Abstract
We tested the hypothesis that the stable isotope [13C]pantoprazole is O-demethylated by cytochrome P450 CYP2C19 and that the 13CO2 produced and exhaled in breath as a result can serve as a safe, rapid, and noninvasive phenotyping marker of CYP2C19 activity in vivo. Healthy volunteers who had been genotyped for the CYP2C19*2, CYP2C19*3, and CYP2C19*17 alleles were administered a single oral dose of [13C]pantoprazole sodium-sesquihydrate (100 mg) with 2.1 g of sodium bicarbonate. Exhaled 13CO2 and 12CO2 were measured by IR spectroscopy before (baseline) and 2.5 to 120 min after dosing. Ratios of 13CO2/12CO2 after [13C]pantoprazole relative to 13CO2/12CO2 at baseline were expressed as change over baseline (DOB). Maximal DOB, DOB15 to DOB120, and area under the DOB versus time curve (AUC0–120 and AUC0–∞) were significantly different among three genotype groups (CYP2C19*1/*1, n = 10; CYP2C19*1/*2 or CYP2C19*1/*3, n = 10; and CYP2C19*2/*2, n = 5) with predicted extensive metabolizers (EMs), intermediate metabolizers (IMs), and poor metabolizers (PMs) of CYP2C19, respectively (Kruskal-Wallis test, p < 0.01); linear regression analysis indicated a gene-dose effect relationship (r2 ranged between 0.236 and 0.522; all p < 0.05). These breath test indices were significantly lower in PMs than IMs (p < 0.05) or EMs (p < 0.01) of CYP2C19. [13C]Pantoprazole plasma exposure showed significant inverse correlation with breath test indices in the respective subjects (Pearson r = -0.74; p = 0.038). These feasibility data suggest that the [13C]pantoprazole breath test is a reliable, rapid, and noninvasive probe of CYP2C19 and seems to be a useful tool to optimize drug therapy metabolized by CYP2C19.
Human cytochrome P450 CYP2C19 is important in the metabolism of several drugs, including proton pump inhibitors (e.g., omeprazole, lansoprazole, and pantoprazole), antidepressants, diazepam, carisoprodol, nelfinavir, clopidogrel, voriconazole, thalidomide, clonazepam, and cyclophosphamide (Ando et al., 2002; Desta et al., 2002; Takada et al., 2004; Hulot et al., 2006). The clearance of drugs metabolized by CYP2C19 varies 5- to 20-fold among individuals and ethnic groups primarily because of effects of genetic polymorphisms (Goldstein, 2001; Desta et al., 2002) but also as a result of nongenetic factors [(e.g., drug interactions) (Desta et al., 2002), age (Ishizawa et al., 2005), pregnancy (McGready et al., 2003), and disease state (Desta et al., 2002; Frye et al., 2006)].
S-Mephenytoin hydroxylase, which was later purified as CYP2C19 (Wrighton et al., 1993), was first reported in 1979 (Kupfer et al., 1979). The molecular basis of this polymorphism was identified later with the cloning of the gene (de Morais et al., 1994; Goldstein and de Morais, 1994). Twenty-one alleles associated with complete loss of enzyme activity (e.g., CYP2C19*2 to *8) (Goldstein, 2001; Desta et al., 2002), decreased activity (e.g., CYP2C19*9, CYP2C19*11, and CYP2C19*13) (Blaisdell et al., 2002), or increased activity (CYP2C19*17) (Sim et al., 2006) have been reported currently (http://www.cypalleles.ki.se/cyp2C19.htm). Based on the ability to metabolize probe drugs, individuals can be categorized as CYP2C19 poor metabolizers (PMs), intermediate metabolizers (IMs), extensive metabolizers (EMs), and ultrarapid metabolizers (UMs) (Desta et al., 2002; Sim et al., 2006). The most common loss-of-function alleles accounting for the majority of PMs are CYP2C19*2 and CYP2C19*3 (Goldstein et al., 1997; Xie et al., 1999; Goldstein, 2001; Desta et al., 2002; Hamdy et al., 2002). The allelic frequency of the CYP2C19*2 allele is 23 to 39%, 11 to 16%, and 13 to 25% in Asian, white, and black subjects, respectively. The frequency of the CYP2C19*3 allele is 5 to 12% in Asians and <2% in white and black subjects. Thus, considerable interethnic differences in the distribution of PMs have been observed: e.g., 2 to 5% in whites, 4 to 7.5% in blacks, 13 to 20% in east Asians, and 38 to 79% in Pacific Islanders (Xie et al., 1999; Goldstein, 2001; Desta et al., 2002). The allelic frequency of the CYP2C19*17 allele (associated with gain in function) ranges from 18 to 32.9% in whites, 4% in Ethiopians, and 1.3% in Japanese (Kurzawski et al., 2006; Rudberg et al., 2008; Sugimoto et al., 2008).
A growing body of evidence suggests that altered CYP2C19 activity is clinically important. Compared with EMs, PMs of CYP2C19: 1) are at increased risk for diazepam and clonazepam adverse effects (Desta et al., 2002), 2) achieve greater exposure of proton pump inhibitors, gastric acid suppression, and eradication of Helicobacter pylori infection (Furuta et al., 1998, 1999, 2005, 2007); and 3) show markedly less clinical response to prodrugs that requires metabolic activation by CYP2C19 (e.g., clopidogrel, cyclophosphamide, and thalidomide) (Takada et al., 2004; Hulot et al., 2006; Li et al., 2007; Gilard et al., 2008). Thus, knowledge of CYP2C19 activity may help optimize therapy and avoid adverse effects of drugs metabolized by this enzyme.
CYP2C19 metabolic status in vivo can be inferred from genotype or by measuring the metabolism of a probe substrate (Desta et al., 2002). Reliable genotyping platforms are currently available, although accurate prediction of phenotype from genotype seems difficult in some cases for similar reasons outlined for CYP2D6 recently (Gaedigk et al., 2008): uncertainty of the functional consequences of certain variants, inability to capture changes in activity caused by nongenetic factors, and the need to genotype for large number of (rare) variants and their combinations. Conventional in vivo CYP2C19 phenotyping tests (e.g., S-mephenytoin 4-hydroxylation or omeprazole 5-hydroxylation) are attractive tools because they can capture changes in CYP2C19 activity caused by both genetic and nongenetic factors (Desta et al., 2002). However, their routine clinical use has been limited because these procedures are time and resource intensive and often invasive. A phenotyping test that overcomes deficiencies of existing approaches would be of great value.
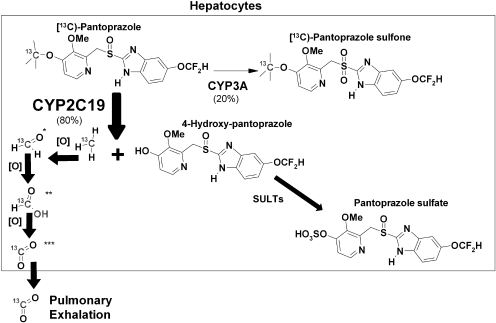
Stable isotope 13C-labeled compounds have been increasingly used as diagnostic probes in a variety of settings (Modak, 2005), including assessment of drug metabolism (Mattison et al., 2004; Leeder et al., 2008). The main purpose of the present study was to determine whether stable isotope [13C]pantoprazole is O-demethylated by CYP2C19, and the 13CO2 produced and exhaled in breath as a result (Fig. 1) can serve as a safe, rapid, and noninvasive phenotyping marker of CYP2C19 activity in vivo. Pantoprazole was selected for study because of its wider clinical use, wide safety margin, extensive metabolism in the liver primarily through CYP2C19-mediated O-demethylation (Fig. 1) (Andersson, 1996; Tanaka et al., 1997a, 2001), and a favorable structural feature for 13C stable-isotope labeling (Fig. 1).
Fig. 1.
Proposed human metabolism of [13C]pantoprazole and production of 3CO2 in breath. This metabolic pathway of 13C labeling is inferred from known metabolism of unlabeled pantoprazole (Tanaka et al., 1997a), assuming that both have similar metabolic pathways. *, formaldehyde; **, formic acid; ***, carbon dioxide.
Materials and Methods
Study Subjects. A total of 25 healthy female and male volunteers, mainly of Asian origin (18–49 years old with body weight of at least 110 pounds and body mass index ≤ 30) and pregenotyped for CYP2C19*2, *3, and *17 alleles, were studied at the outpatient clinic of the Indiana University School of the Medicine General Clinical Research Center (GCRC). This study was approved by the Institutional Review Board of the Indiana University. Investigative Device Exemption application G070004 to conduct the study was also approved by the Food and Drug Administration. This trial is registered at http://www.ClinicalTrials.gov (identifier: NCT00668902). All study subjects provided written informed consent before participation. Subjects were screened for any medical abnormalities within 6 weeks before initiating the breath test study and judged healthy on the basis of medical history, physical examination, vital signs, and standard laboratory tests. Blood samples (approximately 10 ml) were obtained during the screening for DNA analysis. Subjects were asked to refrain from taking any prescription, over-the-counter, or herbal medications and from alcohol consumption 1 week before the start of the study and during the study period. Excluded from the study were those who were tobacco smokers, had a history of intolerance or allergy to pantoprazole or sodium bicarbonate, had donated blood within the last 60 days of the screening visit or had planned to donate blood during the course of the study, had treatment with any investigational drug within the past 30 days, had used illegal drugs within 3 months before enrollment, were females pregnant or lactating, were females taking oral contraceptive birth control pills and who were unwilling or unable to stop oral contraceptives and use a barrier contraceptive method starting from the time of screening phase to the completion of the study, and were unreliable in the opinion of the study physician.
Study Design. This was an open-label, single-dose clinical trial. Eligible subjects were admitted to the GCRC at approximately 7:00 AM after an overnight fast. For female subjects, a urine pregnancy test was conducted before administration of study medications.
One hundred milligrams of [13C]pantoprazole sodium-sesquihydrate (4-O-[methyl-13C]pantoprazole, 99%; CLM-7831-SP; lot no. PR-17177), which was synthesized and supplied by Cambridge Isotope Laboratories, Inc. (Andover, MA) a powder meeting chemical purity specification (>98%), was weighed and placed into a snap-seal plastic container provided by Cambridge Isotope Laboratories, Inc. Because the pharmacokinetics of pantoprazole are linear with dose, and pantoprazole has a wide safety margin, an oral dose of 100 mg of [13C]pantoprazole that represents a higher therapeutic dose range was used in this proof-of-concept study to maximize production and quantification of 13CO2. Sodium bicarbonate (2.1 g) was weighed and transferred into the same snap-seal plastic container that contained [13C]pantoprazole and dissolved with water. Because uncoated pantoprazole is acid labile, sodium bicarbonate was concomitantly dispensed with [13C]pantoprazole to alkalinize the pH and facilitate absorption by preventing its degradation in the gut. This approach has been used successfully previously to prevent degradation of omeprazole (Howden, 2005) and pantoprazole (Ferron et al., 2003) by acid in the stomach. After baseline, breath samples were collected in 1.2-liter aluminum-lined bags (Otsuka Pharmaceuticals, Tokushima, Japan), and the solution containing [13C]pantoprazole and 2.1 g of sodium bicarbonate was then orally administered to each subject. The caps were rinsed three times with water and administered to the subjects. Breath samples were collected at 2.5, 5, 10, 15, 20, 25, 30, 40, 50, 60, 90, and 120 min after dosing. Venous blood samples (10 ml each) were collected before (predose) and at 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 5, 6, 8, 10, and 12 h after [13C]pantoprazole administration. Plasma was separated by centrifugation and stored frozen at -80°C until use. All subjects were allowed to eat regular meals after the last breath test was obtained, which was 120 min (2 h) after [13C]pantoprazole dosing. The subjects were allowed to drink water freely.
CYP2C19 Genotyping. Genomic DNA was extracted at the Indiana University GCRC Biochemistry Core laboratory from human whole blood with the QIAGEN DNA MiniKit (QIAGEN, Valencia, CA). Genotyping for CYP2C19*2 (rs4244285), CYP2C19*3 (rs4986893), and CYP2C19*17 (rs12248560) was performed by use of the predeveloped TaqMan Assay-Reagents Allelic Discrimination Kits (Applied Biosystems, Foster City, CA) according to the supplier's instructions. Their assay identifications were C__ _25986767_70, C__ _27861809_10, and C__ _469857_10, respectively. Three groups of genotypes were identified: homozygous wild type (CYP2C19*1/*1), heterozygous (CYP2C19*1/*2 or CYP2C19*1/*3), and homozygous variant (CYP2C19*2/*2).
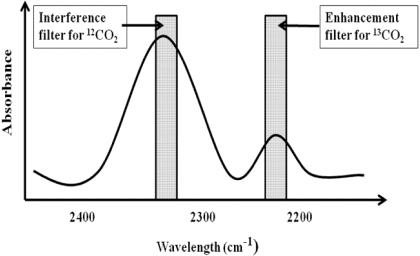
Quantitation of 13CO2. The [13C]pantoprazole breath test exploited the use of the 13C label that is incorporated at the 4-O-methyl site of pantoprazole, which specifically was designed for the hepatic CYP2C19-mediated O-demethylation. The assumption was that because isotope-unlabeled pantoprazole is mainly cleared by CYP2C19-mediated O-demethylation (Tanaka et al., 1997a, 2001), [13C]pantoprazole is O-demethylated by the same enzyme, resulting in stepwise release of 13CO2 and ultimately elimination from the body via pulmonary exhalation (Fig. 1) and that quantification of 13CO2 exhaled in breath will serve as a measure of in vivo hepatic CYP2C19 activity. To test this possibility, we measured the concentrations of 13CO2 and 12CO2 in exhaled breath samples at baseline and after dosing using the UBiT-IR300 IR spectrometry (Meretek Diagnostics, Rockville, MD) equipped with interference filters that are wavelength-selective for the absorbance of 13CO2 and 12CO2 (Fig. 2). The assay was conducted within 3 days of sample collection. The 13CO2 content in breath collection bags stored at room temperature has been shown to be stable up to 210 days (Mattison et al., 2004). Enrichment of 13CO2 in expired air was calculated at each sampling point. The delta over baseline (DOB) in the 13CO2/12CO2 ratio after [13C]pantoprazole relative to predose (baseline) 13CO2/12CO2 ratio was calculated as follows (Mattison et al., 2004; Leeder et al., 2008):
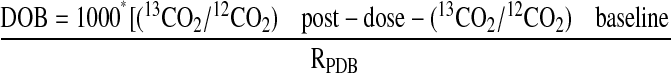 |
(1) |
where DOB was expressed as change per milliliter (0/00) and RPDB = 0.0112372 = 13CO2/12CO2 in the international standard Pee Dee Belemnite. The relative amount of [13C]pantoprazole metabolized and released into the breath as 13CO2 at each sampling time was calculated using the equation described elsewhere (Mattison et al., 2004) and expressed as the cumulative percentage of dose recovered (PDR).
Fig. 2.
Quantitation of 13CO2 and 12CO2 by IR spectroscopy.
Measurement of [13C]Pantoprazole. Plasma concentrations of [13C]pantoprazole were measured using a previously described method (Tanaka and Yamazaki, 1996; Tanaka et al., 1997a), with slight modification. To 100 μl of plasma, internal standard (50 μg/ml phenacetin) was added and deproteinized with 200 μl of acetonitrile. After centrifugation at 3000g for 10 min, the supernatant was evaporated to dryness, the residue was reconstituted in 50 μl of 50 mM sodium perchlorate and acetonitrile [80:20 (v/v)] (solvent A), and 25 μl was injected into a high-performance liquid chromatography system. Separation was performed by a Chiralcel OJ column (5.0 × 150 mm, 5 μm) (Chiral Technologies, Inc., Exton, PA), and a mobile phase was delivered by a gradient pump: 0 to 10 min, 100% solvent A (flow rate, 1.0 ml/min); 10 to 25 min, 100% solvent B [50 μl of 50 mM sodium perchlorate and 70:30 (v/v) acetonitrile]; and 25 to 35 min, 0% solvent A. The UV detector was set at 290 nm.
Analysis of Breath Test Indices and Pharmacokinetics. Breath test indices and pharmacokinetic parameters were determined by fitting the DOB data or plasma concentration data to a standard noncompartmental analysis using WinNonlin professional software (version 5.01; Pharsight, Mountain View, CA).
Statistical Analysis. Continuous variables were summarized by groups using descriptive statistics. Differences in pharmacokinetic parameters and breath test indices [maximal DOB (DOBmax), Tmax, AUC, DOB30min, and PDR] among different genotypes of CYP2C19 were analyzed by the nonparametric Kruskal Wallis test with Dunnett's multiple comparison post test. Linear regression analysis was implemented to determine gene-dose effects. Pearson's correlation analysis was performed to determine relationships between breath indices. All statistical tests were conducted using GraphPad Prism version 5.00 for Windows (GraphPad Software Inc., San Diego, CA). p < 0.05 was considered statistically significant.
Results
A total of 25 subjects of mainly Asian origin [eight Chinese, four Vietnamese, three Taiwanese, two Koreans, two Filipinos, one Japanese, one Indian, and four others (one Japanese/African, one Korean/white, one Chinese/Vietnamese/white, one Filipino/white] genotyped for the CYP2C19*2, *3, and *17 alleles, were studied (Table 1). Ten subjects were carriers of two functional alleles (CYP2C19*1/*1 genotype, n = 9; and CYP2C19*1/*17 genotype, n = 1); 10 carried one loss-of function allele (CYP2C19*1/*2 genotype, n = 9; and CYP2C19*1/*3 genotype, n = 1), and five carried two loss-of function alleles (CYP2C19*2/*2 genotype). The CYP2C19*17 alleles was rare in this population, consistent with the literature (Sim et al., 2006; Sugimoto et al., 2008). However, the frequency of the CYP2C19*3 allele was lower than what would be expected in an Asian population (Desta et al., 2002; Hamdy et al., 2002), probably because of the heterogeneity of the Asian populations studied. A subject with CYP2C19*1/*17 genotype was analyzed, together with the CYP2C19*1/*1 genotype, based on the inferred phenotype. CYP2C19 EMs, IMs, and PMs were inferred from CYP2C19*1/*1 and CYP2C19*1/*17, CYP2C191/*2 and CYP2C19*1/*3, and CYP2C19*2/*2, respectively. In the subsequent texts, EM, IMs, and PMs are used to reflect these specific genotype groups. There was no statistically significant difference in the distribution of demographic characteristics among the three genotype groups, except that female subjects dominated over male in EMs (6:4) and IMs (7:3) of CYP2C19 (Table 1).
TABLE 1.
Subject demographics in CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19*1/*3 (IMs), and CYP2C19*2/*2 (PMs) genotypes Data are presented as mean ± S.D. Note: all subjects were mainly Asian.
|
CYP2C19 Metabolic Status
|
p Value
|
|||
|---|---|---|---|---|
| EM (n = 10) | IM (n = 10) | PM (n = 5) | ||
| Age (years) | 26.2 ± 5.3 | 26.4 ± 6.1 | 24.3 ± 3.3 | 0.86 |
| Height (cm) | 164.4 ± 7.0 | 165.9 ± 9.9 | 163.4 ± 7.1 | 0.97 |
| Weight (kg) | 67.2 ± 11.3 | 70.0 ± 16.1 | 61.5 ± 5.3 | 0.62 |
| BMI | 24.8 ± 3.6 | 24.8 ± 3.5 | 23.0 ± 1.3 | 0.71 |
| Female/male ratio | 6:4 | 7:3 | 2:3 | |
BMI, body mass index
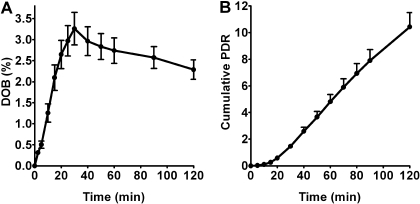
Enrichment of 13CO2 in expired air, expressed as the change or DOB in the 13CO2/12CO2 ratio after [13C]pantoprazole relative to predose (baseline) 13CO2/12CO2 ratio, was determined as a marker of [13C]pantoprazole O-demethylation. When data from all 25 subjects who completed the study were analyzed together, DOB values progressively increased with time, reached a DOBmax value (3.36 ± 1.86%) at 33.8 ± 12.36 min after [13C]pantoprazole dosing, and then declined thereafter (DOB, 2.20 ± 1.2 at t = 120 min after dosing) (Fig. 3A). The relative amount of [13C]pantoprazole metabolized and released into the breath as 13CO2 (expressed as PDR) also progressively increased over time (cumulative PDR at t = 120 min; 10.43 ± 5.42) (Fig. 3B).
Fig. 3.
13CO2 breath patterns (mean ± S.E.) versus time in all healthy volunteers (n = 25) after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate. Breath samples were collected at baseline (before) and up to 120 min after pantoprazole dosing. The amounts of 13CO2 in breath (expressed as DOB) versus time (A) and the cumulative PDR versus time (B) are presented as mean ± S.E.
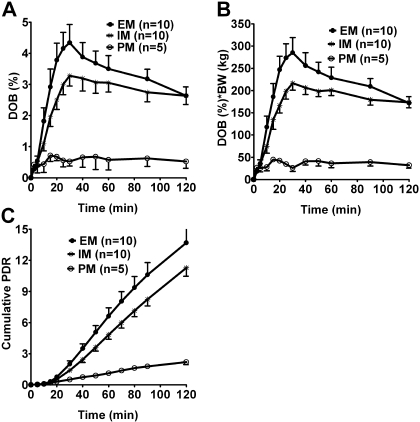
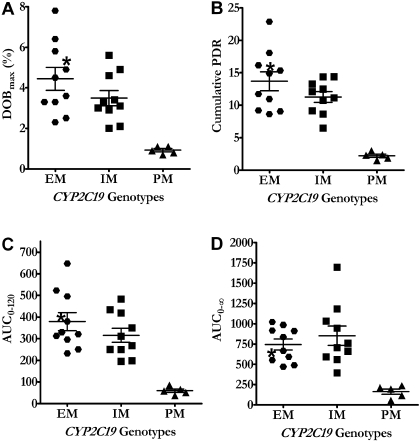
To test the influence of CYP2C19 genetic polymorphisms on [13C]pantoprazole O-demethylation, as measured by 13CO2 in expired air, DOB values were determined in EMs, IMs, and PMs of CYP2C19 at different times (2.5–120 min) after the administration of a fixed dose of [13C]pantoprazole (Fig. 4A). Upon visual inspection, DOB values were lower in PMs than IMs and EMs of CYP2C19. However, previous studies have suggested that body weight might influence breath test indices when fixed doses of phenotyping probes such as [13C]uracil (Mattison et al., 2004) and dextromethorphan (Leeder et al., 2008) were administered. To test this possibility, DOB values at each time point were multiplied by body weight and plotted against time (Fig. 4B), from which DOBmax and area under DOB versus time curve (AUC0–120) were calculated. DOBmax or AUC0–120 values after correcting for body weight significantly correlated with those without correction for body weight (Pearson r = 0.9; p < 0.0001). Therefore, data uncorrected for body weight were used for subsequent analysis. The corresponding breath test parameters are listed in Table 2. DOBmax and AUC0–120 showed a statistically significant difference among the three groups (p <0.01, Kruskal-Wallis test) (Table 2) and with a gene-dose effect (r2 = 0.455–0.485, p < 0.001). The AUC0–∞ showed a statistically significant difference among the three groups (p = 0.0028, Kruskal-Wallis test) (Table 2) and with a gene-dose effect (r2 = 0.225, p = 0.017). Tmax remained comparable between the genotypes (p = 0.51). Post hoc analysis (Dunn's multiple comparison test) revealed that PMs had significantly lower DOBmax and AUC0–120 compared with IMs (p < 0.05) or EMs (p < 0.01) of CYP2C19 (Table 2); the AUC0–120 in PMs (CYP2C19*2/*2 genotype) was ∼5.3-fold lower than in IMs (CYP2C19*1/*2/*1/*3 genotypes) and ∼6.4-fold lower than in EMs (CYP2C19*1/*1 genotype) of CYP2C19. The same trend was observed among the three genotype groups with regard to AUC0–∞. Although the 13CO2 exhaled in breath was lower in genotypes that are associated with partially reduced function (IMs) than those with two fully functional alleles (EMs), this difference did not reach a statistically significant level (Table 2).
Fig. 4.
13CO2 breath patterns (mean ± S.E.) versus time in healthy volunteers with CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19*1/*3 (IMs), and CYP2C19*2/*2 (PMs) genotypes after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate. Breath samples were collected at baseline (before drug administration) and up to 120 min after pantoprazole dosing. The amounts of 13CO2 in breath (expressed as DOB) were multiplied by body weight versus time (A), body weight uncorrected DOB versus time (B), and cumulative PDR (C). Breath test indices calculated from the data in B and C are presented in Table 2.
TABLE 2.
[13C]Pantoprazole breath test indices in healthy volunteers with CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19*1/*3 (IMs), and CYP2C19*2/*2 (PMs) genotypes after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate Breath test indices are presented as mean ± S.D.
| Parameters | EM (n = 10) | IM (n = 10) | PM (n = 5) | p Valuea |
|---|---|---|---|---|
| Tmax (min)b | 30 (20–50) | 30 (25–60) | 40 (10–50) | 0.51 |
| DOBmax | 4.44 ± 1.79** | 3.49 ± 1.18* | 0.92 ± 0.18 | 0.0017 |
| AUC0–120 | 378.9 ± 133.8** | 315.8 ± 101.6* | 59.6 ± 18.0 | 0.0019 |
| AUC0–∞ | 744.2 ± 212.8* | 852.7 ± 378.8** | 163.2 ± 75.8 | 0.0028 |
| PDR | 13.7 ± 4.62** | 11.28 ± 2.58* | 2.21 ± 0.55 | 0.0021 |
p < 0.05, PMs versus IMs
p < 0.01, PMs versus EMs; no statistically significant difference between IMs and EMs
Comparison between the three genotypes was made by Kruskal-Wallis statistic with post hoc analysis using Dunn's multiple comparison test
Median (minimum to maximum)
We also calculated the relative amount of [13C]pantoprazole metabolized and recovered in the breath as 13CO2 at each sampling time from which the cumulative PDR in breath could be estimated in the different genotypes (Fig. 4C). The cumulative PDR values (up to t = 120 min) are shown in Table 2. Consistent with the changes observed with DOB and AUC values, there was a statistically significant difference (Kruskal-Wallis test) in cumulative PDR values among the three genotype groups (p < 0.01). PMs had significantly lower cumulative PDR values (post hoc) compared with IMs (p < 0.05) or EMs (p < 0.01) of CYP2C19 (Table 2), and the effect seen was consistent with a gene-dose effect (r2 = 0.55, p < 0.0001). Differences in cumulative PDR values among IMs and EMs did not reach a statistically significant level.
The differences in 13CO2 breath indices among the different genotypes are more apparent when the individual values are displayed (Fig. 5, A–D). PMs of CYP2C19 had not only significantly lower 13CO2 breath indices, but the indices in PMs did not overlap and were clearly separated from IMs and EMs of CYP2C19. Although the mean (Table 2) and median (Fig. 5, A–D) values were lower in IMs than EMs of CYP2C19, none of the 13CO2 breath indices could segregate the two genotype groups with certainty because the values in the two groups show substantial overlap.
Fig. 5.
Individual values of [13C]pantoprazole breath test indices in healthy volunteers with CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19 *1/*3 (IMs), and CYP2C19*2/*2 (PMs) genotypes after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate. *, data from a subject who carried the CYP2C19*1/*17 genotype. PDR, recovered as 13CO2.
To determine whether [13C]pantoprazole breath test serves as a rapid phenotype marker of CYP2C19 activity in the different genotypes, DOB values at each sampling point (2.5–120 min) were compared among EMs, IMs, and PMs of CYP2C19 by nonparametric analysis of variance and for gene-dose effect relationship by linear regression analysis (Table 3). A statistically significant difference among the three genotype groups was observed as early as 10 min after [13C]pantoprazole dosing, but more robust relationships were seen starting 15 min (peaked at approximately 20–25 min), and this robustness continued until the last breath sampling (t = 120 min), with a significant gene-dose effect (Table 3). A multiple regression analysis was also performed in an attempt to relate AUC0–∞ with DOB values (10–120 min) in all subjects without considering the genotypes. Although a significant association was observed between AUC0–∞ and DOB at the different time points (10–120 min), model-derived r2 was improved from r2 = 0.297 for 20 min or less to r2 = 0.886 for 120 min or less sampling time, suggesting that the last sampling time is more predictive of AUC0–∞.
TABLE 3.
Analysis of DOB values at each sampling time in healthy volunteers with CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19*1/*3 (IMs), and CYP2C19*2/*2 (PMs) genotypes after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate
|
DOB Values at Each Sampling Time
|
Differences among
Genotypesa
(p Values)
|
Differences between
Genotypesb
|
Gene-Dose
Effectc
|
||
|---|---|---|---|---|---|
| PMs versus IMs | PMs versus EMs | r2 | p Values | ||
| DOB2.5 | 0.0455 | * | 0.18 | 0.038 | |
| DOB5 | 0.99 | 0.01 | 0.72 | ||
| DOB10 | 0.017 | * | 0.236 | 0.014 | |
| DOB15 | 0.0017 | * | ** | 0.299 | 0.0048 |
| DOB20 | 0.0008 | * | *** | 0.474 | 0.0001 |
| DOB25 | 0.0016 | * | ** | 0.522 | <0.0001 |
| DOB30 | 0.0044 | * | ** | 0.459 | 0.0003 |
| DOB40 | 0.002 | * | ** | 0.430 | 0.0004 |
| DOB50 | 0.0019 | * | ** | 0.464 | 0.0002 |
| DOB60 | 0.0023 | * | ** | 0.438 | 0.0003 |
| DOB90 | 0.0022 | * | ** | 0.478 | 0.0001 |
| DOB120 | 0.003 | ** | ** | 0.366 | 0.0014 |
p < 0.05
p < 0.01
p < 0.001. No statistically significant difference between IM and EM subjects at any of the time points
Comparison of DOB values at each sampling time among the three genotype groups (EM, IM, and PM subjects) was performed by Kruskal-Wallis test
Post hoc analysis was performed using Dunn's multiple comparison test
Gene-dose effect among the three genotypes was determined by linear regression
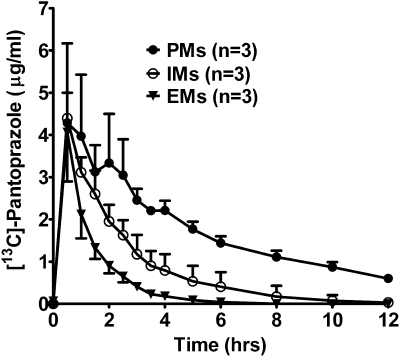
The pharmacokinetic profiles of [13C]pantoprazole from nine subjects (n = 3 in each genotype) are shown in Fig. 6, and the pharmacokinetic parameters derived are listed in Table 4. Significant differences in terminal elimination half-life, AUC0–120, AUC0–∞, and apparent oral clearance (body weight adjusted) were observed among the three genotypes (all p values < 0.05, Kruskal-Wallis test) (Table 4), with post hoc analysis showing that PMs had significantly (p < 0.05) longer t1/2, higher AUC0–12, AUC0–∞, and lower clearance compared with EMs of CYP2C19. The AUC0–∞ of [C]pantoprazole was inversely correlated with the AUC0–∞ obtained from the breath test, and this was statistically significant (Pearson r = 0.74; p = 0.038).
Fig. 6.
[13C]Pantoprazole plasma concentrations (mean ± S.D.) versus time curves in healthy volunteers with CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19*1/*3 (IMs) and CYP2C19*2/*2 (PMs) genotypes after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate.
TABLE 4.
Pharmacokinetic parameters of [13C]pantoprazole (mean ± S.D.) in healthy volunteers with CYP2C19*1/*1 (EMs), CYP2C19*1/*2 or CYP2C19*1/*3 (IMs), and CYP2C19*2/*2 (PMs) genotypes after oral administration of a solution consisting of 100 mg of [13C]pantoprazole sodium-sesquihydrate and 2.1 g of sodium bicarbonate Pharmacokinetic parameters are presented as mean ± S.D.
| Parameters | EM (n = 3) | IM (n = 3) | PM (n = 3) | p Valuea |
|---|---|---|---|---|
| Tmax (h) | 0.5 | 0.5 | 0.5 | |
| Half-life (h) | 0.88 ± 0.14* | 1.60 ± 0.61 | 4.23 ± 0.57 | 0.0439 |
| Cmax (μg/ml) | 4.06 ± 1.16 | 4.39 ± 0.61 | 4.29 ± 1.88 | 0.88 |
| AUC0—120 (h/μg/ml) | 4.99 ± 1.08* | 9.96 ± 3.43 | 21.19 ± 4.25 | 0.0439 |
| AUC0—∞ (h/μg/ml) | 5.09 ± 1.08* | 10.35 ± 3.69 | 24.94 ± 4.08 | 0.0439 |
| Vz/F (liters) | 26.20 ± 8.35 | 22.20 ± 1.48 | 25.10 ± 7.35 | 0.88 |
| Cl (ml/h/kg) | 326.9 ± 97.00* | 159.30 ± 52.00 | 67.78 ± 7.60 | 0.0439 |
AUC, area under the concentration versus time curve, 0 to 120 min or 0 to infinity; Vz/F, apparent volume of distribution; Cl, apparent oral clearance corrected for body weight
p < 0.05, PM versus EM
Comparison between the three genotypes was compared by Kruskal-Wallis statistic with post-hoc analysis using Dunn's Multiple Comparison Test
Discussion
In the present study, we have shown for the first time that: 1) [13C]pantoprazole is effectively O-demethylated in humans, as shown by an increase in 13CO2 in breath; 2) 13CO2 production was dependent on CYP2C19; and 3) enrichment of exhaled 13CO2 through the lung was inversely related to [13C]pantoprazole exposure. These data support the idea that 13CO2 exhaled via the lung as a result of O-demethylation of 13C-labeled pantoprazole is a reliable phenotype probe to identify PM individuals from IMs and EMs of CYP2C19.
Nonisotope-labeled pantoprazole is predominantly cleared by hepatic CYP2C19-mediated O-demethylation and, to some extent, through CYP3A-mediated sulfone formation (Andersson, 1996; Tanaka et al., 1997a, 2001; Furuta et al., 2005). The selection of 13C-labeled pantoprazole for the study was based on the assumption that incorporation of the 13C label at the 4-O-methyl site of the pyridine ring of pantoprazole would not influence the pattern of metabolism. Consistent with this suggestion, we have shown that [13C]pantoprazole is effectively O-demethylated in humans, as shown by a substantial increase in the enrichment of 13CO2 in breath over time (Fig. 3). We also have shown that [13C]pantoprazole O-demethylation is mainly mediated by CYP2C19; the amount of 13CO2 in breath and the cumulative percentage of dose recovered as 13CO2 in breath was significantly lower in subjects being PMs than IMs or EMs with respect to CYP2C19 (Figs. 4 and 5; Tables 2 and 3); and [13C]pantoprazole exposure in plasma (Table 4) was significantly higher in PM than EM subjects. These data suggest that the differences in breath test indices among the genotypes were due to marked reduction in [13C]pantoprazole O-demethylation in PMs and are consistent with previous data that the systemic exposure of nonisotope-labeled pantoprazole is approximately 6-fold higher in PM than in EM subjects (Tanaka et al., 1997a). Therefore, CYP2C19-mediated O-demethylation seems to be the major route of metabolism of both 13C-labeled and -unlabeled pantoprazole; incorporation of the 13C label at the 4-O-methyl site of the pyridine ring does not seem to alter the pattern of pantoprazole metabolism.
The time to DOBmax and Cmax of [13C]pantoprazole was shorter (on the average, 33.8 and 30 min, respectively) as opposed to the time to Cmax of the unlabeled drug, which is ∼2.4 h in pharmacokinetic studies (Tanaka et al., 1997a, 2001). Like other proton pump inhibitors, pantoprazole is acid labile, and the oral formulation of pantoprazole is often administered as an enteric-coated tablet to avoid degradation by gastric acid. In the present study, sodium bicarbonate was used to transiently neutralize the gastric acid and to prevent degradation of pantoprazole. This and the administration of the test compounds as a solution might have enhanced its rapid absorption, allowing a rapid and early time point breath test measurement to effectively distinguish PMs from IMs and EMs of CYP2C19. As shown in Table 3, a statistically significant difference in DOB values among the genotypes was observed as early as 10 min after [13C]pantoprazole administration, and a robust difference was seen at 15 min and thereafter. This test seems to be as reliable as the established phenotyping approaches (Desta et al., 2002). Our feasibility study suggests that the [13C]pantoprazole breath test offers advantages over existing genotype and phenotype approaches in predicting or assessing CYP2C19 activity in vivo, particularly in effectively distinguishing PMs from IMs and EMs of CYP2C19 (Fig. 5, A–D), because it can be performed rapidly and possibly at a single time point in a noninvasive manner. However, the optimal time breath test measurement that effectively distinguishes PMs from IMs and EMs of CYP2C19 awaits further investigation and should take into account factors such as absorption lag time. In addition, a relatively higher dose of [13C]pantoprazole was used in this feasibility study to maximize the 13CO2 signal in expired air, but the utility of the lowest doses of this probe that is appropriate for the phenotyping purpose and avoids any potential adverse effects associated with the use of high dose should be explored in the future.
Although the [13C]pantoprazole breath test is expected to discriminate PMs from UMs and probably IMs from UMs, our sample size (only one subject with the CYP2C19*1/*17 genotype) did not allow us to properly evaluate the performance of this test with respect to UMs versus other phenotype groups. The ability of the [13C]pantoprazole breath test to discriminate IMs from EMs of CYP2C19 with certainty seems to be weak. Although a gene-dose effect relationship was noted regarding the influence of CYP2C19 genotype on the inferred phenotypes (Figs. 4 and 5; Tables 2 and 3), no statistically significant difference in phenotype was observed among those that were predicted to be CYP2C19 IMs and EMs. It is noteworthy that conventional probes such as omeprazole and S-mephenytoin also accurately discriminate PMs from IMs or EMs, but there is often uncertainty in their ability to effectively distinguish subjects with IMs from those with EMs of CYP2C19, despite the fact that a statistically significant difference between the two groups has been reported (Yin et al., 2004; Furuta et al., 2005). Our findings suggest a better separation of IMs from EMs when [13C]pantoprazole exposure is considered relative to the breath test indices. The reasons for this observation are not fully known. The methyl group formed from CYP2C19-mediated [13C]pantoprazole O-demethylation passes through the carbon pool (13CH3 to 13CHO to 13COO and 13CO2) before it eventually traverses to the lung and is exhaled. Differential handlings during these processes might influence the breath test indices. Although we attempted to minimize the impact of baseline 13CO2 on the calculation of enriched 13CO2 by requesting subjects to refrain from activities that increase 13Cinthe body before and during the study (e.g., eating foods enriched with 13C, exercise, alcohol and cigarette consumption, and nonfasting overnight before the study), the level of compliance was difficult to assess. The influence of baseline 13CO2 level was partially corrected because the 13CO2 measured after the administration of [13C]pantoprazole was corrected for baseline values. However, given the small difference and overlapping DOB values among the IM and EM groups, the possibility that baseline 13CO2 might influence the calculation of 13CO2 enrichment cannot be ruled out. Third, pantoprazole is also metabolized to pantoprazole sulfone, which could be O-demethylated and contribute to 13CO2 production. Although factors independent of hepatic CYP2C19 activity could potentially affect 13CO2 measurement, this test still seems to be a noninvasive, safe, and rapid indirect surrogate marker of CYP2C19 activity in vivo.
The CYP2C19*2/*2 genotype associated with PM status is expected to produce no 13CO2 in breath because this genotype is expected to produce nonfunctional CYP2C19 enzymatic activity. Assuming that pantoprazole O-demethylation is exclusively catalyzed by CYP2C19, no production of 13CO2 should have been expected in PMs of CYP2C19. However, as shown in Figs. 3 and 4 and Table 2, small but appreciable enrichment of 13CO2 in breath in PM subjects was observed. These data suggest that, although CYP2C19 is the major enzyme catalyzing O-demethylation of pantoprazole, the contribution of other enzymes cannot be ruled out. Pantoprazole is a chiral drug that is clinically administered as a racemic mixture. In vivo, (+)-pantoprazole has been shown to be more dependent on CYP2C19 than (-)-pantoprazole (Tanaka et al., 1997b, 2001), which seems to be a characteristic of omeprazole and lansoprazole (Andersson and Weidolf, 2008). Thus, it is likely that O-demethylation of (-)-pantoprazole is catalyzed by cytochromes P450 other than CYP2C19, particularly when the activity of CYP2C19 is substantially diminished. The [13C]pantoprazole used in our study is a racemic mixture (approximately 50:50). Because the metabolic profiles of stable isotope-labeled and -unlabeled [13C]pantoprazole seem similar, it is logical to suggest that (+)-[13C]pantoprazole is more dependent on CYP2C19 and that enzymes other than CYP2C19 that are involved in the O-demethylation of (-)-[13C]pantoprazole might have contributed to the residual 13CO2 in the breath of PMs of CYP2C19. The possibility that the sulfone metabolite of pantoprazole might also be O-demethylated to release 13CO2 cannot be excluded.
In summary, given the salient features of nonradioactive 13C labeling, the wide margin of pantoprazole safety, and the noninvasive, inexpensive, and rapid procedures involved, the [13C]pantoprazole breath test seems to offer a useful screening method that can be applied in most clinical settings (e.g., hospitals and physicians' offices) to identify CYP2C19 function before dosing with CYP2C19 substrates. Furthermore, this novel tool should facilitate research and screening of subjects in clinical trials involving CYP2C19. Emerging evidence suggests that interindividual and interethnic differences in CYP2C19 activity influence therapeutic response of drugs, such as proton pump inhibitors, clopidogrel, cyclophosphamide, and thalidomide (Furuta et al., 1998, 2005, 2007; Desta et al., 2002; Takada et al., 2004; Gilard et al., 2008; Hulot et al., 2006; Li et al., 2007). A rapid phenotype test that captures variability of CYP2C19 enzyme activity because of genetic and nongenetic factors and potentially offers greater practical clinical utility than the existing approaches, such as the [13C]pantoprazole breath test described herein, should be an important step to optimize therapy with CYP2C19 substrates or select alternative drugs for the individual patient.
Acknowledgments
We thank the volunteers who participated in this clinical trial and the nurses and technicians at the Indiana University School of Medicine General Clinical Research Center for help with conducting the healthy volunteers study and processing the samples, including DNA extraction.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grants T32GM008425, 1R01GM078501-01A1]; Otsuka Pharmaceuticals Company [Grant-in-aid; Tokyo, Japan]; and the National Institutes of Health National Center for Research Resources [Grant MO1-RR000750].
Z.D., A.M., Y.K., and D.A.F. are co-owners of a patent on a clinical test designed to determine CYP2C19 activity using the [13C]pantoprazole breath test (http://www.wipo.int/pctdb/en/wo.jsp?WO=2008028116&IA=US2007077362&DISPLAY=STATUS).
D.A.F. has served as a paid consultant for Roche Molecular Diagnostics (Indianapolis, IN). The other authors declare no conflict of interest.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.108.147751.
ABBREVIATIONS: PM, poor metabolizer; IM, intermediate metabolizer; EM, extensive metabolizer; UM, ultrarapid metabolizer; GCRC, General Clinical Research Center; DOB, delta over baseline; PDR, percentage dose recovered; DOBmax, maximal DOB; Tmax, time to maximal concentration or DOB; AUC, area under the concentration-time curve or area under DOB-time curve; Cmax, maximal plasma concentration.
References
- Andersson T (1996) Pharmacokinetics, metabolism and interactions of acid pump inhibitors: focus on omeprazole, lansoprazole and pantoprazole. Clin Pharmacokinet 31 9-28. [DOI] [PubMed] [Google Scholar]
- Andersson T and Weidolf L (2008) Stereoselective disposition of proton pump inhibitors. Clin Drug Investig 28 263-279. [DOI] [PubMed] [Google Scholar]
- Ando Y, Fuse E, and Figg WD (2002) Thalidomide metabolism by the CYP2C subfamily. Clin Cancer Res 8 1964-1973. [PubMed] [Google Scholar]
- Blaisdell J, Mohrenweiser H, Jackson J, Ferguson S, Coulter S, Chanas B, Xi T, Ghanayem B, and Goldstein JA (2002) Identification and functional characterization of new potentially defective alleles of human CYP2C19. Pharmacogenetics 12 703-711. [DOI] [PubMed] [Google Scholar]
- de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, and Goldstein JA (1994) The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 269 15419-15422. [PubMed] [Google Scholar]
- Desta Z, Zhao X, Shin JG, and Flockhart DA (2002) Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet 41 913-958. [DOI] [PubMed] [Google Scholar]
- Ferron GM, Ku S, Abell M, Unruh M, Getsy J, Mayer PR, and Paul J (2003) Oral bioavailability of pantoprazole suspended in sodium bicarbonate solution. Am J Health Syst Pharm 60 1324-1329. [DOI] [PubMed] [Google Scholar]
- Frye RF, Zgheib NK, Matzke GR, Chaves-Gnecco D, Rabinovitz M, Shaikh OS, and Branch RA (2006) Liver disease selectively modulates cytochrome P450-mediated metabolism. Clin Pharmacol Ther 80 235-245. [DOI] [PubMed] [Google Scholar]
- Furuta T, Ohashi K, Kamata T, Takashima M, Kosuge K, Kawasaki T, Hanai H, Kubota T, Ishizaki T, and Kaneko E (1998) Effect of genetic differences in omeprazole metabolism on cure rates for Helicobacter pylori infection and peptic ulcer. Ann Intern Med 129 1027-1030. [DOI] [PubMed] [Google Scholar]
- Furuta T, Ohashi K, Kosuge K, Zhao XJ, Takashima M, Kimura M, Nishimoto M, Hanai H, Kaneko E, and Ishizaki T (1999) CYP2C19 genotype status and effect of omeprazole on intragastric pH in humans. Clin Pharmacol Ther 65 552-561. [DOI] [PubMed] [Google Scholar]
- Furuta T, Shirai N, Sugimoto M, Nakamura A, Hishida A, and Ishizaki T (2005) Influence of CYP2C19 pharmacogenetic polymorphism on proton pump inhibitor-based therapies. Drug Metab Pharmacokinet 20 153-167. [DOI] [PubMed] [Google Scholar]
- Furuta T, Sugimoto M, Shirai N, and Ishizaki T (2007) CYP2C19 pharmacogenomics associated with therapy of Helicobacter pylori infection and gastro-esophageal reflux diseases with a proton pump inhibitor. Pharmacogenomics 8 1199-1210. [DOI] [PubMed] [Google Scholar]
- Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, and Leeder JS (2008) The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 83 234-242. [DOI] [PubMed] [Google Scholar]
- Gilard M, Arnaud B, Cornily JC, Le Gal G, Lacut K, Le Calvez G, Mansourati J, Mottier D, Abgrall JF, and Boschat J (2008) Influence of omeprazole on the antiplatelet action of clopidogrel associated with aspirin: the randomized, double-blind OCLA (Omeprazole CLopidogrel Aspirin) study. J Am Coll Cardiol 51 256-260. [DOI] [PubMed] [Google Scholar]
- Goldstein JA (2001) Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol 52 349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JA and de Morais SM (1994) Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 4 285-299. [DOI] [PubMed] [Google Scholar]
- Goldstein JA, Ishizaki T, Chiba K, de Morais SM, Bell D, Krahn PM, and Evans DA (1997) Frequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populations. Pharmacogenetics 7 59-64. [DOI] [PubMed] [Google Scholar]
- Hamdy SI, Hiratsuka M, Narahara K, El-Enany M, Moursi N, Ahmed MS, and Mizugaki M (2002) Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br J Clin Pharmacol 53 596-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden CW (2005) Review article: immediate-release proton-pump inhibitor therapy-potential advantages. Aliment Pharmacol Ther 22 (Suppl 3): 25-30. [DOI] [PubMed] [Google Scholar]
- Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, Aiach M, Lechat P, and Gaussem P (2006) Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood 108 2244-2247. [DOI] [PubMed] [Google Scholar]
- Ishizawa Y, Yasui-Furukori N, Takahata T, Sasaki M, and Tateishi T (2005) The effect of aging on the relationship between the cytochrome P450 2C19 genotype and omeprazole pharmacokinetics. Clin Pharmacokinet 44 1179-1189. [DOI] [PubMed] [Google Scholar]
- Kupfer A, Desmond PV, and Schenker SBR (1979) Family study of a genetically determined deficiency of mephenytoin hydroxylation in man (letter). Pharmacologist 21 173. [Google Scholar]
- Kurzawski M, Gawrońska-Szklarz B, Wrześniewska J, Siuda A, Starzyńska T, and Droździk M (2006) Effect of CYP2C19*17 gene variant on Helicobacter pylori eradication in peptic ulcer patients. Eur J Clin Pharmacol 62 877-880. [DOI] [PubMed] [Google Scholar]
- Leeder JS, Pearce RE, Gaedigk A, Modak A, and Rosen DI (2008) Evaluation of a [13C]-dextromethorphan breath test to assess CYP2D6 phenotype. J Clin Pharmacol 48 1041-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hou J, Jiang H, Wang D, Fu W, Yuan Z, Chen Y, and Zhou L (2007) Polymorphisms of CYP2C19 gene are associated with the efficacy of thalidomide based regimens in multiple myeloma. Haematologica 92 1246-1249. [DOI] [PubMed] [Google Scholar]
- Mattison LK, Ezzeldin H, Carpenter M, Modak A, Johnson MR, and Diasio RB (2004) Rapid identification of dihydropyrimidine dehydrogenase deficiency by using a novel 2–13C-uracil breath test. Clin Cancer Res 10 2652-2658. [DOI] [PubMed] [Google Scholar]
- McGready R, Stepniewska K, Seaton E, Cho T, Cho D, Ginsberg A, Edstein MD, Ashley E, Looareesuwan S, White NJ, et al. (2003) Pregnancy and use of oral contraceptives reduces the biotransformation of proguanil to cycloguanil. Eur J Clin Pharmacol 59 553-557. [DOI] [PubMed] [Google Scholar]
- Modak A (2005) 13C breath tests: transition from research to clinical practice, in Breath Analysis for Clinical Diagnosis and Therapeutic Monitoring (Amann A and Smith D eds) pp 457-478, World Scientific, Singapore.
- Rudberg I, Mohebi B, Hermann M, Refsum H, and Molden E (2008) Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther 83 322-327. [DOI] [PubMed] [Google Scholar]
- Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, and Ingelman-Sundberg M (2006) A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79 103-113. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Uno T, Yamazaki H, and Tateishi T (2008) Limited frequency of the CYP2C19*17 allele and its minor role in a Japanese population. Br J Clin Pharmacol 65 437-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Arefayene M, Desta Z, Yarboro CH, Boumpas DT, Balow JE, Flockhart DA, and Illei GG (2004) Cytochrome P450 pharmacogenetics as a predictor of toxicity and clinical response to pulse cyclophosphamide in lupus nephritis. Arthritis Rheum 50 2202-2210. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ohkubo T, Otani K, Suzuki A, Kaneko S, Sugawara K, Ryokawa Y, Hakusui H, Yamamori S, and Ishizaki T (1997a) Metabolic disposition of pantoprazole, a proton pump inhibitor, in relation to S-mephenytoin 4′-hydroxylation phenotype and genotype. Clin Pharmacol Ther 62 619-628. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ohkubo T, Otani K, Suzuki A, Kaneko S, Sugawara K, Ryokawa Y, and Ishizaki T (2001) Stereoselective pharmacokinetics of pantoprazole, a proton pump inhibitor, in extensive and poor metabolizers of S-mephenytoin. Clin Pharmacol Ther 69 108-113. [DOI] [PubMed] [Google Scholar]
- Tanaka M and Yamazaki H (1996) Direct determination of pantoprazole enantiomers in human serum by reversed-phase high-performance liquid chromatography using a cellulose-based chiral stationary phase and column-switching system as a sample cleanup procedure. Anal Chem 68 1513-1516. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Yamazaki H, Hakusui H, Nakamichi N, and Sekino H (1997b) Differential stereoselective pharmacokinetics of pantoprazole, a proton pump inhibitor in extensive and poor metabolizers of pantoprazole: a preliminary study. Chirality 9 17-21. [DOI] [PubMed] [Google Scholar]
- Wrighton SA, Stevens JC, Becker GW, and VandenBranden M (1993) Isolation and characterization of human liver cytochrome P450 2C19: correlation between 2C19 and S-mephenytoin 4′-hydroxylation. Arch Biochem Biophys 306 240-245. [DOI] [PubMed] [Google Scholar]
- Xie HG, Stein CM, Kim RB, Wilkinson GR, Flockhart DA, and Wood AJ (1999) Allelic, genotypic and phenotypic distributions of S-mephenytoin 4′-hydroxylase (CYP2C19) in healthy Caucasian populations of European descent throughout the world. Pharmacogenetics 9 539-549. [PubMed] [Google Scholar]
- Yin OQ, Tomlinson B, Chow AH, Waye MM, and Chow MS (2004) Omeprazole as a CYP2C19 marker in Chinese subjects: assessment of its gene-dose effect and intrasubject variability. J Clin Pharmacol 44 582-589. [DOI] [PubMed] [Google Scholar]