239) that places the β- and α-hairpins far outside of the tunnel. As shown in Figure 4, Pxlink in the T65D mutant is 0.46 for the β-hairpin and 0.26 for the α-hairpin, each significantly lower than wildtype Pxlink (P = 0.003 and 0.01, respectively). The α-hairpin is more disrupted than the β-hairpin, consistent with accumulated disruption for the more C-terminal location of the α-hairpin. We suggest that propagated disruption from the site of mutation all along the folded T1 interface is responsible for defective quaternary structure formation of the T65D mutant T1 domain. Folding of the α-hairpin depends on the presence and status of the β-hairpin.
DISCUSSION
Subdomain Folding
While it is too cramped inside the tunnel to accommodate the entire T1 domain of Kv channels in its fully folded state1, consistent with previous investigations of other proteins6,26,27 and the geometric analysis of Moore and co-workers28, tertiary interactions do occur in the tunnel, particularly in its last 20Å. This region supports helix formation1–4,29, and specifically folding of the α5 helix of the T1 domain studied herein3. Why is this region permissive for tertiary interactions? First, the dimensions of the tunnel in this region may be wider30 than the 20Å estimated from the crystal structures of the ribosome, which lack both nascent peptides and attendant chaperones. Second, the ribosomal tunnel may be more dynamic than previously thought (whereas Steitz’s estimate of the diameter of the largest sphere that can fit inside a peptide-less tunnel, 13.7Å31, can accommodate the β-hairpin, an increase of < 2-fold is required to accommodate the α-hairpin). Moreover, the entrance of the ribosomal tunnel undergoes conformational changes during peptide elongation32 and the tunnel is believed to have a gate33–35. Third, tertiary and secondary structure formation may be coupled36 and thus potentiated in this distal portion of the tunnel that favors secondary folding29, 3. An α-helix requires less room than a hairpin of similar length and amino acid composition, and thus an α-helix may form along the first 80Å of the tunnel, independent of tertiary folding, but the converse may not hold. For the α4/α5 hairpin, tertiary structures in the last 20Å of the tunnel may be favored by coupled secondary structure formation. Moreover, chaperone proteins hovering at the exit port of the tunnel may facilitate tertiary and/or coupled folding.
Although these two subdomains differ in size and length of the hairpin, and the number of hydrophobic interactions along their respective hairpin interfaces, our crosslinking assay suggests that the ease of folding of the β- and the α-hairpin are not significantly different. The β-hairpin evidently folds despite the absence of the α-hairpin. Similarly, the α-hairpin apparently folds in the absence of the β-hairpin in a truncated N-terminally deleted (ΔN) mutant. However, folding of the α-hairpin is not independent of the β-hairpin. The α-hairpin in the ΔN mutant folds less than the α-hairpin in the full-length peptide that contains both β- and α-subdomains. Moreover, mutation of the β-hairpin residue 65 (T65D) causes a decrease in folding of both the β- and α-hairpins. In the full-length mature Kv1.3, T65D prevents both quaternary and complete tertiary folding of the T1 domain, and consequently, function of the Kv1.3 channel23,25. This is expected because tertiary folding and tetramerization of T1 are coupled25. We now understand that the basis for this defect in tertiary folding involves the propagated disruption of subdomain formation. Our findings underscore the usefulness of this approach in diagnosing the molecular basis for folding defects.
Information regarding the extent of folding at discrete locations along the tunnel is embedded in the crosslinking-accessibility (PF) curves in Figure 3a. The locations are deduced from a cysteine accessibility assay. This assay is based on the monotonic dependence of the fraction pegylated for a given cysteine in an all-extended tape measure on the distance of that cysteine from the PTC2. We have compared the cysteine accessibility results obtained for α-hairpin constructs with those obtained for this all-extended tape measure. The accessibility of the C-terminal cysteine in the tunnel, in α-hairpin constructs, is similar to the accessibility of cysteines in the tape measure with the same ΔPTC, demonstrating that these cysteines are at the same location in the tunnel. This similarity is expected given that the segment of the peptide between the C-terminal cysteine of the α-hairpin (149C) and the PTC is completely extended3. Thus, the measured accessibility of 149C is not an artifact of secondary structure formation in the region downstream from 149C. Nor is it an artifact of blocked access to 149C by the upstream α4 and α5 helices in the tunnel.
Dynamics of the Nascent Peptide-Tunnel Complex
Our results suggest that the nascent peptide-tunnel complex may be more dynamic than originally believed. At one location (ΔPTC 24), the level of crosslinking suggests that the α-hairpin exists in a time-averaged equilibrium between an extended α4/α5 species and a tertiary hairpin. Dynamics are also manifest in the distal portion of the tunnel that provides an entropic window for initial exploration of local peptide regions for folding. This represents a restricted time and space in which the nascent peptide visits potential tertiary conformational states. It is possible that this region, the last 20Å near the exit port, is lined by chaperone proteins, which operationally extend the length of an 80Å tunnel. Estimates of the length of the tunnel vary from 80–112Å2,9,10,12,28. Regardless, this region exhibits a monotonically increasing accessibility with increasing distance from the PTC2,30. If this increased accessibility represents a widening of the tunnel, this will allow an increased number and mobility of water molecules and ions, and therefore an increased driving force for burying hydrophobic residues at a tertiary folded interface (hydrophobic effect37). Both α4/α5 and β1/β2 hairpins contain hydrophobic residues that are in close contact all along their folded interfaces13. The final tertiary hairpin fold is stabilized by constraints of the additional secondary and tertiary interactions of the whole folded T1 domain outside the tunnel.
Our results highlight three principles that are likely to play critical roles in protein folding during biogenesis. The first is that intramolecular tertiary interactions occur before the nascent peptide has fully emerged from the ribosomal tunnel, allowing for a regulatory role of folding by the ribosome and its attendant chaperones. The second is that the nascent peptide in the exit port of the tunnel is structurally dynamic and can therefore explore a myriad of conformations before fully emerging from the ribosome. This may enable the peptide to fold more efficiently than it would in a less confined space. The third is that tertiary interactions are influenced by subdomain folding elsewhere in the nascent peptide. We propose that these three principles constrain and shape the earliest stages of protein folding and may be used to help understand the molecular basis for folding defects that underlie protein malfunction.
METHODS
Constructs and In Vitro Translation
We used standard methods of bacterial transformation, plasmid DNA preparation, and restriction enzyme analysis. The nucleotide sequences of all mutants were confirmed by automated cycle sequencing performed by the DNA Sequencing Facility at the School of Medicine on an ABI 377 Sequencer using Big dye terminator chemistry (A0BI). All mutant DNAs were sequenced throughout the entire coding region. Engineered cysteines, restriction enzyme sites, and N-terminus deletions were introduced into pSP/Kv1.3/cysteine-free22 using QuikChange Site-Directed Mutagenesis Kit as described in Kosolapov and Deutsch23.
We synthesized capped cRNA in vitro from linearized templates using Sp6 RNA polymerase (Promega, Madison, WI). Linearized templates for Kv1.3 translocation intermediates were generated using several restriction enzymes to produce different length DNA constructs lacking a stop codon. cRNAs were translated in vitro with [35S]Methionine (2 µl per 25 µl translation mixture; ~10 µCi µl−1 Express, Dupont/NEN Research Products, Boston, MA) for 1 h at 22°C in a rabbit reticulocyte lysate (2 mM final [DTT] ) according to the Promega Protocol and Application Guide.
Crosslinking assay
As described previously23, we added translation reaction (10–20 µl) to 500 µl phosphate buffered saline (PBS: Ca -free DPBS (GIBCO), containing 4 mM MgCl2, pH 7.3) with 2 mM DTT. The suspension was centrifuged using a TLA100.3 Beckman rotor at 70,000 rpm/20 min/4°C through a sucrose cushion (120 µl, containing 0.5 M sucrose, 100 mM KCl, 50 mM Hepes, 5 mM MgCl2, 2 mM DTT, pH 7.5). The pellet was resuspended in 50 or 500 µl PBS. Ortho-phenyldimaleimide (PDM; Sigma Chemical Co.), 0.5 mM, was added to those samples to be labeled while a control sample was treated identically but in the absence of PDM, at ~0°C for 30 min. No reducing agent is present in these incubations. PDM samples used for subsequent MAL-pegylation were quenched with 10 mM beta-mercaptoethanol at room temperature for 10 min. Control samples, untreated with PDM, were treated identically. A third sample was labeled with PDM but reserved for treatment with PEG-SH. Thiol reducing agents must be avoided in PDM samples subsequently treated with PEG-SH, otherwise free maleimides will be modified and further assay with PEG-SH will be blocked. Samples were centrifuged at 70,000 rpm, 4°C, for 20 min (TLA100.3 Beckman rotor), resuspended in 50 µl PBS containing 1% (w/v) SDS and incubated at room temperature for 20 min. Those samples designated for pegylation with methoxy-polyethylene glycol maleimide (PEG-MAL, MW 5000; Shearwater, Inc.) were treated with 10 mM beta-mercaptoethanol to prevent oxidation, which inhibits pegylation. Samples destined for pegylation with methoxy-polyethylene-thiol (PEG-SH, MW 5000; Shearwater, Inc.) received 50 µl PBS containing SDS only. All SDS-treated samples were diluted with either 50 µl PBS containing PEG-MAL to give a final concentration of PEG-MAL of 20 mM and 5 mM beta-mercaptoethanol or 50 µl PBS containing PEG-SH to give a final concentration of 20 mM PEG-SH. Both the pegylation and PDM reactions reached a maximum, constant level within 60 min and < 15 min, respectively, at 4°C 23.
Single- and double-cysteine constructs were treated identically, as described above. Data derived from single-cysteine constructs serve as controls to calculate the efficiency of PEG-SH pegylation needed to determine the probability of crosslinking/folding in the double-cysteine constructs.
Accessibility Assay
We added translation reaction (10–20 µl) to 500 µl phosphate buffered saline (PBS: Ca -free DPBS (GIBCO), containing 4 mM MgCl2, pH 7.3) with 2 mM DTT. The suspension was centrifuged at 70,000 rpm/20 min/4°C (TLA100.3 Beckman rotor) through a sucrose cushion (120 µl, containing 0.5M sucrose, 100 mM KCl, 50 mM Hepes, 5 mM MgCl2, 2 mM DTT, pH 7.5). The pellet was resuspended in 100 µl PBS with 50 µM beta-mercaptoethanol and treated with 1 mM PEG-MAL (SunBio) at 4°C for 4–6 h. To quench the pegylation reaction each sample was treated with DTT to neutralize PEG-MAL (200:1 ratio), incubated at room temperature for 15 min and centrifuged at 70,000 rpm/20 min/4°C (TLA100.3 Beckman rotor). . The relative accessibility of each C-terminal cysteine indicates whether the subdomain is inside or outside of the tunnel.
Gel Electrophoresis and Fluorography
We treated all samples with RNase (20 µg ml−1) before loading on the gel and performed electrophoresis using the NuPAGE system and precast Bis-Tris 10% or 12% gels and MOPS running buffer. Gels were soaked in Amplify (Amersham Corp., Arlington Heights, IL) to enhance 35S fluorography, dried, and exposed to Kodak X-AR film at −70°C. Typical exposure times were 16–30 hours. We quantified gels using a Molecular Dynamic PhosphorImager (Sunnyvale, CA), which detects cpm that are not necessarily visualized in autoradiograms exposed for 16–30 hours.
Analysis of Pegylation Ladders
For any given construct, radioactive protein incubated with PEG-MAL or PEG-SH was detected as distinct bands on NuPAGE gels and quantified using a PhosphorImager (Molecular Dynamics, Inc). The data were analyzed as follows. For each lane, j, of the gel, the fraction of total protein molecules with exactly i pegylated cysteines was calculated as Wj(i)=cpm(i)/Σcpm(i) [eqn. 1], where cpm(i) is the counts per minute in the ith bin. If each cysteine is assumed to label to the same final extent, the fraction Fj of individual cysteines pegylated in the jth lane is ΣiWj(i)/N [eqn. 2], where N is the total number of cysteines in the protein molecule.
In this study, we apply the analysis described in Kosolapov et al.1 to determine the probability of crosslinking cysteines that reside inside the ribosomal tunnel as well as those residing outside. Specifically, we compare the labeling in single cysteine constructs and double cysteine constructs and can estimate the crosslinking efficiency. The crosslinking efficiency is used to determine the probability of a pair of cysteines being crosslinked by PDM: Pxlink = 1 − (2F3,AB)/(F3,A + F3,B), where F3,A and F3,B are fractions of PDM-treated single cysteine (A or B) mutants labeled with PEG-SH, and F3,AB is the fraction of PDM-treated double cysteine (A and B) mutant labeled with PEG-SH.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Richard Horn, Joachim Frank, and Ulrich Hartl for careful reading of the manuscript and Dr. Richard Horn for helpful discussion. This research was funded by NIH grant GM 52302 to C.D.
A.K. performed the experiments; A.K. and C.D. designed the research, interpreted the results, and wrote the manuscript.
Supported by National Institutes of Health Grant GM
REFERENCES
- 1.Kosolapov A, Tu L, Wang J, Deutsch C. Structure Acquisition of the T1 Domain of Kv1.3 During Biogenesis. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 3.Tu L, Wang J, Deutsch C. Biogenesis of the T1-S1 linker of voltage-gated K+ channels. Biochemistry. 2007 doi: 10.1021/bi700319f. in press. [DOI] [PubMed] [Google Scholar]
- 4.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 5.Mingarro I, Nilsson I, Whitley P, von Heijne G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biology. 2000;1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowarik M, Kung S, Martoglio B, Helenius A. Protein folding during cotranslational translocation in the endoplasmic reticulum. Molecular Cell. 2002;10:769–778. doi: 10.1016/s1097-2765(02)00685-8. [DOI] [PubMed] [Google Scholar]
- 7.Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Progress in Nucleic Acid Research & Molecular Biology. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 8.Matlack KE, Walter P. The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. Journal of Biological Chemistry. 1995;270:6170–6180. doi: 10.1074/jbc.270.11.6170. [DOI] [PubMed] [Google Scholar]
- 9.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 10.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 11.Menetret JF, et al. The structure of ribosome-channel complexes engaged in protein translocation. Molecular Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 12.Beckmann R, et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 13.Minor DL, et al. The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel. Cell. 2000;102:657–670. doi: 10.1016/s0092-8674(00)00088-x. [DOI] [PubMed] [Google Scholar]
- 14.Kreusch A, Pfaffinger PJ, Stevens CF, Choe S. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature. 1998;392:945–948. doi: 10.1038/31978. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Jan YN, Jan LY. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channels. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 16.Shen NV, Chen X, Boyer MM, Pfaffinger P. Deletion analysis of K+ channel assembly. Neuron. 1993;11:67–76. doi: 10.1016/0896-6273(93)90271-r. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Yu W, Jan JN, Jan L, Li M. Assembly of voltage-gated potassium channels. Conserved hydrophilic motifs determine subfamily-specific interactions between the α-subunits. J. Biological Chemistry. 1995;270:24761–24768. doi: 10.1074/jbc.270.42.24761. [DOI] [PubMed] [Google Scholar]
- 18.Gu C, Jan YN, Jan LY. A conserved domain in axonal targeting of Kv1 (Shaker) voltage-gated potassium channels. Science. 2003;301:646–649. doi: 10.1126/science.1086998. [DOI] [PubMed] [Google Scholar]
- 19.Cushman SJ, et al. Voltage dependent activation of potassium channels is coupled to T1 domain structure. Nature Structural Biology. 2000;7:403–407. doi: 10.1038/75185. [DOI] [PubMed] [Google Scholar]
- 20.Kurata HT, et al. Amino-terminal determinants of U-type inactivation of voltage-gated K+ channels. Journal of Biological Chemistry. 2002;277:29045–29053. doi: 10.1074/jbc.M111470200. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Covarrubias M. Voltage-dependent gating rearrangements in the intracellular T1-T1 interface of a K+ channel. J. Gen. Physiol. 2006;127:391–400. doi: 10.1085/jgp.200509442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Robinson JM, Edwards D, Deutsch C. T1-T1 interactions occur in ER membranes while nascent Kv peptides are still attached to ribosomes. Biochemistry. 2001;40:10934–10946. doi: 10.1021/bi010763e. [DOI] [PubMed] [Google Scholar]
- 23.Kosolapov A, Deutsch C. Folding of the voltage-gated K+ channel T1 recognition domain. Journal of Biological Chemistry. 2003;278:4305–4313. doi: 10.1074/jbc.M209422200. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Deutsch C. Pegylation: a method for assessing topological accessibilities in Kv1.3. Biochemistry. 2001;40:13288–13301. doi: 10.1021/bi0107647. [DOI] [PubMed] [Google Scholar]
- 25.Robinson JM, Deutsch C. Coupled Tertiary Folding and Oligomerization of the T1 Domain of Kv Channels. Neuron. 2005 doi: 10.1016/j.neuron.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 26.Kolb VA, Makeyev EV, Spirin AS. Folding of firefly luciferase during translation in a cell-free system. EMBO J. 1994;13:3631–3637. doi: 10.1002/j.1460-2075.1994.tb06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makeyev EV, Kolb VA, Spirin AS. Enzymatic activity of the ribosome-bound nascent polypeptide. FEBS Lett. 1996;378:166–170. doi: 10.1016/0014-5793(95)01438-1. [DOI] [PubMed] [Google Scholar]
- 28.Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J. Mol. Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 31.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 32.Gabashvili IS, et al. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Molecular Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 33.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 34.Berisio R, et al. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nature Structural Biology. 2003;10:366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- 35.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Daggett V, Fersht AR. Is there a unifying mechanism for protein folding? Trends in Biochemical. Sciences. 2003;28:18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 37.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.