Abstract
Mifepristone [RU486; 17β-hydroxy-11β-(4-dimethylaminophenyl)-17α-(1-propynyl)-estra-4,9-dien-3-one] inactivates CYP2B6 in the reconstituted system in a mechanism-based manner. The loss of 7-ethoxy-4-(trifluoromethyl)-coumarin deethylation activity of CYP2B6 is concentration- and time-dependent. The inactivation requires NADPH and is irreversible. The concentration of inactivator required to give the half-maximal rate of inactivation is 2.8 μM, and the maximal rate constant for inactivation at a saturating concentration of the inactivator is 0.07 min-1. Incubation of CYP2B6 with 20 μM RU486 for 15 min resulted in 61% loss of catalytic activity, 60% loss of the reduced cytochrome P450 (P450)-CO complex, and a 40% loss of native heme. The partition ratio is ∼5, and the stoichiometry of binding is ∼0.6 mol RU486/mol P450 inactivated. SDS-polyacrylamide gel electrophoresis and high-pressure liquid chromatography analysis showed that [3H]RU486 was irreversibly bound to CYP2B6 apoprotein. RU486 is metabolized to form three major metabolites and bioactivated to give reactive intermediates by purified P450s in the reconstituted system. After incubation of RU486 with the purified P450s and liver microsomes from rats and humans in the presence of glutathione (GSH) and NADPH, GSH conjugates with MH+ ions at m/z 769, 753, and 751 were detected by liquid chromatography-tandem mass spectrometry. Two GSH conjugates with MH+ ions at m/z 753 are formed from the reaction of GSH with RU486. The adducts are formed after addition of an activated oxygen to the carbon-carbon triple bond of the propynyl moiety. This suggests that oxirene intermediates may be involved in the mechanism of inactivation. It seems that the potential for drug-drug interactions of RU486 may not be limited only to CYP3A4 and should also be evaluated for drugs metabolized primarily by CYP2B6, such as bupropion and efavirenz.
Mifepristone (RU486), a synthetic internal acetylenic steroid with a propynyl group at the 17α-position, is a remarkably effective antiprogesterone and antiglucocorticosteroid agent in humans. It has potential for use in the treatment of breast cancer, prostate cancer, uterine leiomyoma, and Cushing's syndrome (Chasserot-Golaz and Beck, 1992; Cadepond et al., 1997). RU486 is extensively metabolized by demethylation of the C-11 dimethylaminophenyl group and by hydroxylation of the C-17 propynyl group in liver microsomes from rats and humans (Heikinheimo et al., 1990; Chasserot-Golaz and Beck, 1992; Jang et al., 1996). The mechanism-based inactivation of purified CYP3A4 and P450s in liver microsomes from rats and humans has been demonstrated (Jang and Benet, 1998; He et al., 1999; Reilly et al., 1999). Halpert and coworkers have reported that RU486 is a selective inactivator of human CYP3A4, but not of CYP3A5 (Khan et al., 2002). We have recently demonstrated that bergamottin, a relatively potent mechanism-based inactivator of human CYP3A4, is even more effective as a mechanism-based inactivator of CYP2B6 (Lin et al., 2005). Therefore, we have investigated the ability of RU486 to act as a mechanism-based inactivator of CYP2B6. A variety of widely used drugs, including bupropion, efavirenz, methadone, ifosfamide, and cyclophosphamide, are preferentially metabolized or stereoselectively metabolized by CYP2B6 (Faucette et al., 2000; Huang et al., 2000; Ward et al., 2003; Gerber et al., 2004). Moreover, CYP2B6 is expressed in human liver, brain, kidney, and lung and exhibits significant genetic polymorphisms (Gervot et al., 1999; Lang et al., 2001).
The occurrence of heme alkylation versus binding to the P450 apoprotein by an external acetylene compound is believed to be related to the addition of oxygen at the internal carbon versus the terminal carbon in the carbon-carbon triple bond (Ortiz de Montellano and Kunze, 1980; Ortiz de Montellano and Komives, 1985; CaJacob et al., 1988; Chan et al., 1993). Delivery of the activated oxygen to the internal carbon of the acetylene is believed to result in heme alkylation, whereas delivery of the oxygen to the terminal carbon leads to acylation on the protein. However, unlike terminal acetylenes, mechanism-based inactivation by internal acetylene compounds seems to inactivate P450s without the formation of detectable heme adducts (Ortiz de Montellano and Kunze, 1980). Studies with internal acetylenes such as the midchain acetylenic compound dodecynoic acid, acetylenic steroids, modified acetylenic steroids, aryl acetylenes, and RU486 have all suggested that the primary mechanism of inactivation was protein modification rather than heme alkylation (Nagahisa et al., 1983; Olakanmi and Seybert, 1990; Foroozesh et al., 1997; Helvig et al., 1997; He et al., 1999). Moreover, it has been suggested that P450 inactivation by 4-(1-propynyl)biphenyl acetylene, which involves the generation of 2-biphenylpropionic acid, proceeds via a 1,2-methyl shift analogous to the mechanism of mechanism-based inactivation by ethynyl acetylene that proceeds via a 1,2-hydrogen shift (Ortiz de Montellano and Kunze, 1981; Foroozesh et al., 1997). Thus far, the covalent binding of internal acetylenes to P450 proteins is believed to be the primary mechanism for the inactivation.
We have reported previously that after exposure of CYP2E1 to peroxynitrite, the amount of the P450 reduced-CO complex was decreased, but the amount of the prosthetic heme group did not, suggesting that the modified protein has lost some of its ability to bind CO (Lin et al., 2007). Although both of the previous reports on the mechanism-based inactivation of CYP3A4 and rat liver microsomal P450s by RU486 have shown the loss of spectrally detectable cytochrome P450, they have not reported on whether the prosthetic heme group is covalently modified or lost (He et al., 1999; Reilly et al., 1999). High-pressure liquid chromatography (HPLC) analysis was used to determine whether any heme destruction and formation of heme adduct had occurred or whether covalent binding of the heme to apoprotein was responsible for the inactivation. Because the reactive intermediates of internal acetylenes formed by P450s have not been reported previously, it was of interest to determine whether two different regioisomers of glutathione (GSH) conjugates are formed via the addition of oxygen to different carbons of the propynyl moiety during the metabolism of RU486 by P450s. GSH was used to trap the reactive species formed in the presence of NADPH, and the GSH conjugates were then characterized by liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis (Baillie and Davis, 1993; Tang and Miller, 2004). The formation of GSH conjugates of RU486 by human liver microsomes, microsomes from phenobarbital (PB)- or dexamethasone (DEX)-induced rat livers, and purified P450s in the reconstituted system were characterized.
Materials and Methods
Chemicals. NADPH, GSH, catalase, and RU486 were purchased from Sigma-Aldrich (St. Louis, MO). 7-Ethoxy-4-(trifluoromethyl)-coumarin (EFC) was obtained from Invitrogen (Carlsbad, CA). [6,7-3H]RU486, originally obtained from Roussel Uclaf (Romainville, France), was a generous gift from Dr. William B. Pratt at the University of Michigan (Ann Arbor, MI). The specific activity was 38.4 Ci/mmol. All other chemicals and solvents used were the highest purity available from commercial sources.
Purification of Enzymes. The plasmid for CPY2B6 was a generous gift from Dr. James R. Halpert (University of California at San Diego, La Jolla, CA) and was expressed as a His-tagged protein in Escherichia coli TOPP3 cells and purified to homogeneity as described previously (Scott et al., 2001). NADPH-cytochrome P450 reductase, cytochrome b5, and the other P450s were expressed and purified as described previously (Lin et al., 2005).
Preparation of Liver Microsomes. Rat liver microsomes were prepared from male Fisher 344 rats (175-190 g; Harlan, Indianapolis, IN) as described previously (Coon et al., 1978). One group was induced by intraperitoneal injection of 100 mg/kg PB in water once a day for 3 days. Another group was induced by intraperitoneal injection of 50 mg/kg DEX in corn oil once a day for 3 days. Animals were fasted for 18 h after the last dose and sacrificed. Frozen human liver samples were a gift from Dr. F. Peter Guengerich (Vanderbilt University, Nashville, TN). Microsomes were prepared as described previously with minor modifications (Guengerich, 1994; Teiber and Hollenberg, 2000).
Enzyme Assay and Inactivation of P450s. To assess catalytic activity, CYP2B6 was reconstituted with reductase and cytochrome b5 at 22°C for 30 min, as described previously (Lin et al., 2005). The primary reaction mixture contained 1 nmol CYP2B6, 2 nmol NADPH-cytochrome P450 reductase, 1 nmol cytochrome b5, 100 units of catalase, and 2 mM GSH in 1 ml of 100 mM potassium phosphate buffer, pH 7.7. Different concentrations of RU486 were added to portions of the primary reaction mixtures, and the reactions were initiated by adding 1 mM NADPH. At the times indicated, aliquots (10 μl) were removed, and the EFC O-deethylation activity of the CYP2B6 was measured to determine activity remaining as described previously (Lin et al., 2005).
Partition Ratio. RU486 at final concentrations ranging from 0.5 to 150 μM was added to the primary reaction mixture containing 1 μM CYP2B6. The reactions were initiated by addition of 1 mM NADPH, and the reaction mixtures were incubated at 30°C for 1 h to allow the inactivation to reach completion. Duplicate aliquots were removed and assayed for EFC O-deethylation activity as described above.
Spectral and HPLC Analyses. After incubation of the primary reaction mixture of CYP2B6 with 20 μM RU486 in the control sample (-NADPH) or the inactivated samples (+NADPH) at 30°C for 15 min, the reduced CO difference spectra of 0.2 nmol aliquots of each sample were determined after bubbling with CO and reduction by sodium dithionite by scanning from 400 to 500 nm on a UV-2501PC spectrophotometer (Shimadzu, Kyoto, Japan). Aliquots were also removed from each sample to determine the catalytic activity. To test whether the inactivation by RU486 was irreversible, control and inactivated samples were dialyzed overnight at 4°C against 1 liter each of 100 mM potassium phosphate buffer, pH 7.5, containing 20% glycerol and 0.1 mM EDTA. The samples were then reanalyzed for enzymatic activity and reduced CO difference spectrum. To investigate whether any heme loss occurred during the inactivation of CYP2B6 by RU486, 0.1 nmol of the control and inactivated sample was analyzed by HPLC under acidic conditions to separate the heme and apoprotein of CYP2B6. The HPLC column was a C4 reverse phase column (5 μm, 4.6 × 250 mm, 300 Å; Phenomenex, Torrance, CA). The solvent system consisted of solvent A [0.1% trifluoroacetic acid (TFA) in water] and solvent B (0.05% TFA in acetonitrile). The column was eluted with a linear gradient from 30 to 80% B over 30 min at a flow rate of 1 ml/min. The absorption spectra of the native and RU486-modified heme were recorded using a diode-array detector.
Stoichiometry and Specificity of Binding. After incubating the control and inactivation samples of CYP2B6 prepared as indicated previously with 10 μM [3H]RU486 at 30°C for 20 min, 0.1 nmol samples were mixed with 10 mg of bovine serum albumin and precipitated by adding a 5-fold volume of 5% sulfuric acid in methanol (Chan et al., 1993). After repeating the washing and precipitation several times until the radioactivity in the supernatants was essentially at background levels, the pellets were dissolved in 1 N NaOH, incubated at 60°C for 1 h, and counted in a liquid scintillation counter. For SDS-polyacrylamide gel electrophoresis (PAGE) analysis, aliquots containing 50 pmol P450 from the control and inactivated samples were resolved by electrophoresis on a 10% polyacrylamide gel, and the gel was then dried on 3-mm chromatography paper. The dried gel was exposed to Kodak Biomax MS film (Eastman Kodak, Rochester, NY) at -80°C for 2 weeks before developing.
HPLC Analysis with [3H]RU486. Reaction mixtures containing 0.1 nmol CYP2B6 were incubated with 10 μM [3H]RU486 and then analyzed by HPLC under acidic conditions using a shallow solvent gradient to separate the radiolabeled RU486 and its metabolites from the apoprotein and heme. The solvent system consisted of solvent A (0.1% TFA in water) and solvent B (0.05% TFA in acetonitrile). The samples were analyzed on a C4 reverse phase column (10 μm, 4.6 × 250 mm, 300 Å; Phenomenex). The column was eluted with a linear gradient from 40 to 70% B over 40 min, and the final concentration of B was increased to 95% linearly over another 10 min at a flow rate of 1 ml/min. The fractions were collected and counted by liquid scintillation counting (Econo-Safe; Research Products International, Mt. Prospect, IL).
Metabolism of RU486. One nanomole CYP2B1, CYP2B6, CYP3A4, or CYP3A5 in the reconstituted system was incubated with 20 μM RU486 at 37°C for 30 min in a final volume of 1 ml. The samples were then extracted with 4 ml of ethyl acetate, dried under N2, and dissolved in 200 μl of 50% acetonitrile/0.1% acetic acid. Half of the metabolites (100 μl) were separated by HPLC on a C8 column (Zorbax, 5 μm, 4.6 × 250 mm; MAC-MOD Analytical, Chadds Ford, PA) with a solvent system consisting of solvent A (0.1% acetic acid in water) and solvent B (0.1% acetic acid, 29.9% methanol, and 70% acetonitrile) starting with 35% B for 5 min, followed by a linear gradient from 35 to 60% B for 25 min and then increasing to 95% B for an additional 10 min at a flow rate of 1 ml/min. The eluate was monitored using a 490E multiwavelength detector (Waters, Milford, MA) set at 310 nm. The other half of the metabolites were analyzed by LC-MS/MS (LCQ mass analyzer; Thermo Fisher Scientific, Waltham, MA) using the same column and solvent gradient. The electrospray ionization (ESI) conditions were: sheath gas flow rate, 90 arbitrary units; auxiliary gas, 30 arbitrary units; source voltage, 4.5 kV; capillary temperature, 170°C; capillary voltage, 30 V; and tube lens offset, 25 V. Data were acquired in the positive ion mode using Excalibur software (Thermo Fisher Scientific) with one full scan followed by two data-dependent scans of the most intense and the second most intense ions.
LC-MS/MS Analysis of GSH Conjugates. The purified CYP2B1, CYP2B6, CYP3A4,and CYP3A5 in the reconstituted system, human liver microsomes, and rat liver microsomes (each containing 1 nmol P450) were incubated with 10 mM GSH and 20 μM RU486 in the presence of NADPH at 37°C for 30 min. The samples were then acidified with 60 μl of 10% TFA and then applied to a 1-ml AccuBond ODS-C18 solid phase extraction cartridge (Agilent Technologies, Santa Clara, CA). The cartridge was previously washed with 2 ml of methanol followed by 2 ml of water. After the samples were loaded, the cartridges were washed with 2 ml of water and then eluted with 2 ml of methanol followed by 0.3 ml of acetonitrile. The eluted samples were dried under N2 gas and resuspended in 80 μl of 1:1 solvent A (0.1% acetic acid in H2O) and solvent B (0.1% acetic acid in acetonitrile). The samples were then analyzed on a C18 reverse phase column (Luna, 3 μm, 4.6 × 100 mm; Phenomenex) using a gradient of 20 to 30% B for 5 min followed by a gradient to 40% B over 15 min and then increasing linearly to 90% B over 30 min at a flow rate of 0.3 ml/min. The column effluent was directed into the ESI source of a LCQ mass spectrometer (Thermo Fisher Scientific). The ESI conditions were: sheath gas flow rate, 90 arbitrary units; auxiliary gas, 30 arbitrary units; spray voltage, 4.5 kV; capillary temperature, 170°C; capillary voltage, 30 V; and tube lens offset, 25 V. Data were acquired in positive ion mode using eXcalibur software as described above.
Docking RU486 to CYP3A4 Crystal Structure. RU486 was docked into the active site of CYP3A4 using the energy-based docking software of AutoDock (version 4.0) (Morris et al., 1996). The coordinates of CYP3A4 (PDB ID 1TNQ) were obtained from the Protein Data Bank (1TNQ), whereas the coordinates and the lowest energy conformation of RU486 were obtained with MOPAC 2007 interfaced with ChemBioOffice 2008 (CambridgeSoft Corporation, Cambridge, MA). The flexible RU486 was docked to the rigid CYP3A4 with the Lamarckian Genetic Algorithm approach of AutoDock 4 with the following parameters: mutation rate = 0.02, crossover = 0.80, maximal number of generations = 2.7 × 104, and local search frequency = 0.06. The conformers with the lowest energy are depicted in the figure.
Results
Inactivation of CYP2B6 by RU486. The time courses for the inactivation of CYP2B6 by various concentrations of RU486 are shown in Fig. 1. The loss of EFC deethylation activity exhibited pseudo-first order kinetics with respect to time and required NADPH. Linear regression analysis of the time course data were used to determine the initial rate constants for inactivation (kobs) at various concentrations of RU486. From the double reciprocal plot (inset) of the values for kobs and the concentration of RU486, the values for the concentration of inactivator required to give the half-maximal rate of inactivation, the maximal rate constant for inactivation at a saturating concentration of the inactivator, and t½ were determined to be 2.8 μM, 0.07 min-1, and 26 min, respectively.
Fig. 1.
Time- and concentration-dependent inactivation of CYP2B6 by RU486. The inactivation of the EFC deethylation activity of CYP2B6 in the reconstituted system that was incubated with 0 (○), 2 (•), 3 (▿), 5 (▵), 10 (□), and 25 μM (▪) RU486 is shown. Aliquots were removed at the times indicated and assayed for residual activity as described under Materials and Methods. Inset, double reciprocal plots of the initial rates of inactivation as a function of the RU486 concentrations. The kinetic constants were determined from the double reciprocal plot. The data shown represent the average of three separate experiments done in duplicate that did not differ by more than 10%.
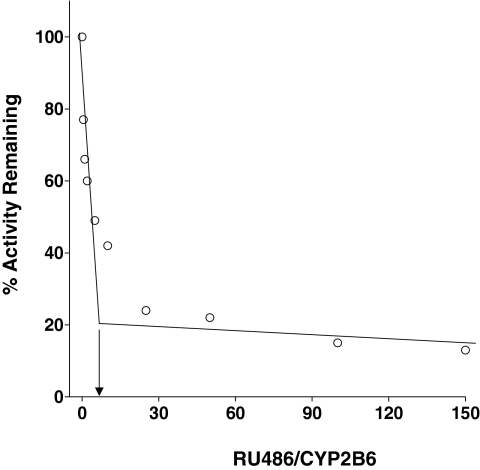
Partition Ratio. CYP2B6 was incubated with various concentrations of RU486, and the inactivation was allowed to progress for 1 h until the inactivation was essentially complete. The percentage of activity remaining was plotted as a function of the molar ratio of RU486 to CYP2B6 (Fig. 2). The partition ratio was estimated from the intercept of the linear regression line obtained from lower ratios of RU486 to CYP2B6, with the straight line derived from the higher ratios of RU486 to CYP2B6 as described previously (Silverman, 1996). With this method, a partition ratio of ∼5 was estimated.
Fig. 2.
Partition ratio determination for the inactivation of CYP2B6 by RU486. The percentage of CYP2B6 catalytic activity remaining was determined as a function of the molar ratio of RU486 to P450. Samples were incubated with various concentrations of RU486 for 1 h in the presence of NADPH until the inactivation reaction was essentially complete. The partition ratio was estimated from the intercept of the linear regression line from the lower ratios of RU486 to CYP2B6 and the straight line obtained from higher ratios of RU486 to CYP2B6.
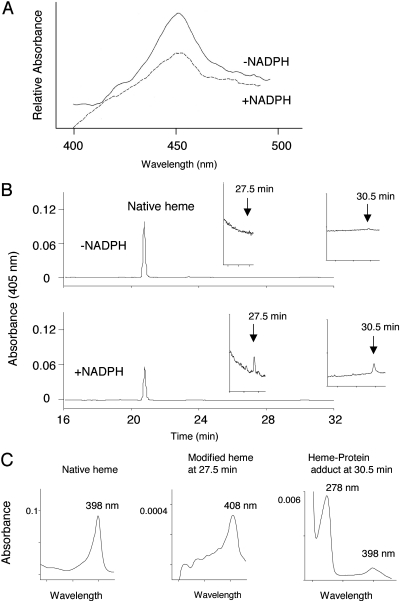
Loss of Native Heme during Inactivation. When CYP2B6 was incubated with 20 μM RU486 at 30°C for 15 min, the ∼60% loss in EFC catalytic activity was accompanied by a ∼61% loss in the spectrally detectable reduced CO spectrum for the inactivated samples compared with the control samples. Representative reduced CO difference spectra for the inactivated and control P450s are shown in Fig. 3A.
Fig. 3.
Effect of inactivation by RU486 on the reduced CO difference spectrum and prosthetic heme remaining. CYP2B6 was incubated with 20 μM RU486 at 30°C for 15 min in the absence (control) or presence of NADPH (inactivated) as described under Materials and Methods. A, reduced CO difference spectra for control and inactivated P450s. B, HPLC elution profiles monitored at 405 nm for the prosthetic heme. Insets, magnified views of the control and inactivated samples eluting from 24 to 32 min. C, absorption spectra of the native heme eluting at 21 min, the dissociable heme eluting at 27.5 min and the crosslinked heme and protein eluting at 30.5 min as analyzed using a diode-array detector.
The control and inactivated samples were also analyzed by HPLC. The column eluate was monitored at 405 nm for detection of the heme moiety. As shown in Fig. 3B, there was a ∼40% loss of native heme in the inactivated sample as measured by the area under the peak. Although the presence of a heme adduct is not obvious in the original elution profile, the expanded view of the profile from a retention time of 24 to 32 min revealed two modified heme peaks at 27.5 and 30.5 min in the NADPH-treated sample, but not in the control sample. The absorption spectra of the native heme and the modified heme are shown in Fig. 3C. The maximal absorbance of the native heme is at 398 nm. A Soret peak at 408 nm for the heme peaklet at 27.5 min indicates that it is a dissociable modified heme. Because the CYP2B6 apoprotein eluted at 30.5 min, the absorbance spectrum shown in Fig. 3C with peaks at 278 and 398 nm indicates that cross-linking of the native heme to the apoprotein has occurred. Under the same experimental conditions, the loss of the heme prosthetic group was also observed during the inactivation of CYP2B1 and CYP3A4 by RU486.
The removal of any unbound RU486 by extensive dialysis of the control and inactivated samples did not lead to any significant recovery of catalytic activity, the reduced CO difference spectrum, or the heme content as measured by HPLC compared with the values obtained for the samples before dialysis (data not shown). Thus, the inactivation of CYP2B6 by RU486 is irreversible. The addition of 10 mM GSH to the reaction mixture during the inactivation had no significant effect on the extent of inactivation (data not shown).
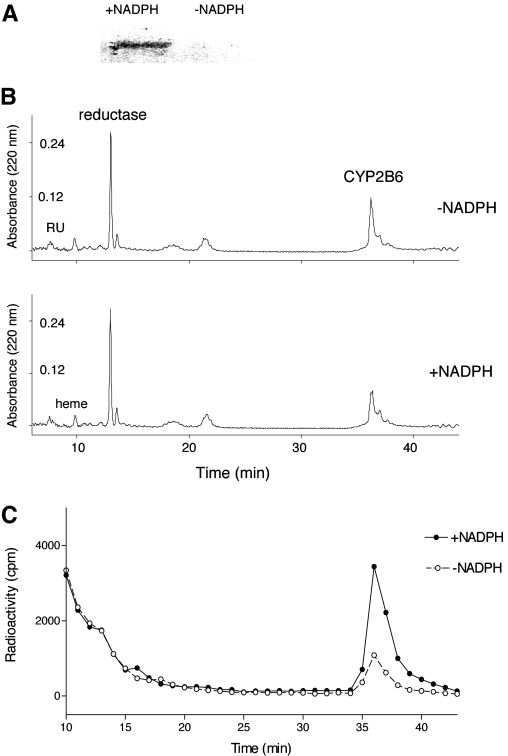
Covalent Binding of RU486 to CYP2B6 Apoprotein. The amount of RU486 that was irreversibly bound to the P450 2B6 protein was determined by precipitating the protein from the control and the RU486-inactivated samples that had been incubated with [3H]RU486 using acidified methanol as described previously (Chan et al., 1993). The stoichiometry for binding was ∼0.6, suggesting that approximately 0.6 mol RU486-reactive intermediate bound per mole of P450 inactivated. Two other approaches were used to investigate binding of RU486 to CYP2B6. After incubating the reaction mixtures with [3H]RU486, 50 pmol of the control and inactivated samples was subjected to SDS-PAGE analysis. As shown in Fig. 4A, significant radioactivity was associated with the CYP2B6 apoprotein in the +NADPH sample but not in the -NADPH sample. This result also demonstrates that the reactive intermediate of RU486 irreversibly binds to the P450 apoprotein. The control and inactivated samples were also analyzed using reverse phase HPLC. Figure 4B shows the elution profile monitored at 220 nm for protein. The peak areas for the reductase eluting at 14 min were similar between two conditions, whereas the peak for the P450 apoprotein eluting at ∼37 min under these conditions showed some change in the peak for the inactivated sample compared with the control sample. When the HPLC fractions were collected and analyzed by liquid scintillation counting, there was a marked increase in the radioactivity associated with the CYP2B6 apoprotein in the inactivated samples compared with the control sample (Fig. 4C). Although some radioactivity was observed in the control sample, the labeling associated with the protein eluting at ∼37 min in the NADPH-treated sample was 5-fold greater than that in the control sample. In this HPLC profile, RU486 and the P450 heme eluted at 7 and 10 min, respectively. The radioactivity eluting before 14 min observed in Fig. 4C was from trailing [3H]RU486, where the great majority of the counts eluted with the parent RU486 at 7 min (data not shown).
Fig. 4.
Covalent binding of RU486 to apoprotein. A, SDS-PAGE separation of CYP2B6 apoprotein incubated with [3H]RU486 in control (-NADPH) and inactivated (+NADPH) samples. B, HPLC elution profiles of the control (-NADPH) and inactivated (+NADPH) samples monitored for protein at 220 nm. C, liquid scintillation counting of the HPLC fractions from B. The experimental conditions were as described under Materials and Methods.
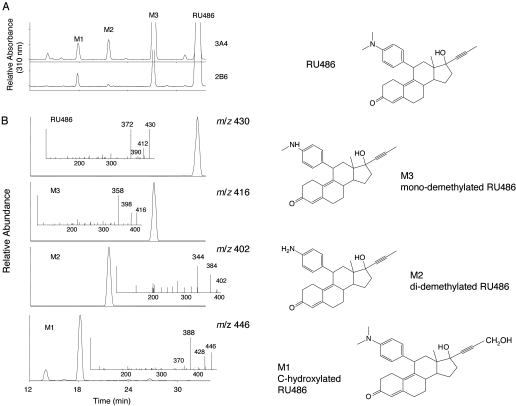
LC-MS and MS/MS Analysis of the Metabolites of RU486. The metabolism of RU486 by human liver microsomes and by the CYP3A4 and CYP3A5 reconstituted system has been reported previously (Heikinheimo et al., 1990; Jang et al., 1996; Khan et al., 2002). Based on these previous studies, the primary metabolites formed in our studies were identified by LC-MS/MS analysis. At least three major metabolites, M1, M2, and M3, were produced by CYP3A4 and CYP2B6 based on HPLC analysis with spectral monitoring in the UV at 310 nm (Fig. 5A). The metabolite profile for rat CYP2B1 is similar to that for human CYP2B6 (data not shown). It is interesting that M3 is the only metabolite formed by 3A5 (data not shown). Figure 5B shows the extracted ion chromatograms of CYP2B6 for the ions at m/z 446, 402, 416, and 430, which correspond to M1, M2, M3, and RU486. The MS/MS spectrum for each of these precursor ions is displayed in the inset. All of the major fragments are derived from the loss of the propynyl side chain, water, or the combination of the propynyl side chain and the water. RU486 elutes at ∼33 min, with a MH+ ion at m/z 430 and a major fragment ion at m/z 372. M3, eluting at ∼28 min with MH+ ion at m/z 416 and exhibiting a major fragment ion at m/z 358, is the mono-demethylated RU486. M2, eluting at ∼22 min with a MH+ ion at m/z 402 and a major fragment ion at m/z 344, is the di-demethylated RU486. M1, eluting at ∼18 min with a MH+ at m/z 446 and a major fragment ion at m/z 388, is a C-hydroxylated metabolite of RU486. The extracted ion chromatogram for the ions at m/z 446 shows that in addition to a major peak at 18 min, CYP2B6 also catalyzes the formation of another metabolite with MH+ ion at m/z 446, which eluted at ∼14 min. CYP3A4 leads to three minor peaks eluting at 14, 16, and 24 min (data not shown) that have precursor ions at m/z 446, indicating that they are also hydroxylated metabolites of RU486, possibly on the steroid moiety. The exact positions of the hydroxyl groups in these metabolites cannot be determined from the MS/MS data. The structures of RU486 and the three major metabolites formed by CYP2B6 and CYP3A4 are illustrated in Fig. 5 (right).
Fig. 5.
HPLC and ESI-LC-MS/MS analysis of the metabolites of RU486 formed by CYP3A4 and CYP2B6 in the reconstituted system. A, CYP3A4 and CYP2B6 were reconstituted with reductase and incubated with RU486 and NADPH. The metabolites were extracted and analyzed by HPLC as described under Materials and Methods. The three major metabolites are M1, M2, and M3. B, extracted ion chromatograms of M1 (m/z 446), M2 (m/z 402), M3 (m/z 416), and RU486 (m/z 430) from CYP2B6. The MS/MS spectra of RU486 and its metabolites are displayed in the insets. For each compound, cleavage of the propynyl group leads to the formation of the major fragment ion. Right, structures of RU486, M1, M2, and M3.
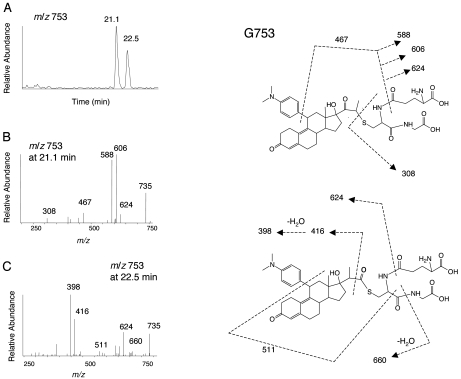
LC-ESI-MS/MS Analysis of the GSH Conjugates of RU486 Formed by P450s. After incubation of RU486 with the purified P450s and liver microsomes in the presence of NADPH and GSH, LC-MS/MS analysis revealed the formation of two metabolites with MH+ ions at m/z 753, which will be referred to as G753, one major metabolite with a MH+ ion at m/z 769, which will be referred to as G769, and one metabolite with a MH+ ion at m/z 751, which will be referred to as G751. Essentially similar GSH conjugates were observed from the incubations with purified P450s and for liver microsomes from rats pretreated with PB or DEX and human liver microsomes. Figure 6A shows the extracted ion chromatograms for the two G753 conjugates, eluting at 21.1 and 22.5 min. The masses of the two G753 peaks are equivalent to the sum of RU486 plus one oxygen and GSH. Figure 6, B and C, shows the MS/MS spectra of these two G753 conjugates. The G753 conjugates may result from the initial formation of an oxirene intermediate followed by Michael addition of GSH, leading to two products. In principle, we cannot assign definitive structures for these conjugates solely based on MS/MS spectra. However, the MS/MS fragmentation patterns for these two conjugates are very different, indicating that two different isomers are formed. On the basis of the heme destruction and covalent binding to the apoprotein during the inactivation of RU486 and the hypothesis that the mechanism-based inactivation of P450s by propynylaryl acetylenes proceeds via a 1,2-methyl shift (Foroozesh et al., 1997), two structures are tentatively proposed for G753, as shown in Fig. 6 (right). For the isomer eluting at 21.1 min, the fragment ion at m/z 308 is the protonated GSH moiety, and the fragment ion at m/z 735 is due to the loss of water from the parent ion. The fragment ions at m/z 624 and 606 are products of neutral loss of Glu (129 Da) from the precursor ion m/z 753 and the fragment ion at m/z 735, respectively. The loss of water from the fragment ion m/z 606 will form the fragment ion m/z 588, and further loss of the di-methylaminophenyl moiety (121 Da) will form the fragment ion at m/z 467. For the isomer eluting at 22.5 min, the fragment ion at m/z 660 is a product of neutral loss of Gly (75 Da) from the ion at m/z 735. The combination of the losses of CO (28 Da) and the di-methylaminophenyl moiety from the ion at m/z 660 will form the ion at m/z 511. The fragment ion at m/z 416 is likely to be from a cleavage between carbon-1 (the carbon closet to the steroid moiety) and carbon-2 (middle carbon) of the 17α-propynyl moiety, and the further loss of water will form the ion at m/z 398. Thus, the fragment ions in the MS/MS spectra of both isomers indicate that RU486 and GSH are components of the G753 conjugates.
Fig. 6.
LC-MS/MS analysis of the RU486-GSH conjugates G753 isolated from human liver microsomes. Incubation conditions and extraction/analysis procedures were as described under Materials and Methods. A, extracted ion chromatogram of the MH+ ions at m/z 753. B, MS/MS spectrum of G753 eluting at 21.1 min. C, MS/MS spectrum of G753 eluting at 22.5 min. The spectra were obtained in positive mode and analyzed using the eXcalibur software. Right, proposed structures. The sites of fragmentation are indicated by dashed lines and explained in the text.
The extracted ion chromatograms for G769 and G751, which are more hydrophilic than G753 and elute at 11.8 and 12.2 min, respectively, are shown in Fig. 7A. The molecular mass of G769 is equivalent to the sum of hydroxylated RU486 plus one oxygen and GSH. The steroid moiety of the hydroxylated RU486 may undergo oxidation to an epoxide intermediate at the 4,5-position followed by the addition of GSH. The further loss of water from G769 will yield G751. The MS/MS spectra of G769 and G751 are displayed in Fig. 7B. The possible structures of G769 and G751 are tentatively proposed and shown in Fig. 7C. For G769, fragment ions at m/z 694 and 640 are from the neutral loss of the Gly and Glu moieties of GSH, respectively. The loss of water from the ion at m/z 640 will form an ion with a m/z 622, and the further cleavage of the 11β-dimethylaminophenyl moiety will produce the ion with m/z 501. Cleavage of the C-S bond will form the ion with m/z 462, indicating that the C-hydroxylated RU486 is a component of G769. Further loss of water will form the ion with m/z 444. For G751, the fragment ions at m/z 676 and 622 are from the neutral loss of Gly and Glu, respectively. The loss of water from the ions at m/z 751 and 622 will form ions at m/z 733 and 604, respectively. Cleavage of the thioether bond within the GSH moiety will form the ion at m/z 478, and the further loss of water will form the ion at m/z 460. The fragment ion at m/z 444 is formed from the cleavage between the hydroxylated RU486 and the GSH moiety. Again, the fragment ions in these MS/MS spectra suggest that GSH and C-hydroxylated RU486 are components of the G769 and G751 conjugates.
Fig. 7.
LC-MS/MS analysis of the RU486-GSH conjugates G769 and G751 formed by liver microsomes from PB-treated rats. A, extracted ion chromatograms at m/z 769 and 751. B, MS/MS spectra of G769 and G751 eluting at 11.8 and 12.2 min, respectively. C, right, proposed structures of G769 and G751. Dashed lines, sites of fragmentation as described in the text.
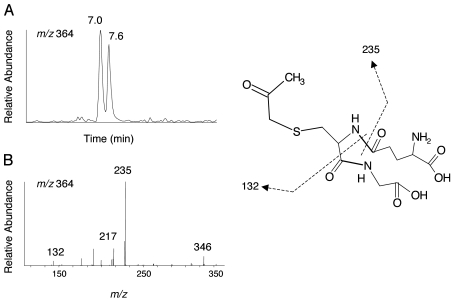
Cleavage Products of the Propynyl Moiety. Two metabolites with the MH+ ion at m/z 364 were formed by the purified P450s and mammalian liver microsomes after incubation with RU486. The masses of these two cleavage metabolites, referred to as G364, correspond to the sum of one oxygen, propynyl moiety, and GSH. Figure 8A shows the extracted ion chromatogram for the two G364 moieties, which elute at 7.0 and 7.6 min. Because the MS/MS spectra of the two G364 conjugates are very similar, only the spectrum for the peak at 7.0 min is shown in Fig. 8B. The fragment ion at m/z 235 would be produced by a neutral loss of 129 Da, and further losses of 75 Da and CO would form a fragment ion with m/z 132, indicating that G364 exhibits the features of a GSH adduct. Oxygenation at either carbon-1 or carbon-2 of the 17α-propynyl moiety will form two isomers of G364. The structure proposed for G364 is shown in Fig. 8 (right).
Fig. 8.
LC-MS/MS analysis of G364 formed by liver microsomes from DEX-treated rats. A, extracted ion chromatogram of the ions at m/z 364, eluting at 7.0 and 7.6 min. B, MS/MS spectrum of ion with m/z 364 eluting at 7.0 min. One possible structure is illustrated in the right. Dashed lines, sites of fragmentation.
Discussion
The inactivation of CYP2B6 by RU486 is concentration- and time-dependent. The inactivation is irreversible and requires the presence of NADPH. The apparent concentration of inactivator required to give the half-maximal rate of inactivation is 2.8 μM, its maximal rate constant for inactivation at a saturating concentration of the inactivator is 0.07 min-1, the t½ is 26 min, and the partition ratio is ∼5. The loss in the catalytic activity is associated with a decrease in the spectrally detectable P450-CO complex, heme loss, and labeling of the apoprotein. In addition, dissociable modified heme adduct and covalent cross-linking of the prosthetic heme to the apoprotein are observed.
Earlier studies have demonstrated that the mechanism of inactivation by both terminal and internal acetylenic compounds can be attributed to the initial oxygenation of the carbon-carbon triple bond. Some examples include: the inactivation of rat liver microsomes by several internal and terminal acetylenes (Ortiz de Montellano and Kunze, 1980; CaJacob et al., 1988; Foroozesh et al., 1997), the inactivation of P450scc by acetylenic steroids (Nagahisa et al., 1983; Olakanmi and Seybert, 1990), the inactivation of Vicia sativa microsomes by 11-dodecynoic acid (Helvig et al., 1997), the inactivation of CYP3A4 by RU486 (He et al., 1999), the inactivation of CYP1A1 by 1-ethynylpyrene and phenylacetylene (Chan et al., 1993), and the inactivation of CYP2B1, CYP2B6, CYP3A4, and CYP3A5 by 17α-ethynylestradiol (Kent et al., 2002; Lin and Hollenberg, 2007). The findings from these studies can be summarized as follows: 1) heme adducts can be detected with terminal acetylenes, but not with internal acetylenes; and 2) by using radiolabeled compounds, covalent modification of the apoprotein can be observed for both internal and terminal acetylenes. By analogy with the terminal acetylenic compounds, it has been proposed that internal acetylenes may be oxidized and converted via a 1,2-methyl shift to a ketene, which subsequently can react with a nucleophile in the active site of P450. However, except for biphenyl propyne, no acidic derivatives have been detected so far with any other internal acetylene (Foroozesh et al., 1997). When we analyzed CYP2B1, CYP2B6, and CYP3A4 that had been inactivated by RU486 using LC-MS, the signal/noise ratio of the deconvoluted mass spectrum was not good enough to allow for the mass determination of the modified apoprotein. However, another laboratory has reported a covalently adducted protein having a mass shift equivalent to one RU486 molecule per molecule of CYP3A4 protein inactivated by utilizing quadrupole time-of-flight mass spectrometry (Yukinaga et al., 2005). Thus, quadrupole time-of-flight mass spectrometry has provided direct confirmation of the formation of an internal acetylenic compound-protein adduct without the use of a radiolabeled compound.
Although the involvement of oxirene-reactive intermediates has been proposed previously for acetylenes, the formation of such reactive intermediates has not been detected yet with internal acetylenes. Halpert and coworkers have reported previously that RU486 selectively inactivates CYP3A4, but not CYP3A5 (Khan et al., 2002). We have taken advantage of this observation to try to identify the reactive intermediate responsible for inactivation. When the GSH conjugates of RU486 that were trapped and identified in the CYP3A5-reconstituted system containing NADPH and GSH were compared with CYP3A4, it was found that G769 and G751 were detected, but G753 was not (data not shown). The fact that the G753 conjugates could only be detected during metabolism by P450s such as CYP2B1, CYP2B6, CYP3A4, and liver microsomes from humans and PB- or DEX-induced rats, all of which show inactivation by RU486, has led us to suggest that the reactive intermediates responsible for forming the G753 conjugates, but not the G769 and G751 conjugates, may play a key role in the inactivation. Although the structure of the reactive intermediate has not been definitely determined yet, the detection of the two G753 GSH conjugates with different MS/MS fragmentation patterns (Fig. 6) suggests that bioactivation most probably involves the formation of an oxirene-reactive intermediate of the 17α-propynyl moiety followed by the addition of GSH, with the oxygen being transferred onto either the carbon-1 or carbon-2. The reactive species that reacts with GSH also may react with either the heme or the protein and thereby contribute to the inactivation.
Previous studies have suggested that binding of GSH to the internal carbon of the reactive oxirene would enhance cleavage of the C-C bond between the steroid and 17α-ethynyl moiety, leading to loss of the ethynyl group (Helton et al., 1977). After incubation of RU486 with the purified P450s in the reconstituted system or with mammalian liver microsomes in the presence of GSH, two G364 metabolites resulting from the cleavage of the C-C bond after binding of GSH to the oxygenated propynyl moiety were detected. To obtain two different structures for the G753 conjugates with GSH binding to the carbon-2 of propynyl moiety, the first metabolite may be formed with oxygen inserted at the carbon-1, and the second metabolite may be formed with oxygen being inserted at the carbon-2, as shown in Fig. 6. The second metabolite is formed via a 1,2-methyl rearrangement analogous to the mechanism of suicide inhibition by ethynyl acetylenes that proceed via a ketene intermediate formed by a 1,2-hydrogen shift (Ortiz de Montellano and Kunze, 1981; Foroozesh et al., 1997). Thus, by using LC-MS/MS analysis, the detection of two G364 and two G753 conjugates from the bioactivation pathway of RU486 in P450s supports the hypotheses proposed by Helton et al. (1977), Ortiz de Montellano and Kunze (1981), and Foroozesh et al. (1997).
In addition to G753, there are two more hydrophilic GSH conjugates of RU486, G769, and G751. The masses of G769 and G751 and the MS/MS spectra suggest that C-hydroxylated RU486 is a component of G769. We postulate that the epoxidation may occur at the double bond joining the 4,5-carbons in the A ring. Subsequent nucleophilic attack of GSH on the epoxide intermediate would generate G769, and subsequent loss of water would form G751. This reaction sequence has been documented previously (Guengerich, 2003; Tang and Miller, 2004; Chen et al., 2006).
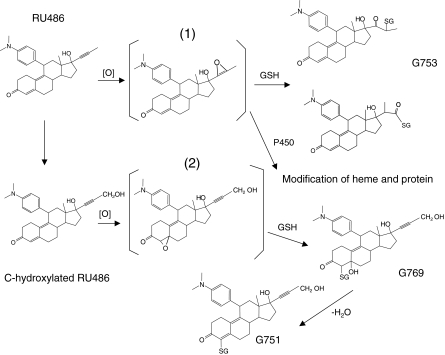
Previous studies on acetylenic steroids that structurally resemble RU486 have led to the suggestion that an oxirene may be the reactive intermediate responsible for the inactivation of P450scc (Nagahisa et al., 1983; Olakanmi and Seybert, 1990). Recently, we have identified two GSH conjugates of 17α-ethynylestradiol during the inactivation of P450s where the structure appears to result from the insertion of oxygen to either one of the acetylenic carbons followed by the addition of GSH into the carbon-carbon triple bond (Lin and Hollenberg, 2007). The results presented here suggest that the mechanism responsible for the inactivation by RU486 may follow the same route as previously reported for 17α-ethynylestradiol. A proposed scheme describing the metabolic activation of RU486 by P450s is shown in Fig. 9. However, we cannot rule out the formation of an oxirene species followed by rearrangement to a ketene intermediate (Ortiz de Montellano and Kunze, 1981). This scheme suggests that the same reactive intermediate that is responsible for forming G753, but not G769 and G751, may be involved in the mechanism of inactivation. However, the reactive intermediate responsible for forming G769 and G751 from the bioactivation of RU486 by P450s may also covalently modify proteins and cause toxicity or affect cellular functions.
Fig. 9.
Proposed scheme for the metabolic bioactivation of RU486 by P450s leading to heme and apoprotein modification and four GSH conjugates: 1) 17α-oxirene related reactive species and 2) the 4,5-epoxide reactive species.
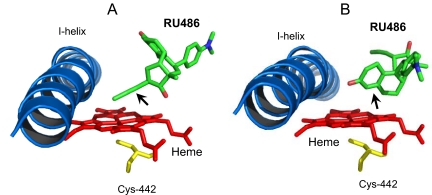
In the attempt to better understand the binding of RU486 in the active site of CYP3A4 and to identify potential sites for bioactivation, docking of RU486 to CYP3A4 was performed. The results of these modeling studies are shown in Fig. 10. The lowest energy of conformers with the 17α-propynyl moiety of RU486 pointing to pyrrole ring C of the heme is displayed in Fig. 10A, and the second lowest energy of the conformer with the 4,5-carbon positions facing the heme plane is displayed in Fig. 10B. The modeling studies suggest that bioactivation might occur at the carbon-carbon triple bond of the 17α-position and at the double bond of 4,5-positions. Thus, the reactive intermediates leading to the formation of the GSH conjugates proposed in Figs. 6 and 7 are in the agreement with the docking and modeling studies for the binding of RU486 to CYP3A4.
Fig. 10.
RU486 is docked in the active site of CYP3A4. The figures show RU486, I-helix, residue Cys442, and the heme. A, in the lowest energy conformation, the 17α-propynyl moiety (indicated by arrow) points to the pyrrole ring C of the heme. B, in the second lowest energy conformation, the 4,5-carbon double bond (indicated by arrow) faces the heme plane.
In conclusion, we have demonstrated that RU486 is a very efficient mechanism-based inactivator of CYP2B6 and that the RU486-dependent inactivation of CYP2B6 occurs by two mechanisms: heme destruction and covalent binding of a reactive intermediate of RU486 to the apoprotein. It is most likely that oxygenation of the carbon-carbon triple bond of the propynyl group leads to the formation of the same reactive intermediates that have been trapped with GSH to form the two G753 conjugates may contribute to the mechanism of inactivation. Thus, the potential for drug-drug interactions after long-term use of RU486 may not be limited to substrates for CYP3A4 alone and should also be evaluated for drugs that are metabolized primarily by CYP2B6. Because CYP2B6 exhibits genetic polymorphisms and has a wide tissue distribution, the involvement of CYP2B6 in the metabolism of RU486 and its inactivation by RU486 could have important implications for adverse effects with a variety of drugs that are metabolized primarily by CYP2B6, including the antidepressant bupropion and the human immunodeficiency virus protease inhibitor efavirenz.
Acknowledgments
We thank Drs. Ute M. Kent and Kathleen Noon for suggestions and advice for the LC-MS/MS analysis. We give special thanks to Dr. Abdul Mutlib (Wyeth, Collegeville, PA) for help with the LC-MS/MS analysis and informative discussions.
This work was supported by the National Institutes Health National Cancer Institute [Grant CA16954].
doi:10.1124/jpet.108.148536.
ABBREVIATIONS: RU486, mifepristone, 17β-hydroxy-11β-(4-dimethylaminophenyl)-17α-(1-propynyl)-estra-4,9-dien-3-one; P450, cytochrome P450; HPLC, high-pressure liquid chromatography; GSH, glutathione; LC, liquid chromatography; MS/MS, tandem mass spectrometry; PB, phenobarbital; DEX, dexamethasone; EFC, 7-ethoxy-4-(trifluoromethyl)-coumarin; TFA, trifluoroacetic acid; PAGE, polyacrylamide gel electrophoresis; ESI, electrospray ionization.
References
- Baillie TA and Davis MR (1993) Mass spectrometry in the analysis of glutathione conjugates. Biol Mass Spectrom 22 319-325. [DOI] [PubMed] [Google Scholar]
- Cadepond F, Ulmann A, and Baulieu EE (1997) RU486 (Mifepristone): mechanisms of action and clinical uses. Annu Rev Med 48 129-156. [DOI] [PubMed] [Google Scholar]
- CaJacob CA, Chan WK, Shephard E, and Ortiz de Montellano PR (1988) The catalytic site of rat hepatic lauric ω-hydroxylase: protein versus prosthetic heme alkylation in the ω-hydroxylation of acetylenic fatty acids. J Biol Chem 263 18640-18649. [PubMed] [Google Scholar]
- Chan WK, Sui Z, and Ortiz de Montellano PR (1993) Determinants of protein modification versus heme alkylation: inactivation of cytochrome P450 1A1 by 1-ethynylpyrene and phenylacetylene. Chem Res Toxicol 6 38-45. [DOI] [PubMed] [Google Scholar]
- Chasserot-Golaz S and Beck G (1992) How the potency of the steroid RU486 is related to P450 activities induced by dexamethasone and phenobarbital in rat hepatoma cells. J Steroid Biochem Mol Biol 41 653-657. [DOI] [PubMed] [Google Scholar]
- Chen Q, Doss GA, Tung EC, Liu W, Tang YS, Braun MP, Didolkar V, Strauss JR, Wang RW, Stearns RA, et al. (2006) Evidence for the bioactivation of zomepirac and tolmetin by an oxidative pathway: identification of glutathione adducts in vitro in human liver microsomes and in vivo in rats. Drug Metab Dispos 34 145-151. [DOI] [PubMed] [Google Scholar]
- Coon MJ, van der Hoeven TA, Dahl SB, and Haugen DA (1978) Two forms of liver microsomal cytochrome P450, P450 LM2 and P450 LM4 (rabbit liver). Methods Enzymol 52 109-117. [DOI] [PubMed] [Google Scholar]
- Faucette SR, Hawke RL, Lecluyse EL, Shord SS, Yan B, Laethem RM, and Lindley CM (2000) Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28 1222-1230. [PubMed] [Google Scholar]
- Foroozesh M, Primrose G, Guo Z, Bell LC, Alworth WL, and Guengerich FP (1997) Aryl acetylenes as mechanism-based inhibitors of cytochrome P450-dependent monooxygenase enzymes. Chem Res Toxicol 10 91-102. [DOI] [PubMed] [Google Scholar]
- Gerber JG, Rhodes RJ, and Gal J (2004) Stereoselective metabolism of methadone N-demethylation by cytochrome P450 2B6 and 2C19. Chirality 16 36-44. [DOI] [PubMed] [Google Scholar]
- Gervot L, Rochat B, Gautier JC, Bohnenstengel F, Kroemer H, de Berardinis V, Martin H, Beaune P, and de Waziers I (1999) Human CPY2B6: expression, inducibility and catalytic activities. Pharmacogenetics 9 295-306. [PubMed] [Google Scholar]
- Guengerich FP (1994) Analysis and characterization of enzymes, in Principles and Methods of Toxicology (Hayes AW ed), Raven Press, New York, pp 1259-1313.
- Guengerich FP (2003) Cytochrome P450 oxidations in the generation of reactive electrophiles: epoxidation and related reactions. Arch Biochem Biophys 409 59-71. [DOI] [PubMed] [Google Scholar]
- He K, Woolf TF, and Hollenberg PF (1999) Mechanism-based inactivation of cytochrome P450 3A4 by mifepristone (RU486). J Pharmacol Exp Ther 288 791-797. [PubMed] [Google Scholar]
- Heikinheimo O, Ylikorkala O, Turpeinen U, and Lähteenmäki P (1990) Pharmacokinetics of the antiprogesterone RU486: no correlation to clinical performance of RU486. Acta Endocrinol 123 298-304. [DOI] [PubMed] [Google Scholar]
- Helton ED, Williams MC, and Goldziecher JW (1977) Oxidative metabolism and de-ethynylation of 17α-ethynylestradiol by baboon liver microsomes. Steroids 30 71-83. [DOI] [PubMed] [Google Scholar]
- Helvig C, Alayrac C, Mioskowski C, Koop D, Poullain D, Durst F, and Salaün JP (1997) Suicide inactivation of cytochrome P450 midchain and terminal acetylenes: a mechanistic study of inactivation of a plant lauric acid ω-hydroxylase. J Biol Chem 272 414-421. [DOI] [PubMed] [Google Scholar]
- Huang Z, Roy P, and Waxman DJ (2000) Role of human microsomal CYP3A4 and CYP2B6 in catalyzing N-dechloroethylation of cyclophosphamide and ifosfamide. Biochem Pharmacol 59 961-972. [DOI] [PubMed] [Google Scholar]
- Jang GR and Benet LZ (1998) Antiprogestin-mediated inactivation of cytochrome P450 3A4. Pharmacology 56 150-157. [DOI] [PubMed] [Google Scholar]
- Jang GR, Wrighton SA, and Benet LZ (1996) Identification of CYP3A4 as the principal enzyme catalyzing mifepristone (RU 486) oxidation in human liver microsomes. Biochem Pharmacol 52 753-761. [DOI] [PubMed] [Google Scholar]
- Khan KK, He YQ, Correia MA, and Halpert JR (2002) Differential oxidation of mifepristone by cytochromes P450 3A4 and 3A5: selective inactivation of P450 3A4. Drug Metab Dispos 30 985-990. [DOI] [PubMed] [Google Scholar]
- Kent UM, Mills DE, Rajnarayanan RV, Alworth WL, and Hollenberg PF (2002) Effect of 17-α-ethynylestradiol on activities of cytochrome P450 2B (P4502B) enzymes: characterization of inactivation of P4560s 2B1 and 2B6 and identification of metabolites. J Pharmacol Exp Ther 300 549-558. [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, and Zanger UM (2001) Extensive genetic polymorphism in human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11 399-415. [DOI] [PubMed] [Google Scholar]
- Lin HL and Hollenberg PF (2007) The inactivation of cytochrome P450 3A5 by 17α-ethynylestradiol is cytochrome b5 dependent: metabolic activation of ethynyl moiety for the formation of glutathione, heme adducts and covalent binding to apoprotein. J Pharmacol Exp Ther 321 276-287. [DOI] [PubMed] [Google Scholar]
- Lin HL, Kent UM, and Hollenberg PF (2005) The grapefruit juice effect is not limited to cytochrome P450 (P450) 3A4: evidence for bergamottin-dependent inactivation, heme destruction, and covalent binding to protein in P450s 2B6 and 3A5. J Pharmacol Exp Ther 313 154-164. [DOI] [PubMed] [Google Scholar]
- Lin HL, Myshkin E, Waskell L, and Hollenberg PF (2007) Peroxynitrite inactivation of human cytochrome P450s 2B6 and 2E1: heme modification and site-specific nitrotyrosine formation. Chem Res Toxicol 20 1612-1622. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Huey R, and Olson AJ (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des 10 293-304. [DOI] [PubMed] [Google Scholar]
- Nagahisa A, Spencer RW, and Orme-Johnson WH (1983) Acetylenic mechanism-based inhibitors of cholesterol side chain cleavage by cytochrome P450scc. J Biol Chem 258 6721-6723. [PubMed] [Google Scholar]
- Olakanmi O and Seybert DW (1990) Modified acetylenic steroids as potent mechanism-based inhibitors of cytochrome P450scc. J Steroid Biochem 36 273-280. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR and Kunze KL (1980) Self-catalyzed inactivation of hepatic cytochrome P450 by ethynyl substrates. J Biol Chem 255 5578-5585. [PubMed] [Google Scholar]
- Ortiz de Montellano PR and Kunze KL (1981) Shift of the acetylenic hydrogen during chemical and enzymatic oxidation of the biphenylacetylene triple bond. Arch Biochem Biophys 209 710-712. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR and Komives EA (1985) Branchpoint for heme alkylation and metabolite formation in the oxidation of arylacetylenes by cytochrome P450. J Biol Chem 260 3330-3336. [PubMed] [Google Scholar]
- Reilly PE, Gomi RJ, and Mason SR (1999) Mechanism-based inhibition of rat liver microsomal diazepam C3-hydroxylase by mifepristone associated with loss of spectrally detectable cytochrome P450. Chem Biol Interact 118 39-49. [DOI] [PubMed] [Google Scholar]
- Scott EE, Spatzenegger M, and Halpert JR (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility and decreased aggregation while retaining function. Arch Biochem Biophys 395 57-68. [DOI] [PubMed] [Google Scholar]
- Silverman RB (1996) Mechanism-based enzyme inactivation, in Contemporary Enzyme Kinetics and Mechanisms (Purich DL ed) pp 291-335, Academic Press, San Diego, CA.
- Tang W and Miller RR (2004) Thiol conjugation in in vitro drug metabolism. in Methods in Pharmacology and Toxicology, Optimization in Drug Discovery: In vitro Methods (Yan Z and Cladwell GW eds) pp 369-383, Humana Press Inc., Totowa, NJ.
- Teiber JF and Hollenberg PF (2000) Identification of the human liver microsomal P450s involved in the metabolism of N-nitrosodi-n-propylamine. Carcinogenesis 21 1559-1566. [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, and Desta Z (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306 287-300. [DOI] [PubMed] [Google Scholar]
- Yukinaga H, Murayama N, Okzaki O, and Sudo K (2005) Structural analysis of covalent adducts to cytochrome P450 utilizing quadrupole time-of-flight mass spectrometry. Drug Metab Rev 37 158-159. [Google Scholar]