Abstract
Purpose
Advanced prostate cancer is first treated with androgen deprivation therapy. However, tumors become resistant to and grow despite castrate levels of testosterone. Growth and proliferation of castration-resistant prostate cancer (CRPC) is mediated by gain-of-function changes in the androgen receptor (AR) and AR reactivation. Expression of manganese superoxide dismutase (SOD2), which regulates cellular reactive oxygen species (ROS), is markedly downregulated in CRPC when compared to hormone responsive tumors.
Experimental Design
Here, we knocked down SOD2 expression in AR-expressing LNCaP prostate cancer cells and determined gene expression changes, transcription factor binding and AR transcription activity in SOD2 knockdown cells.
Results
SOD2 knockdown results in an increase in ROS. Gene expression changes induced by SOD2 knockdown results in the upregulation of genes which are also androgen responsive and 46% of genes upregulated two-fold by the androgen ligand R1881 are also upregulated to the same extent with SOD2 knockdown. The induction of many of these genes with SOD2 knockdown, such as VEGFA and FKBP5, is reversible with the antioxidant N-acetylcysteine (NAC), suggesting that this mechanism is directly linked to ROS. Furthermore, an array for transcription factor DNA binding activity shows that SOD2 knockdown induces DNA binding by several transcription factors, including AR. SOD2 knockdown-induced AR activation was confirmed by electrophoretic mobility shift assay (EMSA) and luciferase activity and both were readily reversible with NAC.
Conclusions
These findings show that downregulation of SOD2 induces AR activity in a ROS-dependent manner, and suggests that there may be a role for antioxidant therapy in CRPC.
Introduction
The androgen receptor (AR) is expressed in almost all prostate cancers and the growth and proliferation of prostate cancer cells is initially an androgen-dependent process (1). The time-honored observation by Huggins and Hodges in the 1940’s that prostate cancer regresses after castration was the first illustration of the activity of androgen deprivation therapy (ADT), which is the standard frontline therapy for advanced prostate cancer today (2,3). Although prostate cancer generally responds to ADT, the majority of tumors will eventually grow and is termed castration-resistant prostate cancer (CRPC). Despite castrate levels of testosterone, AR is reactivated in CRPC and drives the proliferation and survival of these tumors. AR reactivation occurs via gain-of-function changes, through AR gene amplification, mutations in AR, changes in expression of steroid receptor coactivators, activation through growth factor receptors and other phosphorylation-dependent mechanisms and local tumor-driven synthesis of androgens (4–8). Gene expression profiling of hormone responsive tumors versus CRPC has revealed some of these mechanisms. These studies also suggest that in the transition to CRPC, manganese superoxide dismutase (SOD2) is one of the most downregulated genes (9). SOD2 regulates reactive oxygen species (ROS) by converting superoxide to a less reactive species. The mechanism of gene expression for steroid hormone receptors depends on an oxidative process, which occurs as a result of localized histone demethylation (10). Therefore, we hypothesized that SOD2 downregulation in CRPC is mechanistically linked to AR reactivation.
Here, we examine changes in the expression of androgen responsive genes after knocking down SOD2 in an AR-expressing prostate cancer cell line. We show that knocking down SOD2 leads to the expression of androgen responsive genes. The expression of these genes is reversible upon treatment with N-acetylcysteine (NAC). DNA binding to androgen response elements is induced by SOD2 knockdown in a NAC-reversible manner both in a Panomic’s transcription factor array and by electrophoretic mobility shift assay (EMSA). Furthermore, SOD2 knockdown also induces AR transcription activity, which is reversible with NAC. These findings suggest that SOD2 downregulation in CRPC directly affects AR activity and AR DNA binding in a ROS-dependent manner. Furthermore, the ROS-dependent mechanism of AR reactivation in CRPC suggests that antioxidants may have therapeutic value for CRPC.
Materials and Methods
Cell Lines and drug treatments
The LNCaP cell line was maintained in phenol-red free RPMI-1640 containing 10% fetal bovine serum, 2mM L-glutamine and antibiotics. For the gene expression studies, cells were washed with PBS, supplemented with RPMI-1640 containing 10% charcoal-stripped fetal bovine serum, 2mM L-glutamine with antibiotics and treated with 10 nM R1881 (Sigma, St. Louis, MO) and 10 mM N-acetylcysteine (NAC; Sigma) for 6 hours. In all other studies, cells were treated with 10 nM R1881 for 1 hour and with 5 mM N-acetylcysteine for 3 hours.
Generation of shSOD2 LNCaP cells
The shRNA oligos used for knock-down of SOD2 are as follows: 5’-GATCCCGGGGTTGGCTTG-GTTTCAATATTCAAGAGATATTGAAACCAAGCCAACCCCTTTTTA-3' and 5'-AGCTTAAAAAGGGGTTGGCTTGGTTTCAATATCTCTTGAATATTGAAACCAAG CCAACCCCGG-3'. These oligos were annealed and cloned into the BglII/HindIII sites of pRetroSuper (11). Phoenix amphotrophic retroviral packaging cells (generously provided by Gary Nolan) were transfected with either 2 micrograms of this plasmid or separately with empty pRetroSuper plasmid. LNCaP cells were infected with viral supernatant, supplemented with 10 microgram/mL polybrene, and selected with 0.2 microgram/mL puromycin.
Western Blot
Total protein was isolated from SOD2 shRNA and vector-infected LNCaP cells using lysis buffer and quantitated using the Pierce (Rockford, IL) bicinchoninic acid (BCA) kit. Forty micrograms of protein was loaded and run on 4–20% tris-glycine gels (Invitrogen, Carlsbad, CA). Protein was transferred to a polyvinylidene fluoride membrane, blocked in 1% fish gelatin, incubated with primary (anti-SOD2, ab16954; Abcam, Cambridge, MA) and secondary (LI-COR Biotechnology, Lincoln, NE) antibodies and scanned with the LI-COR Odyssey Infrared Imaging System.
Measurement of superoxide dismutase activity
Two million cells were lysed in 20 mM HEPES buffer containing 1mM EGTA, 210 mM mannitol and 70 mM sucrose using Calbiochem’s Superoxide Dismutase Assay Kit II (EMD Chemicals, Gibbstown, NJ) according to manufacturer’s directions. Ten microliters of lysate was mixed with 200 µl tetrazolium salt, the radical detector. The reaction also contained 1 mM potassium cyanide and 20 µl xanthine oxidase to inhibit SOD1 and SOD3 activities. The reaction was incubated at room temperature for 20 minutes and read at 450 nm using a Genios plate reader (Tecan, Palm Springs, CA). One unit of SOD activity is defined as the amount of enzyme needed to exhibit 50% dismutation of the superoxide radical.
Measurement of cellular ROS
Cellular ROS was measured by the hydrolysis of the acetate esters of 5-(and-6)-chloromethyl-2',7'-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Invitrogen, Carlsbad, CA). One million cells per reaction were washed and resuspended in pre-warmed (37°C) PBS containing 5 µM CM-H2DCFDA and incubated at 37°C for 15 minutes to load the dye into the cells. Following the incubation, cells were washed with pre-warmed (37°C) growth media and re-suspended in 10 mls growth media/million cells and incubated at 37°C for 30 minutes to allow for recovery and conversion of the acetate esters. Cells were washed and re-suspended in PBS+1% FBS and read by flow cytometry immediately.
Microarray analysis
Total RNA was isolated from 1 million cells using Trizol (Invitrogen, Carlsbad, CA). Twenty micrograms of RNA was used in reverse transcription reaction containing either Cy3- or Cy-5 dUTP, oligo dT primer, and Superscript III, as described previously (12). Each experimental sample was labeled with Cy5-dUTP and Human Universal RNA (Stratagene, Cedar Creek, TX) was labeled with Cy3-dUTP. Following reverse transcription RNA was hydrolysed using NaOH, the Cy3 and Cy5 reactions were mixed and the probe was cleaned up using a microcon YM-30 column (Millipore, Billerica, MA). The probe was hybridized to the National Cancer Institute oligonucleotide arrays and analyzed as previously described (12).
Real-Time PCR
RNA was extracted using Trizol (Invitrogen, Carlsbad, CA). Ten micrograms of RNA was used in a reverse transcription reaction using an oligo dT primer and Superscript III (Invitrogen, Carlsbad, CA). One hundred ng of cDNA was then used in 20 µl taqman assay using Universal Master Mix and gene specific taqman probes both from Applied Biosystems (Foster City, CA). The gene specific taqman mixes used are as follows: FKBP5 Hs00188025_m1, Aprin Hs00971886_m1, VEGF Hs00900054_m1, AKR1C3 Hs00366267_m1, HSD17B14 Hs00212233_m1, HSD17B8 Hs00157993_m1, HSD17B2 Hs00157993_m1, PSA Hs02576345_m1, TMPRSS2 Hs00237175_m1, 18S Hs99999901_m1. PCR was performed using a One-Step Real-Time PCR machine (Applied Biosystems, Foster City, CA).
DNA/Protein Arrays
Transcription factor binding site analysis was performed using Panomics’ DNA/protein Combo arrays containing 345 consensus binding sites (Panomics, Fremont, CA) according to manufacturer’s instructions. Fifteen micrograms of each nuclear protein lysate, prepared using Panomics’ nuclear protein extraction kit, were used in the binding reaction. Following binding and separation of unbound from bound biotinylated TF binding sites, the probe was hybridized at 45°C overnight. The blot was then washed and detected using a streptavidin horse radish peroxidase conjugate. Data was quantitated using ImageQuant (GE Healthcare, Piscataway, NJ ) and a binding site was considered to be regulated only if it showed reciprocal regulation in the shSOD2 and shSOD2 + NAC cell lines.
Electrophoretic Mobility Shift Assay
Nuclear protein extracts were prepared from cell lines using Panomic’s nuclear extraction kit (Panomics, Freemont, CA), following manufacturer’s instructions. The nuclear protein was quantified using the BCA colorometric assay (Pierce). Eight micrograms of nuclear extract was mixed with 50 nM infrared-labeled androgen responsive element (ARE; Li-Cor Biosciences, Lincoln, NE) in a 20 microliter reaction containing 10 mM Tris, 50 mM KCl, 1mM DTT, 1 µg poly (dI-dC), 2.5 mM DTT/0.25% Tween-20, and 5 mM MgCl2 and incubated for 30 minutes at room temperature. Following incubation, the reaction was run on a 6% polyacrylamide gel in tris-glycine buffer and scanned using an Odyssey infrared scanner (Li-Cor Biosciences, Lincoln, NE).
Luciferase Assay
Empty vector-expressing and shSOD2-expressing LNCaP cells were seeded at 20,000 cells/well and transfected with TK-luc-ARE3 (150 ng) to measure AR activity and pGL4.75 (15 ng) (Promega, Madison WI) as a transfection control using Lipofectamine 2000 (Invitrogen, Carlsbad CA). Twenty-four hours post transfection, media was changed to phenol red free, serum free media containing 10 nM R1881 or 5 mM NAC, as indicated. Firefly and Renilla luciferase activity was measured using the Dual Glo Luciferase Assay System (Promega) twenty-four hours later. Firefly luciferase measurements were normalized using the Renilla luciferase measurements. All measurements were done in quadruplicate.
Results
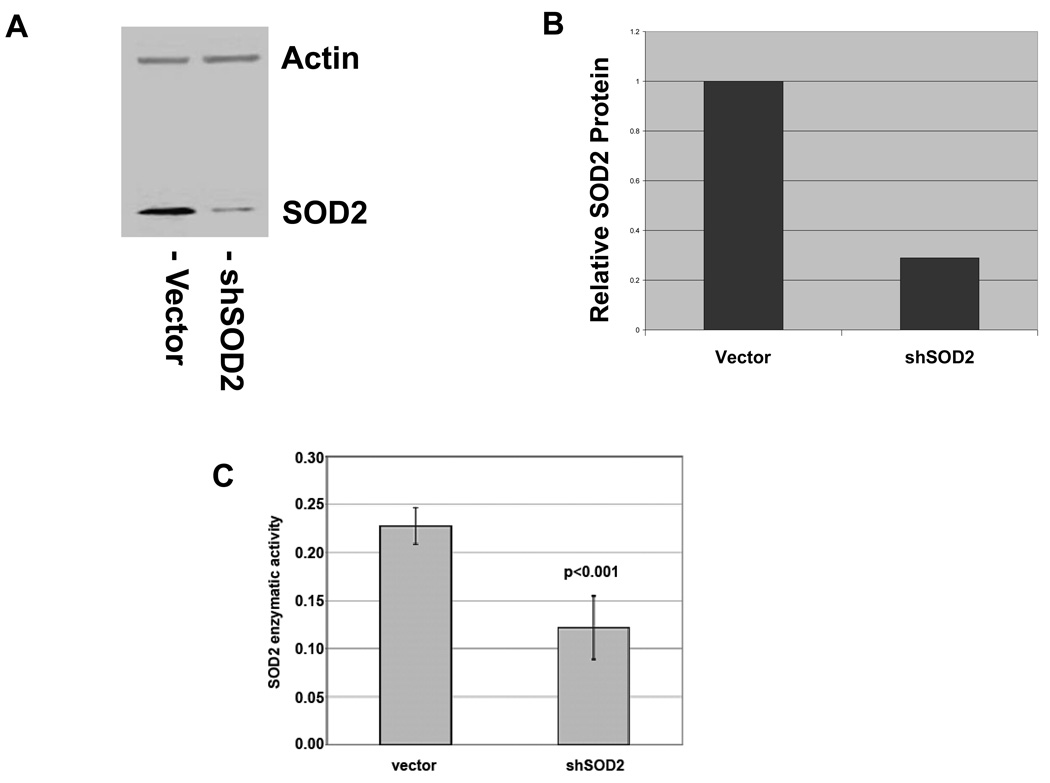
In order to determine the consequences of SOD2 downregulation, we infected LNCaP cells with a construct expressing a SOD2 shRNA and a puromycin resistance gene (shSOD2). shSOD2 expressing LNCaP cells had a 71% decrease in SOD2 protein expression when compared to cells expressing the empty vector (Figure 1A and 1B). The function of SOD2 is to neutralize reactive superoxide radicals by converting them to hydrogen peroxide. To determine the degree to which shSOD2 expressing LNCaP cells have an altered ability to neutralize superoxide, we assayed these cells for enzymatic function specific for SOD2. LNCaP cells expressing shSOD2 showed a 2-fold decrease in SOD2 enzymatic activity as compared to control cells (Figure 1C).
Figure 1.
Expression of shSOD2 decreases SOD2 protein and SOD2 enzymatic activity in LNCaP cells. A. Western blot of vector control and shSOD2-expressing LNCaP cells with actin as a loading control. B. Quantitation of decrease in SOD2 protein normalized to actin. C. Decrease in SOD2 enzymatic activity in shSOD2 expressing cells.
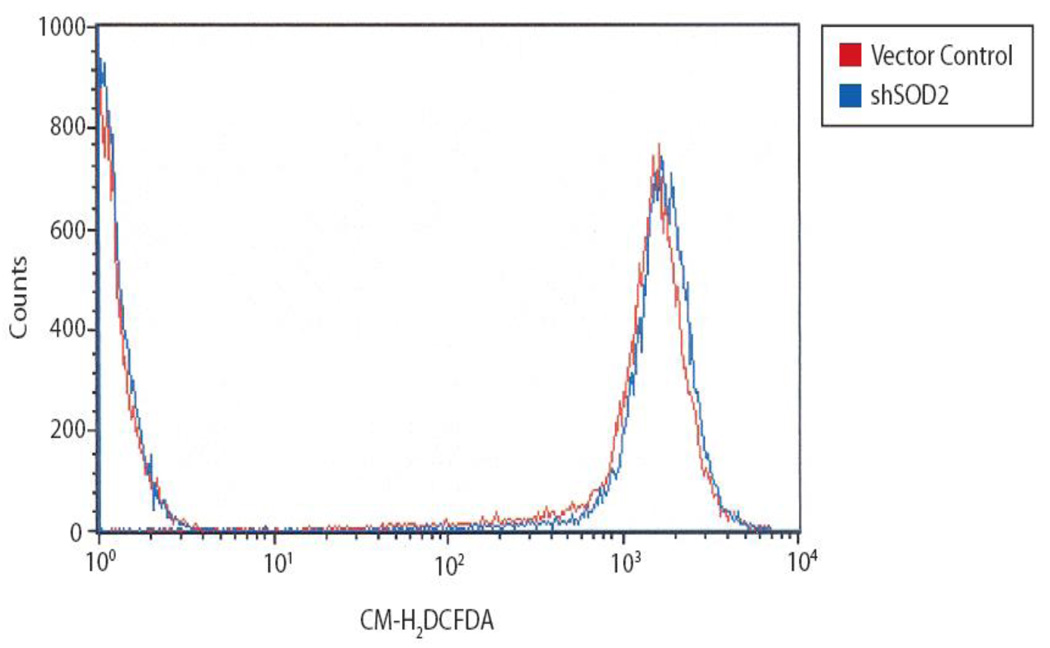
Downregulation of SOD2 protein and enzymatic activity is expected to increase levels of ROS. To determine the change in cellular ROS induced by SOD2 knockdown, we measured ROS activity as determined by hydrolysis of acetate esters susceptible to ROS-mediated cleavage using flow cytometry. The distribution of cells with higher levels of cleaved acetate esters is significantly greater (p<0.05) in SOD2 knockdown cells compared with control cells (Figure 2). These results show that SOD2 downregulation in LNCaP prostate cancer cells leads to lower levels of SOD2 enzymatic activity and consequently higher levels of ROS activity.
Figure 2.
Reactive oxygen species (ROS) are increased in the shSOD2 cell line compared with vector control cells. ROS activity was measured in triplicate with the intensity shifting from a mean of 1497 in the vector sample to a mean of 1681 in the shSOD2 sample (p<0.05) by flow cytometry.
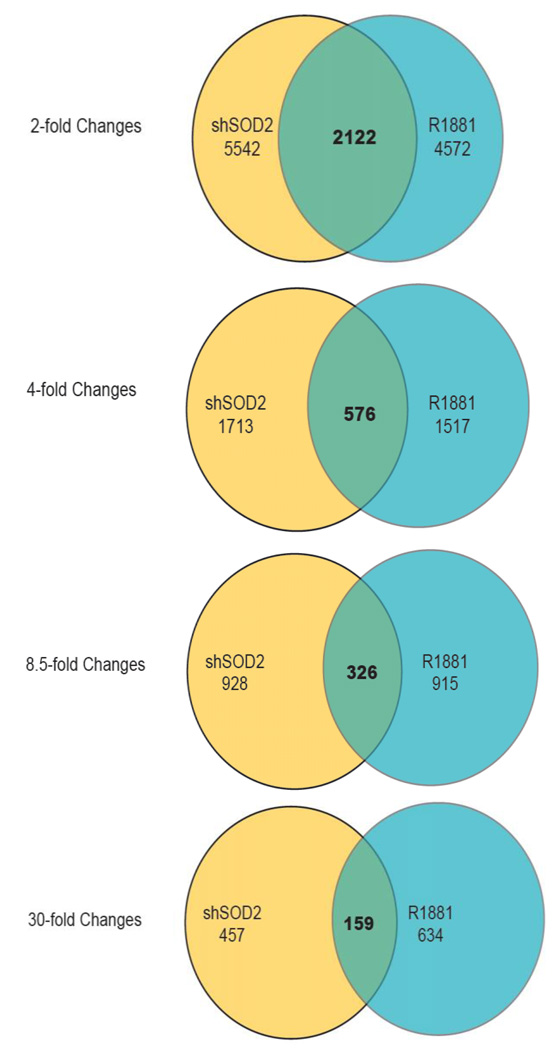
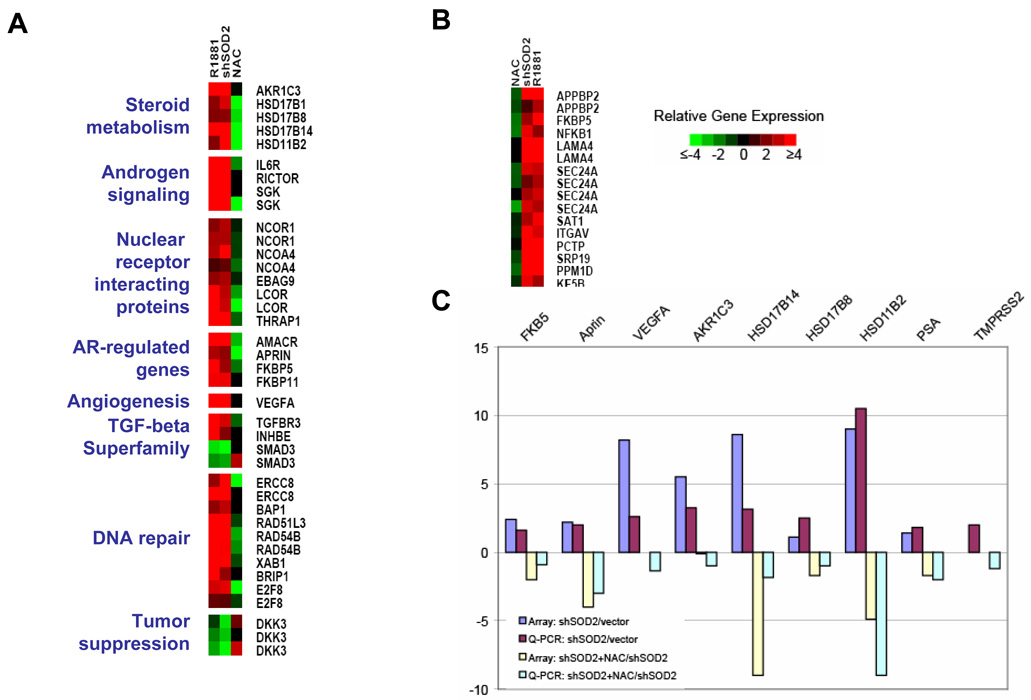
Next, we determined gene expression changes in LNCaP induced by 1) direct activation of AR with R1881 2) SOD2 knockdown 3) SOD2 knockdown cells treated with NAC (Figure 3). Gene expression changes that occur with SOD2 knockdown and are ROS-mediated, would be reversed with NAC treatment. We found that 46% and 38% of genes that are induced two-fold and four-fold, respectively, by R1881 are also induced to the same extent with SOD2 downregulation. Examination of the shared genes that are induced with R1881 exposure and SOD2 downregulation reveals both potential mechanisms through which AR may be reactivated and a set of well-recognized AR-responsive genes that are also activated through SOD2 knockdown. Furthermore, many of the gene expression changes that are common to both direct AR activation with R1881 and SOD2 knockdown are reversed in SOD2 knockdown cells by treatment with NAC, suggesting that activation of these androgen regulated genes in SOD2 knockdown cells is ROS-mediated (Figure 4A). Several genes induced by SOD2 knockdown, such as aldo-keto reductase family 1, member C3 (AKR1C3), 17-beta-hydroxysteroid dehydrogenase B (HSD17B) -1, -8, -14 and HSD11B2 are intimately involved in steroid metabolism. Some of these genes have been implicated in AR reactivation by converting adrenal androgens to testosterone in the setting of CRPC (13). A subset of genes induced by SOD2 downregulation are coregulators of steroid hormone receptors. Some of these coregulators, such as NCOA4, specifically activate AR by direct protein-protein interaction (14). Another subset of genes including IL-6 receptor and RICTOR are involved in AR activation in the presence of low levels of testosterone through phosphorylation-dependent mechanisms. Numerous studies have shown that IL-6 is implicated in castration-resistant growth and activates AR through a STAT3-dependent mechanism (15,16). RICTOR overexpression upregulates the mTOR/Akt pathway (17), which results in AR activation (18).
Figure 3.
Genes regulated by SOD2 show a large overlap with androgen signaling. A. Venn diagrams showing the number of genes regulated by SOD2 and R1881 as well as the intersection of these genes.
Figure 4.
Androgen-responsive genes are regulated by SOD2 downregulation. A. The expression of a sub-set of genes that are regulated concordantly by at least two-fold by both the knockdown of SOD2 (shSOD2) and by treatment with R1881 (R1881). Also shown is the expression of these genes when the shSOD2 cells are treated with NAC (NAC). The functional categories of these genes are shown on the left side. B. A subset of genes found to be highly androgen regulated in another study (21), that is also regulated by both shSOD2 and R1881 that is also reversible in shSOD2 cells that were treated with NAC. C. Quantitative PCR (Q-PCR) shows a similar level of regulation of the genes assayed. SOD2 knockdown-inducible and NAC-reversible expression of PSA and TMPRSS2 by QPCR is also shown.
Widely studied AR-regulated genes, such as FK506-binding protein 5 (19,20), alpha-Methylacyl-CoA racemase (AMACR) (21) and APRIN (22,23) were also upregulated in a NAC-reversible manner in these microarray studies. To further validate our findings, we analyzed a selected group of genes found to be induced by R1881 in a previously published cDNA microarray study of prostate cancer cells (21). A subset of these genes is also induced by SOD2 knockdown in a NAC-reversible manner (Figure 4B). Next, for selected genes, we confirmed gene expression changes that were induced by SOD2 knockdown and reversible with NAC treatment using quantitative PCR (Figure 4C). All seven genes we tested – FKBP5, APRIN, VEGFA, AKR1C3, HD17B8, HSD17B14 and HSD11B2 – were induced by SOD2 knockdown and repressed with NAC treatment. Furthermore, we also tested prostate-specific antigen (PSA), which is an androgen-regulated gene that changed less than two-fold with SOD2 knockdown and TMPRSS2, which was not on our microarray but is known to drive oncogene expression through an AR-driven regulatory region with a chromosomal translocation (24). Both of these genes are induced by SOD2 knockdown in a NAC-reversible fashion. Notably, we found that several nuclear receptors were among the genes regulated by SOD2 knockdown in a NAC-reversible manner, but not by R1881. Estrogen receptor-alpha, estrogen related receptor-beta and RXR-alpha are downregulated, and progesterone receptor is upregulated with SOD2 knockdown. These changes are all reversible with NAC treatment (Supplemental Figure 1).
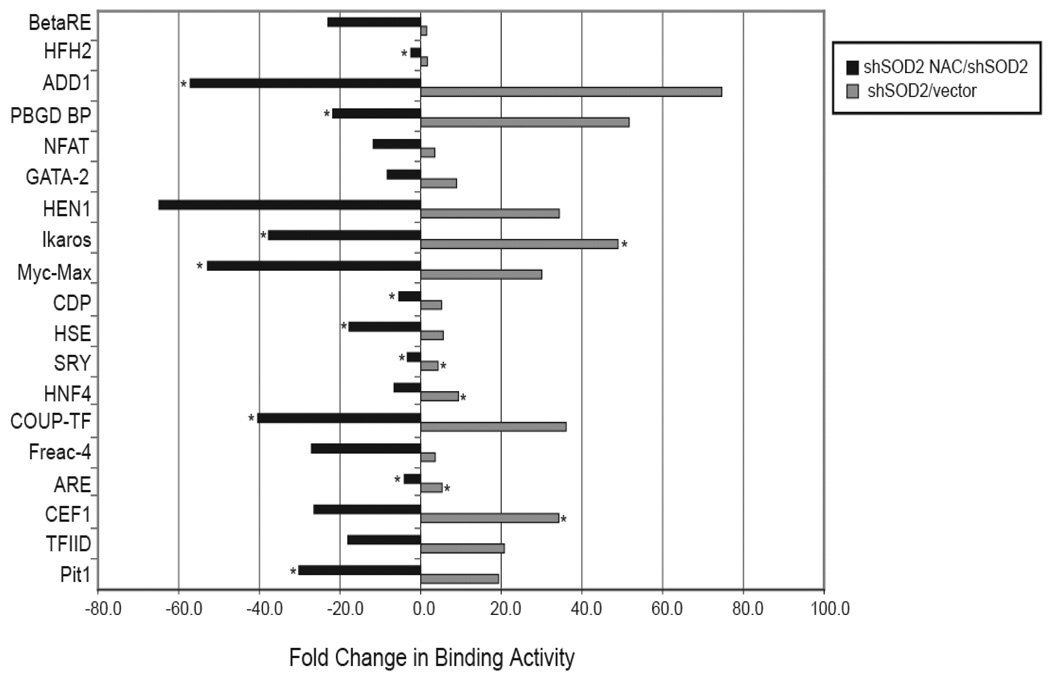
Although we found that induction of androgen-regulated gene expression changes with SOD2 knockdown were largely reversible with NAC, suggesting a ROS-mediated mechanism of AR activation, we wished to further confirm that this mechanism is indeed ROS-mediated. Therefore, we examined DNA binding changes of transcription factors induced by SOD2 downregulation that were also NAC-reversible, using a Panomics’ array, which detects the relative activation status of transcription factors (Figure 5). Nineteen of 345 (5.5%) transcription factors had an increase in DNA binding with SOD2 knockdown that was reversible with NAC treatment. Binding to the androgen response element (ARE) increased 5.3-fold with SOD2 knockdown and decreased 4.1-fold with NAC treatment of SOD2 knockdown cells. Importantly, binding to GATA-2 elements increased 8.9–fold with SOD2 knockdown and decreased 8.3–fold with NAC treatment. GATA-2 activity has recently been shown to be absolutely required to drive the expression of AR-regulated genes (25). Binding to the myc-max DNA binding element in our Panomics’ array is also consistent with ROS-regulation. However, myc is an AR-regulated gene and this effect is probably downstream of AR (26).
Figure 5.
Changes in transcription factor binding in response to SOD2 knockdown. Binding of transcription factors was measured using a Panomic’s array and the fold change in binding activity is shown. An asterisk represents a low level of binding in the control sample, and the value for quantitation was set to 100 over background.
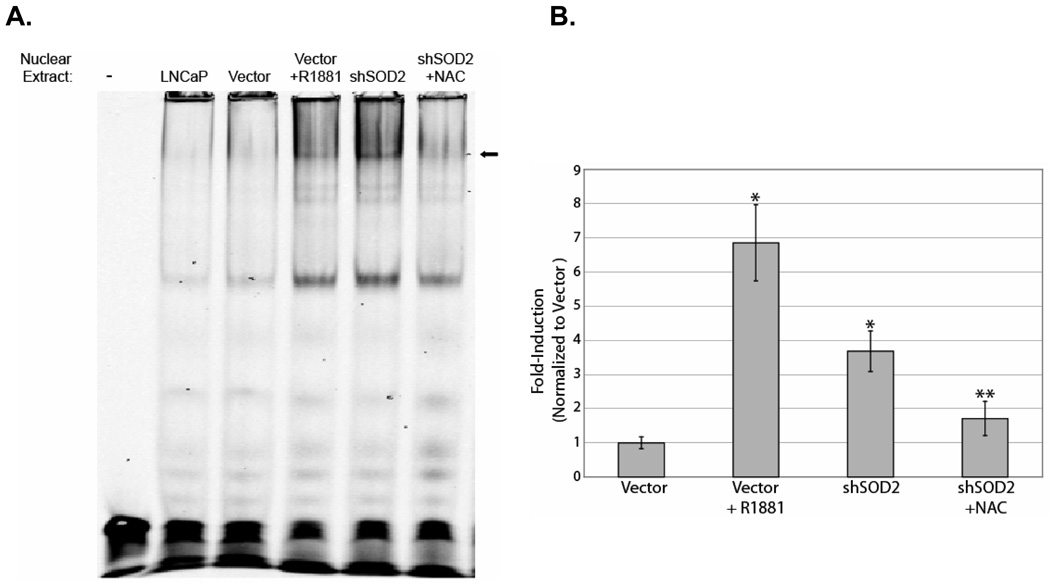
To further investigate the effect of SOD2 downregulation and ROS on AR activity, we performed EMSA using ARE oligos (Figure 6A). Treatment with 10 nM R1881 and SOD2 knockdown induced ARE binding to a similar extent. Treatment of SOD2 knockdown cells with NAC reversed ARE binding, consistent with a ROS-mediated mechanism. These EMSA results in conjunction with the results of the Panomics’ array show that SOD2 knockdown induces ARE binding in a NAC-reversible manner. This suggests that the induction of androgen-responsive genes we observed are due to direct activation of AR via a ROS-mediated mechanism.
Figure 6.
SOD2 knockdown induces NAC-reversible AR DNA binding and transcription activity. A. An electrophoretic mobility shift assay (EMSA) showing androgen receptor (AR) binding. An increase in AR binding occurs in both cells treated with R1881 (R1881) as well as in cells with SOD2 knockdown (shSOD2). Binding of AR is lost when shSOD2 cells are treated with NAC (shSOD2+NAC). B. AR luciferase activity is increased in SOD2 knockdown cells. Luciferase activity was measured in vector and SOD2 knockdown (shSOD2) cells after the indicated treatments and was normalized to Renilla luciferase. The graph shows the fold-increase in luciferase activity for each of the treatments over vector control cells. An asterisk (*) represent p<0.001 with respect to the control. Two asterisks (**) represent p<0.01 for the comparison of shSOD2+NAC with shSOD2.
Finally, we directly tested the effect of SOD2 downregulation on AR transcriptional activity using the luciferase assay (Figure 6B). Treatment of empty vector-infected LNCaP with R1881 induced ARE-luciferase activity to 7-fold above baseline, whereas SOD2 knockdown cells have activity that is 4-fold over the baseline of empty vector-expressing LNCaP. Furthermore, ARE-luciferase activity is largely reversible in SOD2 knockdown cells upon treatment with NAC.
Discussion
Although by definition testosterone is depleted with CRPC, the growth and progression of prostate cancer in this form is still dependent on AR (1,6). Defining exactly how AR is reactivated in CRPC is the first step in determining how AR should be targeted (27). SOD2 is one of the most potently downregulated genes in CRPC when compared to hormone responsive tumors (9). Here, we show that SOD2 downregulation is directly responsible for AR reactivation in prostate cancer and that this occurs through a ROS-mediated mechanism. SOD2 knockdown induces AR binding to ARE followed by gene expression changes that overlap with gene expression changes that occur with ligand-mediated activation of AR. AR activation that occurs with SOD2 knockdown is reversible with NAC and is therefore mediated directly through ROS.
Our studies reveal several ways in which ROS activates AR. First, we found that several genes involved in steroid metabolism, including AKR1C3, are regulated by SOD2 knockdown. One mechanism of AR reactivation in CRPC is the local conversion of androgen precursors to ligands, such as testosterone, which effectively activate AR. AKR1C3 is overexpressed in CRPC and reduces adrenal androstenedione to testosterone (13). Such gene expression changes lead to an increase in the local concentration of androgens in CRPC (8). We found that all four genes involved in steroid metabolism that are induced by SOD2 knockdown and that we tested by real time PCR, reverse gene expression with NAC.
Second, AR reactivation can be induced by altering the balance of steroid receptor coregulators (28). Five nuclear receptor coregulators are induced in a ROS-reversible manner. Although the ultimate effect of the combination of these expression changes is difficult to discern, the overexpression of one these genes, NCOA4, has been shown to directly result in an increase in AR activity (14).
Third, multiple signaling pathways have been indicted in ligand-independent or ligand-sensitizing AR activation (29). One of the most widely studied of these is the IL-6 pathway (15). IL-6 activates AR in a STAT3-dependent manner and antibodies to IL-6 reverses castration-resistance (16,30). Furthermore, levels of IL-6 receptor are predictive of biochemical recurrence and metastasis (31,32). Our finding that IL-6 receptor is induced by SOD2 downregulation in a NAC-reversible manner, suggests that treatment with antioxidants may downregulate this pathway.
Whether ROS reactivates the expression of androgen responsive genes through increasing the local availability of androgen ligand, altering the distribution of coregulators to more favorable levels, or through signaling mechanisms such as the IL-6 pathway, our data suggests that it directly activates AR, results in increased binding to ARE and stimulation of AR-driven transcription. Interestingly, we also found that binding to GATA2 DNA motifs also increases with SOD2 knockdown and is reversed with NAC treatment. GATA2 directly interacts with AR and the GATA2 transcription factor and DNA binding motif are absolutely required for optimal expression of androgen regulated genes (25,33). Therefore, GATA2 is another way in which ROS may activate AR in the castration-resistant setting.
We found that VEGFA was strongly induced by both R1881 and SOD2, and was reversible with NAC by quantitative PCR. ROS may therefore mediate an angiogenic switch that also occurs through AR. One possibility we considered is that since VEGFA is activated through hypoxia-inducible factor-1 (HIF-1) (34) and HIF-1 activity may be reversible with antioxidants (35), some of what we have observed may be due to HIF-1 activation. However, HIF-1 activity did not increase with SOD2 knockdown in our Panomics’ array, nor by EMSA (data not shown). Furthermore, it has been shown that VEGFA is located in an AR binding region (36), further supporting that ROS-mediated VEGFA activation in our experiments indeed occurs through AR. Others have shown that SOD2 has tumor suppressor activity in prostate cancer cells that do not express AR (37). However, the vast majority of clinical prostate cancers do express AR and the significance changes in SOD2 expression in prostate tumors that do not express AR is unclear.
One caveat is that our studies focus on a single AR-expressing cell line, LNCaP. Although further work is required to determine if these results can be generalized to other prostate cancer cell lines, the LNCaP growth rate and relatively high levels of AR expression make this the most amenable model to test our hypothesis. Furthermore and importantly, it should be noted that the hypothesis that SOD2 downregulation is involved AR activation originated from observations made from clinical tumors, not cell lines.
In conclusion, we have found that SOD2 downregulation, a gene expression change which occurs clinically in patients with CRPC, is responsible for AR reactivation. AR activation in this setting is ROS-dependent and results in the expression of genes such as AKR1C3, NCOA4 and IL-6 receptor, which are known to lead to AR activation. Furthermore, AR DNA binding is increased and results in expression of androgen responsive genes. Finally, AR reactivation in cells with low SOD2 expression is reversible with antioxidant treatment. In general, re-expression of genes that have lost function in cancer for therapeutic purposes is problematic and has not been feasible. However, SOD2 downregulation in CRPC may be overcome simply by treatment with an antioxidant and may result in silencing AR reactivation. Further work is warranted to determine if this strategy will be of therapeutic value clinically.
Supplementary Material
Acknowledgments
This publication has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract No. N01-CO-12400. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Statement of Clinical Relevance
This body of work shows that SOD2 downregulation, which has been shown to occur in castration-resistant prostate cancer (CRPC), induces androgen receptor (AR) activity and the expression of AR-responsive genes. It is known that the transition from androgen-responsive to castration-resistant prostate cancer is mediated by a gain-of-function in AR. Therefore, reversing this process would provide a new avenue of therapy for prostate cancer. SOD2 downregulation leads to an increase in reactive oxygen species (ROS) and this work also shows that the mechanism of AR activation in this setting is mediated through ROS and furthermore, that reversal of increased ROS with an antioxidant reverses AR activity. These findings have clear implications for clinical therapy and suggest that antioxidants may have a role in the treatment of patients with CRPC.
References
- 1.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Hodges CV. Studies on prostatic cancer, I: the effect of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer research. 1941;1:293–297. [Google Scholar]
- 3.Sharifi N, Gulley JL, Dahut WL. Androgen deprivation therapy for prostate cancer. Jama. 2005;294:238–244. doi: 10.1001/jama.294.2.238. [DOI] [PubMed] [Google Scholar]
- 4.Mostaghel EA, Montgomery RB, Vessella R, et al. Correlating androgen levels and steroidogenic transcript expression in castration resistant prostate cancer metastases; American Society for Clinical Oncology, Genitourinary Cancers Symposium; 2008; San Francisco, CA. 2008. [Google Scholar]
- 5.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 6.Sharifi N, Farrar WL. Androgen receptor as a therapeutic target for androgen independent prostate cancer. American journal of therapeutics. 2006;13:166–170. doi: 10.1097/00045391-200603000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Mostaghel EA, Montgomery RB, Lin DW. The basic biochemistry and molecular events of hormone therapy. Current urology reports. 2007;8:224–232. doi: 10.1007/s11934-007-0010-z. [DOI] [PubMed] [Google Scholar]
- 8.Titus MA, Schell MJ, Lih FB, Tomer KB, Mohler JL. Testosterone and dihydrotestosterone tissue levels in recurrent prostate cancer. Clin Cancer Res. 2005;11:4653–4657. doi: 10.1158/1078-0432.CCR-05-0525. [DOI] [PubMed] [Google Scholar]
- 9.Best CJ, Gillespie JW, Yi Y. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–6834. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perillo B, Ombra MN, Bertoni A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science (New York, NY. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 11.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 12.Hurt EM, Thomas SB, Peng B, Farrar WL. Integrated molecular profiling of SOD2 expression in multiple myeloma. Blood. 2007;109:3953–3962. doi: 10.1182/blood-2006-07-035162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanbrough M, Bubley GJ, Ross K, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer research. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 14.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70,for the androgen receptor in human prostate cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Wang LH, Farrar WL. Interleukin 6 activates androgen receptor-mediated gene expression through a signal transducer and activator of transcription 3-dependent pathway in LNCaP prostate cancer cells. Cancer research. 2000;60:2132–2135. [PubMed] [Google Scholar]
- 16.Wallner L, Dai J, Escara-Wilke J, et al. Inhibition of interleukin-6 with CNTO328,an anti-interleukin-6 monoclonal antibody, inhibits conversion of androgen-dependent prostate cancer to an androgen-independent phenotype in orchiectomized mice. Cancer research. 2006;66:3087–3095. doi: 10.1158/0008-5472.CAN-05-3447. [DOI] [PubMed] [Google Scholar]
- 17.Masri J, Bernath A, Martin J, et al. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer research. 2007;67:11712–11720. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- 18.Gao H, Ouyang X, Banach-Petrosky WA, Gerald WL, Shen MM, Abate-Shen C. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14477–14482. doi: 10.1073/pnas.0606836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Febbo PG, Lowenberg M, Thorner AR, Brown M, Loda M, Golub TR. Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. The Journal of urology. 2005;173:1772–1777. doi: 10.1097/01.ju.0000155845.44729.ba. [DOI] [PubMed] [Google Scholar]
- 20.Mostaghel EA, Page ST, Lin DW, et al. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Cancer research. 2007;67:5033–5041. doi: 10.1158/0008-5472.CAN-06-3332. [DOI] [PubMed] [Google Scholar]
- 21.DePrimo SE, Diehn M, Nelson JB, et al. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geck P, Maffini MV, Szelei J, Sonnenschein C, Soto AM. Androgen-induced proliferative quiescence in prostate cancer cells: the role of AS3 as its mediator. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10185–10190. doi: 10.1073/pnas.97.18.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy S, Agoulnik IU, Weigel NL. Androgen receptor signaling and vitamin D receptor action in prostate cancer cells. The Prostate. 2005;64:362–372. doi: 10.1002/pros.20251. [DOI] [PubMed] [Google Scholar]
- 24.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Molecular cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva IS, Morsch DM, Urnauer L, Spritzer PM. Androgen-induced cell growth and c-myc expression in human non-transformed epithelial prostatic cells in primary culture. Endocrine research. 2001;27:153–169. doi: 10.1081/erc-100107177. [DOI] [PubMed] [Google Scholar]
- 27.Sharifi N, Dahut WL, Figg WD. Secondary hormonal therapy for prostate cancer: what lies on the horizon? BJU international. 2008;101:271–274. doi: 10.1111/j.1464-410X.2007.07236.x. [DOI] [PubMed] [Google Scholar]
- 28.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocrine reviews. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 29.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 30.Lee SO, Lou W, Hou M, de Miguel F, Gerber L, Gao AC. Interleukin-6 promotes androgen-independent growth in LNCaP human prostate cancer cells. Clin Cancer Res. 2003;9:370–376. [PubMed] [Google Scholar]
- 31.Kattan MW, Shariat SF, Andrews B, et al. The addition of interleukin-6 soluble receptor and transforming growth factor beta1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J Clin Oncol. 2003;21:3573–3579. doi: 10.1200/JCO.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 32.Shariat SF, Andrews B, Kattan MW, Kim J, Wheeler TM, Slawin KM. Plasma levels of interleukin-6 and its soluble receptor are associated with prostate cancer progression and metastasis. Urology. 2001;58:1008–1015. doi: 10.1016/s0090-4295(01)01405-4. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Stable CM, Pozas A, Roos BA. A role for GATA transcription factors in the androgen regulation of the prostate-specific antigen gene enhancer. Molecular and cellular endocrinology. 2000;167:43–53. doi: 10.1016/s0303-7207(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 34.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Jr, Dervan PB. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao P, Zhang H, Dinavahi R, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes & development. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkataraman S, Jiang X, Weydert C, et al. Manganese superoxide dismutase overexpression inhibits the growth of androgen-independent prostate cancer cells. Oncogene. 2005;24:77–89. doi: 10.1038/sj.onc.1208145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.