Abstract
The organic cation/ergothioneine transporter OCTN1 (SLC22A4) and the high-affinity carnitine transporter OCTN2 (SLC22A5), play an important role in the disposition of xenobiotics and endogenous compounds. Here, we analyzed the sequence of the proximal promoter regions of OCTN1 and OCTN2 in four ethnic groups and determined the effects of the identified genetic variants on transcriptional activities and mRNA expression. Six variants were found in the proximal promoter of OCTN1, one of which showed high allele frequency ranging from 13 to 34% in samples from individuals with ancestries in Africa, Europe, China, and Mexico. OCTN1 haplotypes had similar activities as the reference in luciferase reporter assays. For OCTN2, three of the seven variants identified in the proximal promoter showed allele frequencies greater than 29.5% in all populations, with the exception of -207C>G (rs2631367) that was monomorphic in Asian Americans. OCTN2 haplotypes containing -207G, present in all populations, were associated with a gain of function in luciferase reporter assays. Consistent with reporter assays, OCTN2 mRNA expression levels in lymphoblastoid cell lines (LCLs) from gene expression analysis were greater in samples carrying a marker for -207G. This SNP seems to contribute to racial differences in OCTN2 mRNA expression levels in LCLs. Our study with healthy subjects (n = 16) homozygous for either -207C or -207G, showed no appreciable effect of this SNP on carnitine disposition. However, there were significant effects of gender on carnitine plasma levels (p < 0.01). Further in vivo studies of OCTN2 promoter variants on carnitine disposition and variation in drug response are warranted.
Many genes that influence drug disposition are known to have polymorphisms that may play a critical role in determining interindividual variability in pharmacokinetics (Eichelbaum et al., 2006; Giacomini et al., 2007; Cropp et al., 2008). For example, recent studies have demonstrated that nonsynonymous SNPs in genes encoding membrane transporters such as organic anion/cation transporters contribute to interindividual variation in pharmacokinetics and drug response (Ieiri et al., 2006; Urban et al., 2007; Shu et al., 2008; Sissung et al., 2008). SNPs in noncoding regulatory regions have also been found to contribute to interindividual variation in the pharmacokinetics of various drugs (de Jong et al., 2006; Poonkuzhali et al., 2008; Wang et al., 2008).
The present study focused on the proximal promoter region of the human organic cation/carnitine transporters OCTN1 (SLC22A4) and OCTN2 (SLC22A5), members of the solute carrier 22 family (SLC22A), which includes multispecific transporters of organic cations, anions, and zwitterions. OCTN1 is strongly expressed in kidney, skeletal muscle, bone marrow, and trachea and weakly expressed in many other organs (Bleasby et al., 2006), whereas OCTN2 is expressed ubiquitously with high expression in the kidney and considerable expression in skeletal muscle, placenta, testis, prostate, small intestine, and heart (Nishimura and Naito, 2005). In the kidney, OCTN1 and OCTN2 have a bifunctional role, transporting selected xenobiotics in the secretory direction into the tubular lumen and particular endogenous compounds such as l-carnitine in the reabsorptive direction (Ohashi et al., 2001; Koepsell and Endou, 2004).
Genetic variants of OCTN1 and OCTN2 have been associated with human disease. For example, systemic carnitine deficiency (SCD), marked by cardiac and skeletal myopathy, hypoketotic hypoglycemia, hyperammonemia, encephalopathy, and, in some cases, acute liver failure (Lahjouji et al., 2001; Tein, 2003), is caused by mutations in the coding region of OCTN2. Mice carrying a mutation in the mouse Octn2 exhibit a phenotype similar to human SCD (Kuwajima et al., 1998) and also exhibit altered drug disposition (Ohashi et al., 2001). In addition, an intronic SNP of OCTN1 has been associated with rheumatoid arthritis in Japanese (Tokuhiro et al., 2003), and a nonsynonymous OCTN1 SNP (OCTN1-L503F) showing strong linkage disequilibrium with an SNP in the promoter region of OCTN2 (-207G>C) is associated with Crohn's disease (Peltekova et al., 2004). However, a causal role of OCTN1 and OCTN2 for this polymorphism in the pathogenesis of rheumatoid arthritis and Crohn's disease remains controversial (Silverberg et al., 2007).
Because of the critical role of OCTN1 and OCTN2 in human disease and drug response, a study of common polymorphisms in these two genes is warranted. In this study, we analyzed genetic variation in the proximal promoter region of OCTN1 and OCTN2 in a large ethnically diverse sample of healthy volunteers (African-, European-, Asian- and Mexican-American, n = 272). We examined the functional effects of variants in the promoters using luciferase reporter assays. We further studied the relationship between expression level of OCTN2 and a common promoter variant by associating mRNA expression levels of OCTN2 retrieved from the Gene Expression Omnibus (GEO) database, with a variant in strong linkage disequilibrium with the common promoter variant of OCTN2. Finally, we determined the effect of a common variant in the proximal promoter of OCTN2 on carnitine disposition in a small sample of healthy volunteers. Our data suggest that an SNP in the promoter region of OCTN2 may explain interindividual differences in the expression level of this transporter in lymphoblastoid cell lines but that further studies are needed to demonstrate this effect in vivo.
Materials and Methods
Materials. Custom oligonucleotides for cloning of OCTN1 and OCTN2 proximal promoters were synthesized by Integrated DNA Technologies (Coralville, IA). PfuUltra high-fidelity DNA polymerase was purchased from Stratagene (La Jolla, CA). HindIII, NheI, and DpnI restriction enzymes were purchased from New England Biolabs (Danvers, MA). Lipofectamine LTX, Opti-MEM I reduced serum medium, Taq polymerase, MAX Efficiency DH5Alpha competent cells, and the TA cloning kit were purchased from Invitrogen (Carlsbad, CA). The luciferase reporter vectors pGL4.10[luc2] and pGL4.74[hRluc/TK] and the Dual-Luciferase Reporter Assay system were purchased from Promega (Madison, WI). Plasmid Mini kit was supplied by QIAGEN (Valencia, CA). Cell culture supplies were purchased from the Cell Culture Facility (University of California San Francisco, San Francisco, CA). ACHN, human renal cell adenocarcinoma cell line, was purchased from American Type Culture Collection (Manassas, VA). The human hepatoblastoma cell line HepG2 and the human colorectal carcinoma cell line HCT-116 were supplied by the Cell Culture Facility (University of California San Francisco).
Identification of OCTN1 and OCTN2 Promoter Variants. Genomic DNA samples were collected from unrelated healthy persons in the San Francisco Bay Area as part of the Studies of Pharmacogenetics in Ethnically Diverse Populations (SOPHIE) project. OCTN1 and OCTN2 promoter variants were identified by direct sequencing of genomic DNA from an ethnically diverse population of 272 individuals: 68 African-Americans, 68 European-Americans, 68 Chinese-Americans, and 68 Mexican-Americans. Primers for genotyping were designed to cover chromosome 5, ranging from the genomic position 131657365 to 131658198 (833 bp) for OCTN1 and from 131732980 to 131733464 (484 bp) for OCTN2. Primer sequences for cloning of proximal promoter region were 5′-CCTGTTTTCCCTGTGCAAGATGAGG-3′ (sense) and 5′-GCCGCTCCGAAACTTGCAACTAC-3′ (antisense) for OCTN1 and 5′-GACCCTGGGCCAGTGACTTTCT-3′ (sense) and 5′-CAGCAGGCGACCCAAGACC-3′ (antisense) for OCTN2. Materials and methods for sequencing were carried out as described previously by our group (Leabman et al., 2003).
Cloning of Promoter Region in Reporter Constructs. The OCTN1 and OCTN2 genomic fragments were -247 to +87 (334 bp) and -253 to +89 (342 bp) of the translational start site, respectively. These were amplified from HepG2 genomic DNA. Cooper et al. (2006) have demonstrated that functional proximal promoter regions of randomly selected subset of 45 human promoters were, on average, the sequence -300 to -50 bp of the transcriptional starting site using deletion analyses. The amplification product was digested with the restriction enzymes HindIII and NheI and subsequently cloned into these same sites within the multiple cloning site of the pGL4.10[luc2] vector (Promega). Site-directed mutagenesis was performed to obtain the variant constructs by using PfuUltra high-fidelity DNA polymerase (Stratagene). After mutagenesis, the complete OCTN1 and OCTN2 inserts were sequenced using specific forward primers for pGL4.10[luc2] vector to verify the sequence of the clone. The mutagenized amplicon was excised from the pGL4.10[luc2] vector using the restriction enzymes NheI and HindIII and recloned into these same sites within the pGL4.10[luc2] vector.
Cell Culture, Transfection, and Luciferase Reporter Assay. HepG2, HCT-116, and ACHN cells were cultured in Dulbecco's modified Eagle's medium and Dulbecco's modified Eagle's medium-Ham's F-12 50:50 mixture supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin and maintained in a humidified incubator at 37°C in an atmosphere of 5% CO2. Cultures were never allowed to grow beyond 80% confluence. For transfection studies, cells were seeded into 96-well culture dishes, and 24 h later they were transfected with 0.25 μl/well Lipofectamine LTX, 0.1 μl/well of Plus, 100 ng of pGL4.10[luc2] vector with or without the promoter region constructs, and 2.5 ng each of pGL4.74[hRluc/TK] vector in Opti-MEM I reduced serum medium. pGL4.74[hRluc/TK] vector was cotransfected with pGL4.10[luc2] vector to control for transfection efficiency. After incubation for 6 h at 37°C, transfection medium was replaced with normal culture medium, and cells were incubated for an additional 18 h. Cell lysates were prepared from each transfection and were assayed for luciferase and Renilla activities in a 96-well plate luminometer (Tecan, Durham, NC) according to the protocol in the Dual-Luciferase Reporter Assay system kit (Promega). Luciferase to Renilla ratios were determined and expressed as relative luciferase activity. Data are reported as the mean ± S.D. of at least three determinations performed with two or three independent experiments.
Association of OCTN2 mRNA Expression with OCTN1 Genotype in Subjects of Diverse Ethnicity. The OCTN1 1507T>C (rs1050152) genotype from the four different ethnic groups—1) CEPH, Utah residents with ancestry from northern and western Europe (CEU); 2) Yoruba in Ibadan, Nigeria (YRI); 3) Han Chinese (CHB); and 4) Japanese (JPT) descent—was retrieved from the HapMap database (www.hapmap.org; HapMap Data Release 23a/phase II March 2008). The OCTN2 mRNA expression level of the individual lymphoblastoid cell lines was obtained from the GEO database (accession numbers GSE5859 and GSE7761). The gene expression values (log2) reported in these data sets are based on two different gene expression platforms, namely, the Affymetrix Human Genome Focus Array and the Affymetrix GeneChip Human Exon 1.0ST Array.
Association of an OCTN2 Promoter Variant with Carnitine Disposition in Unrelated Healthy Individuals. Individual subjects in the SOPHIE cohort were screened by direct sequencing of 5′-untranslated region of OCTN2 to identify individuals homozygous for -207G/G or -207C/C (Urban et al., 2006). Because this polymorphism had previously only been described in individuals of European ancestry, this screen was restricted to the European American subset of SOPHIE. Subjects homozygous for -207G/G or -207C/C were recruited into a clinical study to evaluate the influence of -207G/G and -207C/C in the OCTN2 promoter on plasma carnitine levels and carnitine clearance in healthy subjects. Subjects in the study group were between the ages of 18 and 40 years and were selected as healthy by medical history questionnaire and screening blood work (complete blood count and comprehensive metabolic panel). Subjects were taking no regular medications and had normal renal function. Subjects were excluded from participation if they were pregnant, had a new history indicating they are no longer healthy, were taking a medication that could confound study results, or did not consent to participate in the study. Individuals with anemia (hemoglobin, <12 g/dl), an elevation in liver enzymes (alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, and γ-glutamyl-transferase) to higher than double the respective normal value, or elevated creatinine concentrations (men, ≥1.5 mg/dl; women, ≥1.4 mg/dl) were excluded.
In all subjects, plasma from each individual was analyzed for a panel of biochemical markers, including carnitine, acylcarnitines, and creatinine at 0 and 2 h using standard assays. Carnitine concentrations (including carnitine and acylcarnitines) in a 2-h urine collection were determined. Renal clearance of carnitine was estimated by dividing the urinary excretion rate by the average plasma concentration (CLr = Ae,0–2 h/(2 h × (Cp, t = 0 h + Cp, t = 2 h)/2). Differences between subject groups were determined using Student's t test, with p < 0.05 as the threshold for significance. Multiple regression was performed to test for independent effects of the various genetic and demographic measures recorded. Informed consent was obtained from each participant of the study, and the study design was approved by the University of California San Francisco Committee on Human Research.
Sequence and Data Analysis. DNA sequence data were analyzed using Finch TV software (Geospiza, Seattle, WA). Sequences were scanned for potential transcription factor recognition sequences using the Matinspector software (Genomatix, Ann Arbor, MI) and TFSearch (http://mbs.cbrc.jp/papia/papiaJ). Based upon the observed frequencies of each SNP, haplotype was estimated using the PHASE algorithm (Stephens and Donnelly, 2003). Functional differences between reference and variant haplotypes were evaluated using Dunnett's multiple comparison test. A p < 0.05 was considered significant.
Results
Identification of Genetic Variants in the Proximal Promoter Regions of OCTN1 and OCTN2. The variants identified in the region between -415 and -85 bp and between -594 and -224 bp upstream of the translational start sites of OCTN1 and OCTN2, respectively, are described in Tables 1 and 2. In total, six variant sites were identified in the proximal promoter of OCTN1 with an allelic frequency ranging from 0.7 to 33.8%. One of the variants, -248C (rs460271), had a high allele frequency (>13%) across all ethnic groups. In contrast, the other five variants in the OCTN1 proximal promoter had low allele frequencies and have not been reported previously. Four of the five new variants in the OCTN1 promoter were singletons, found on only one chromosome (-337insCGGG, -286T, -215G, and -184G).
TABLE 1.
Identity and frequency of SNPs in the proximal promoter region of SLC22A4 (OCTN1)
| Position from Translational Starting Site | Genomic Position | Nucleotide Change | Frequency in AA, n = 136 | Frequency in EA, n = 136 | Frequency in AS, n = 136 | Frequency in ME, n = 134a | rs ID |
|---|---|---|---|---|---|---|---|
| –337 | 131657872 | C → CGGGC | 0.007 | 0b | 0 | 0 | |
| –286 | 131657923 | A → T | 0.007 | 0 | 0 | 0 | |
| –256-1 | 131657953 | C → T | 0 | 0.007 | 0 | 0 | |
| –256-2 | 131657953 | C → G | 0 | 0 | 0.015 | 0 | |
| –248 | 131657961 | G → C | 0.132 | 0.338 | 0.324 | 0.306 | rs460271 |
| –215 | 131657994 | A → G | 0.007 | 0 | 0 | 0 | |
| –184 | 131658025 | A → G | 0 | 0.007 | 0 | 0 |
AA, African-American; ME, Mexican-American; AS, Asian-American; EA, European-American
Two of the samples have no data
A “0” implies that the SNP was not detected in a particular ethnic group
TABLE 2.
Identity and frequency of SNPs in the proximal promoter region of SLC22A5 (OCTN2)
| Position from Translational Starting Site | Genomic Position | Nucleotide Change | Frequency in AA, n = 136 | Frequency in EA, n = 132a | Frequency in AS, n = 128a | Frequency in ME, n = 112a | rs ID |
|---|---|---|---|---|---|---|---|
| –446 | 131733118 | C → T | 0b | 0 | 0.031 | 0 | rs4646298 |
| –399 | 131733165 | G → C | 0.316 | 0.356 | 0.438 | 0.304 | rs2631369 |
| –392 | 131733172 | C → T | 0 | 0 | 0 | 0.009 | rs57961304 |
| –368 | 131733196 | T → G | 0.324 | 0.356 | 0.438 | 0.295 | rs2631368 |
| –319 | 131733245 | C → A | 0 | 0.023 | 0 | 0 | rs60978556 |
| –234 | 131733330 | C → G | 0 | 0 | 0.094 | 0.018 | rs4646300 |
| –207 | 131733357 | C → G | 0.618 | 0.500 | 1.00 | 0.679 | rs2631367 |
AA, African-American; ME, Mexican-American; AS, Asian-American; EA, European-American
Some of samples have no data
A “0” implies that the SNP was not detected in a particular ethnic group
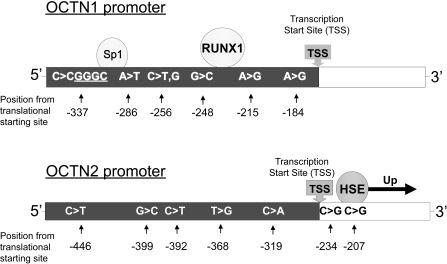
The promoter of OCTN2 exhibited greater variability than that of OCTN1. Six SNPs were identified in the OCTN2 proximal promoter region with minor allele frequencies ranging from 1.8 to 43.8%. One SNP in OCTN2 promoter, -207G>C SNP is major allele, has allele frequencies >50% for all ethnic groups. However, we defined -207C allele as reference based on UCSC Genome Bioinformatics (http://genome.ucsc.edu/). Four SNPs, namely, -399G>C, -368T>G, -234C>G, and -207G>C, had previously been reported in the dbSNP database as rs2631369, rs2631368, rs4646300, and rs2631367, respectively. The minor allele frequencies of two of the SNPs (-399G>C and -368T>G) were more than 29.5% in all ethnic groups. Interestingly, despite its high frequency in all other ethnic groups, the -207C allele was not detected in the Asian sample. The -234G allele was found in Asians and Mexicans at frequencies of 9.4 and 1.8%, respectively. Three other novel variants (-446T, -392T, and -319A) were identified but had frequencies less than 3.1%. Position -207 is not located in the basal promoter region; however, it is +58 bp from the transcription start site, whereas the other SNPs that are described in the OCTN2 basal promoter region are located within the region -250 and +50 bp from the transcription start site.
Haplotype Analysis of OCTN1 and OCTN2 Promoters in Four Ethnic Groups. Haplotypes were estimated using the PHASE algorithm (Stephens and Donnelly, 2003). This method identified eight haplotypes of OCTN1 and nine haplotypes of OCTN2 (Tables 3 and 4). Two major haplotypes in the basal promoter of SLC22A4 (OCTN1), namely, SLC22A4-P*1 and SLC22A4-P*2, accounted for more than 98% of the total haplotypic variation in each of the four ethnic groups. For SLC22A5 (OCTN2), three major haplotypes, namely, SLC22A5-P*1, SLC22A5-P*2, and SLC22A5-P*3, accounted for >98% of the total variation in African-, Mexican-, and European-Americans. Two of these three haplotypes, SLC22A5-P*1 and SLC22A5-P*2, accounted for 88% of the Asian sample, in which the SLC22A5-P*3 haplotype was not present. Two haplotypes (SLC22A5-P*4 and SLC22A5-P*5), were also identified in Asians but were not observed in the other three ethnic groups. These haplotypes accounted for 9.8% (SLC22A5-P*4) and 3.1% (SLC22A5-P*5) of total variation in the Asian sample. All other haplotypes (SLC22A5-P*6, -P*7, -P*8, and -P*9) had frequencies less than 2.3% and were present in only one ethnic group.
TABLE 3.
Haplotypes in the proximal promoter of SLC22A4 (OCTN1)
|
Name
|
Haplotype
|
No. of Chromosomes
|
Haplotypes
|
Ethnicity
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | EA | AS | ME | AA | EA | AS | ME | |||
| % | ||||||||||
| SLC22A4-P*1 | CACGAA | 388 | 85.4 | 65.4 | 66.2 | 69.4 | 30.3 | 22.8 | 23.1 | 23.8 |
| SLC22A4-P*2 | CACCAA | 147 | 11.8 | 33.8 | 32.4 | 30.6 | 12.1 | 30.9 | 29.5 | 27.5 |
| SLC22A4-P*3 | CAGGA | 2 | 0.0 | 0.0 | 1.5 | 0.0 | 0.0 | 0.0 | 100 | 0.0 |
| SLC22A4-P*4 | CATGAA | 1 | 0.0 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| SLC22A4-P*5 | CTCGAA | 1 | 0.7 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 | 0.0 |
| SLC22A4-P*6 | CACGGA | 1 | 0.7 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 | 0.0 |
| SLC22A4-P*7 | CACCAG | 1 | 0.0 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| SLC22A4-P*8 | CGGGCACCAA | 1 | 0.7 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 | 0.0 |
AA, African-American; ME, Mexican-American; AS, Asian-American; EA, European-American
TABLE 4.
Haplotypes in the proximal promoter of SLC22A5 (OCTN2)
|
Name
|
Haplotype
|
No. of Chromosomes
|
Haplotypes
|
Ethnicity
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | EA | AS | ME | AA | EA | AS | ME | |||
| % | ||||||||||
| SLC22A5-P*1 | CGCTCCG | 166 | 29.4 | 12.1 | 53.1 | 38.9 | 24.1 | 9.6 | 41.0 | 25.3 |
| SLC22A5-P*2 | CCCGCCG | 165 | 31.6 | 35.6 | 34.4 | 28.7 | 26.1 | 28.5 | 26.7 | 18.8 |
| SLC22A5-P*3 | CGCTCCC | 151 | 38.2 | 50.0 | 0.0 | 30.6 | 34.9 | 42.3 | 0.0 | 22.8 |
| SLC22A5-P*4 | CCCGCGG | 12 | 0.0 | 0.0 | 9.4 | 0.0 | 0.0 | 0.0 | 100 | 0.0 |
| SLC22A5-P*5 | TGCTCCG | 4 | 0.0 | 0.0 | 3.1 | 0.0 | 0.0 | 0.0 | 100 | 0.0 |
| SLC22A5-P*6 | CGCTACG | 3 | 0.0 | 2.3 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 |
| SLC22A5-P*7 | CGCGCCG | 1 | 0.7 | 0.0 | 0.0 | 0.0 | 100 | 0.0 | 0.0 | 0.0 |
| SLC22A5-P*8 | CCCTCCG | 1 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 | 0.0 | 0.0 | 100 |
| SLC22A5-P*9 | CGTTCCC | 1 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 | 0.0 | 0.0 | 100 |
AA, African-American; ME, Mexican-American; AS, Asian-American; EA, European-American
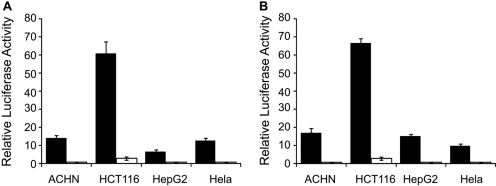
Identification of Host Cells for Luciferase Assays of Promoter Region Variants of OCTNs. We cloned the OCTN1 and OCTN2 proximal promoter regions and tested them for promoter activity in four different cell lines (ACHN, HCT-116, HepG2,, and HeLa) using transient transfection reporter assays. The luciferase activities of the OCTN1 and OCTN2 promoter in these cell lines were determined by the ratio between the firefly luciferase and Renilla luciferase activity. As shown in Fig. 1, luciferase activities of OCTN1 and OCTN2 constructs were highest in HCT-116. Because of the high luciferase activities and the fact that OCTN1 and OCTN2 are highly expressed in the human intestine and kidney (Nishimura and Naito, 2005; Meier et al., 2007), HCT-116 and ACHN were selected as the cell lines to evaluate the promoter variants of the two transporters.
Fig. 1.
Luciferase activity in four cell lines transfected with reporter constructs of the proximal promoter of SLC22A4 (OCTN1; A) and SLC22A5 (OCTN2; B). Results are expressed as relative activity of firefly luciferase normalized for Renilla luciferase. Data are shown as the mean ± S.D. from one experiment performed in quadruplet. The closed and open boxes indicate reference promoter region and empty vector constructs, respectively.
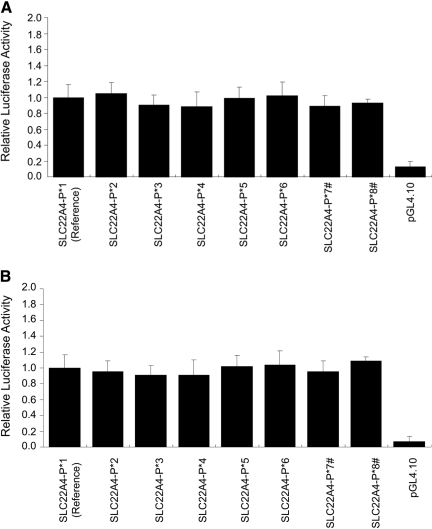
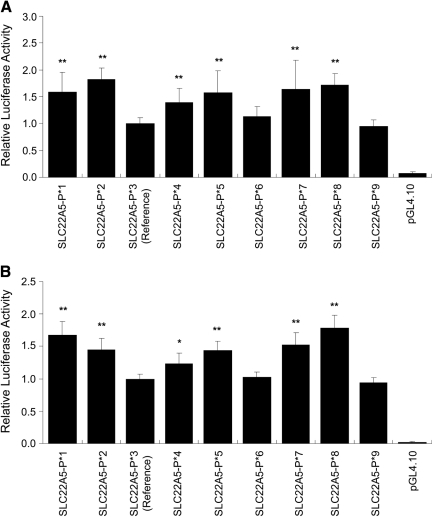
Functional Activity of OCTN1 and OCTN2 Promoter Variants. Luciferase activities of the OCTN1 promoter variant constructs in ACHN and HCT-116 cells are shown in Fig. 2. Compared with the reference haplotype SLC22A4-P*1 (the major haplotype), haplotypes SLC22A4-P*2, -P*3, -P*4, -P*5, -P*6, -P*7, and -P8* demonstrated similar luciferase activities in ACHN cells (Fig. 2A) and HCT-116 cells (Fig. 2B). In contrast to OCTN1 promoter haplotypes, there were notable differences in reporter activities across the various OCTN2 promoter haplotypes. In ACHN cell lines (Fig. 3A), the OCTN2 haplotypes SLC22A5-P*1, -P*2, -P*4, -P*5, -P*7, and -P*8 showed significant increases in reporter activities compared with the reference haplotype SLC22A5-P*3 (although this has a minor haplotype frequency, but we defined SLC22A5-P*3 as reference based on UCSC Genome Bioinformatics March 2006 hg18; http://genome.ucsc.edu/). Likewise, in HCT-116 cell lines (Fig. 3B), there were significant increases in luciferase activities of the OCTN2 promoter haplotypes SLC22A5-P*1, -P*2, -P*4, -P*5, -P*7, and -P*8 (68, 45, 23, 44, 53, and 79%, respectively), relative to the reference haplotype SLC22A5-P*3.
Fig. 2.
Luciferase activity in ACHN (A) or HCT-116 (B) cell lines transfected with reporter constructs containing variants in the proximal promoter of SLC22A4 (OCTN1). The OCTN1 promoter construct (100 ng) was cotransfected with the Renilla construct (2.5 ng) into each cell line. Results are expressed as activity of firefly luciferase (normalized for Renilla luciferase) relative to the reference promoter. Data are shown as the mean ± S.D. of three separate experiments performed in triplicate. Dunnett's multiple comparison test was used to determine statistical differences. A p < 0.05 was used as the threshold for significance. #, reporter constructs of haplotypes with -248C variant.
Fig. 3.
Luciferase activity in ACHN (A) or HCT-116 (B) cell lines transfected with reporter constructs of haplotypes in the proximal promoter of SLC22A5 (OCTN2). The OCTN2 promoter construct (100 ng) was cotransfected with Renilla construct (2.5 ng) into each cell line. Results are expressed as activity of firefly luciferase (normalized for Renilla luciferase) relative to the activity of the reference promoter. Data are shown as the means ± S.D. of three separate experiments performed in triplicate. Dunnett's multiple comparison test was used to determine statistical differences. A p < 0.05 was used as the threshold for significance (*, p < 0.05; **, p < 0.01).
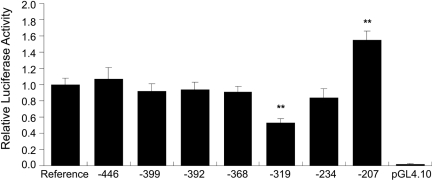
Functional Activity of Individual OCTN2 Promoter Variants. To determine which SNP may have caused the observed altered luciferase activities of the OCTN2 haplotypes (Fig. 2), the luciferase activity of promoter constructs containing each of the individual SNPs in the OCTN2 proximal promoter was investigated in HCT-116 cell lines. As shown in Fig. 4, the -319A SNP produced a 50% decrease in luciferase activity (p < 0.01) and the -207G produced a 60% increase (p < 0.01) in luciferase activity in HCT-116 cell lines. The -207G in the OCTN2 promoter has been reported previously to create a binding site for a heat shock element (Fig. 7) (Peltekova et al., 2004). However, in repeated experiments, we did not observe an effect of heat shock on the OCTN2 promoter activity in either HCT-116 or ACHN cell lines (data not shown).
Fig. 4.
Luciferase activity in HCT-116 cell lines transfected with reporter constructs containing individual variants in the proximal promoter of SLC22A5 (OCTN2). The OCTN2 promoter construct (100 ng) was cotransfected with Renilla construct (2.5 ng) into HCT-116 cells. Results are expressed as activity of firefly luciferase (normalized for Renilla luciferase) relative to the activity of the reference promoter. Data are shown as the means ± S.D. of two separate experiments performed in triplicate. Dunnett's multiple comparison test was used to determine statistical differences. A p < 0.05 was used as the threshold for significance (*, p < 0.05; **, p < 0.01).
Fig. 7.
Schematic model of the predicted transcription factor binding sites and location of SNPs in the OCTN1 and OCTN2 promoters. Transcription binding factors of OCTN1 promoter RUNX1 and Sp1 have been reported previously by Maeda et al. (2007). There were no SNPs in these binding sites of the OCTN1 promoter. The -207G in the OCTN2 promoter has been reported previously to create a binding site for a heat shock element (HSE) (Peltekova et al., 2004). A change from C to G at position -207 creates the HSE binding site, resulting in an increase in transcriptional activity.
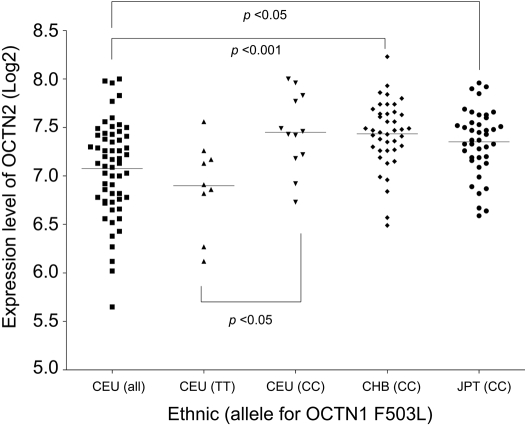
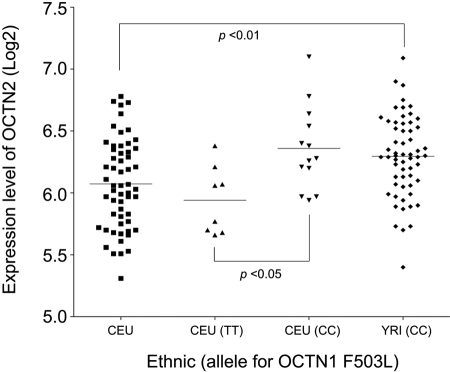
Ethnic Differences in Expression of OCTN2 in Lymphoblastoid Cell Lines. A C>T substitution in OCTN1 exon 9 (1507C>T; nucleotide position is relative to the “A” in the ATG start codon) that results in a nonsynonymous variant (L503F; rs1050152) and the OCTN2 promoter SNP (-207G>C; rs2631367) are in linkage disequilibrium in the Caucasian population and as a result create a different two-allele haplotypes TC or CG. However, they are not in perfect linkage disequilibrium, because the allele frequencies are different. The 1507T allele is almost always found with -207C, but the 1507C allele is usually linked to -207G, although 1507C allele can also be found with -207C (Peltekova et al., 2004). Because the genotype for the OCTN2 promoter variant, -207G>C, was not available in the HapMap data, we associated the OCTN1-1507C>T genotype in the lymphoblastoid cell lines derived from unrelated individuals from CEU with OCTN2 mRNA expression level using two different data sets in GEO (GSE5859 and GSE7761). As shown in Figs. 5 and 6, the OCTN2 mRNA expression levels in cell lines from individuals homozygous for OCTN1-1507 (C/C), which is in linkage disequilibrium with OCTN2-207 (G/G), was significantly higher than those for OCTN1-1507 (T/T), which is linked to OCTN2-207 (C/C). However, this linkage disequilibrium is not perfectly complete. The overall OCTN2 mRNA expression levels in the different ethnic groups were compared. Figure 5 shows that OCTN2 expression levels in lymphoblastoid cell lines are lower in the CEU group compared with the other three ethnic groups; however, OCTN2 mRNA expression levels in CEU subjects with the OCTN1-1507 (C/C) genotype (who are predicted to almost all have the G/G or G/C genotype at OCTN2-207) were not significantly different from OCTN2 mRNA levels in lymphoblastoid cell lines from Chinese and Japanese, all of whom are homozygous for OCTN2-207(G/G). These data suggest that the OCTN2-207G variant may explain ethnic differences in mRNA expression levels of OCTN2 in lymphoblastoid cell lines between Caucasians and Chinese or Japanese.
Fig. 5.
Association of OCTN1 genotype with mRNA expression levels of OCTN2 in lymphoblastoid cell lines derived from unrelated individuals from HapMap populations. Genotypes for OCTN1 (1507T>C, F503L, rs1050152) were obtained from the HapMap, and the mRNA expression levels of OCTN2 (log2) were obtained from the GEO site (accession code GSE5859). There is significantly higher expression of OCTN2 mRNA in individuals who are homozygous CC (n = 12) compared with individuals who are homozygous TT (n = 9). Dunn's multiple comparison test was used to evaluate differences by genotype and p < 0.05 was used as the threshold for significance.
Fig. 6.
Association of ethnicity with OCTN2 mRNA expression levels in lymphoblastoid cell lines derived from unrelated individuals from CEU and YRI. The information of the individual genotype for OCTN1 (1507T>C; F503L; rs1050152), C/C, C/T, or T/T were obtained from the HapMap and the mRNA expression levels of OCTN2 (log2) were obtained from the GEO site (accession code GSE7761). There is significantly higher expression OCTN2 mRNA in individual (CEU) who are homozygous for CC (n = 13) compared with individual who are homozygous for TT (n = 8). Dunn's multiple comparison test was used to determine statistical differences p < 0.05 was used as the threshold for significance.
Effects of OCTNs Haplotype on Carnitine Disposition in the Healthy Volunteers. To study the effect of -207C>G genotype on the disposition of carnitine, we enrolled a total of 16 healthy adult volunteers into a 2-h study of carnitine renal clearance, including seven subjects homozygous for the C/G* haplotype (OCTN1-1507C, OCTN2-207G) and nine homozygous for the T/C* haplotype (OCTN1-1507T, OCTN2-207C). The subjects were recruited to achieve gender balance between haplotype groups. The demographic and clinical characteristics of the subjects are shown in Table 5. The two haplotype groups did not differ with respect to gender, age, height, or weight. The primary study measures are summarized in Table 6. There were no significant differences between haplotype groups in terms of carnitine clearance (CLR) (free and total) or plasma carnitine concentration. The mean CLR of free carnitine was a small fraction (<5%) of the creatinine clearance, reflecting the efficient OCTN2-mediated reabsorption of carnitine. Likewise, subjects did not differ between haplotype groups with respect to plasma free carnitine or acylcarnitines.
TABLE 5.
Demographic characteristics of study sample by haplotype Healthy subjects aged 18–40 years and of European descent were recruited for the study based on haplotype at the OCTN locus. Subjects were homozygous for either the common C/G* haplotype [OCTN1-1507C, OCTN2-(–207G)] or the T/C* haplotype [OCTN1-1507T, OCTN2-(–207C)]. Values are shown as mean ± S.D. Baseline differences between subject groups were assessed using a two-tailed unpaired t test. Gender differences were assessed by Fisher's exact test.
|
Total Sample
|
Haplotype
|
p Value
|
||
|---|---|---|---|---|
| C/G* (n = 7) | T/C* (n = 9) | |||
| Gender (male/female) | 7/9 | 4/3 | 3/6 | 0.614 |
| Age (yr) | 31.9 ± 5.1 | 30.1 ± 5.4 | 33.3 ± 4.6 | 0.223 |
| Height (cm) | 176 ± 11 | 181 ± 12 | 172 ± 8.8 | 0.184 |
| Weight (kg) | 74.4 ± 13.1 | 79.4 ± 15.1 | 70.2 ± 10.8 | 0.270 |
| Body mass index | 23.7 ± 2.5 | 24.0 ± 3.4 | 23.5 ± 1.9 | 0.749 |
TABLE 6.
Summary of carnitine disposition by haplotype After a 12-h fast, blood was drawn from subjects homozygous for either the C/G* haplotype [OCTN1-1507C, OCTN2-(–207G); n = 7] or T/C* haplotype [OCTN1–1507T, OCTN2-(–207C); n = 9] at the OCTN locus. Urine was collected over the next 2 h, and a final blood draw was collected at 2 h after initiating the study. Plasma and urine samples were analyzed for free (nonesterified) carnitine and total (free plus esterified) carnitine as well as creatinine. The mean analyte concentration (from the baseline and 2-h blood draws) for each subject was used in the analysis. Values are shown as mean ± S.D. Differences between haplotype groups were assessed using a two-tailed unpaired t test.
|
Carnitine
|
Total Sample
|
Haplotype
|
p Value
|
||
|---|---|---|---|---|---|
| C/G* (n = 7) | T/C* (n = 9) | ||||
| Plasma carnitine levels (μM) | Free | 31.2 ± 5.3 | 32.5 ± 5.9 | 30.2 ± 4.9 | 0.410 |
| Esters | 8.35 ± 2.45 | 7.90 ± 1.51 | 8.71 ± 3.04 | 0.532 | |
| Total | 39.6 ± 5.7 | 40.4 ± 6.2 | 38.9 ± 5.6 | 0.622 | |
| Carnitine CLR (ml/min) | Free | 2.77 ± 2.25 | 3.20 ± 2.97 | 2.44 ± 1.61 | 0.527 |
| Total | 5.77 ± 3.24 | 5.78 ± 3.58 | 5.76 ± 3.17 | 0.993 | |
| Creatinine CL (ml/min) | 138 ± 34 | 145 ± 40 | 132 ± 30 | 0.478 | |
We found that gender was significantly associated with plasma concentrations of carnitine in our sample, with male subjects having significantly higher plasma carnitine levels than female subjects (34.8 ± 4.6 μM for males versus 28.5 ± 4.2 μM for females; mean ± S.D.; p = 0.01). This difference was not explained by decreased carnitine clearance in male subjects. It is noteworthy that carnitine plasma levels were not significantly influenced by carnitine renal clearance in either univariate or multiple regression analyses (i.e., after correcting for gender effects).
Discussion
In relation to the widely studied functional analysis of nonsynonymous SNPs in membrane transporters, there have been comparably few studies analyzing the functional effects of genetic polymorphisms in noncoding regions. In the present study, we performed genetic and functional analysis of proximal promoter variants of in the SLC22A family, OCTN1 and OCTN2. Ethnic differences in the OCTN2 expression levels were also examined across the populations (CEU, CHB, and JPT). In addition, the effect of OCTN2 genotype (-207G>C) on the disposition of carnitine was examined in healthy volunteers. Promoter region variants of other SLC22A family members, i.e., organic cation transporters and organic anion transporters identified in Japanese have been characterized recently (Ogasawara et al., 2008).
Although six variants and eight haplotypes were found in the proximal promoter of OCTN1, two major haplotypes accounted for >98% of total chromosomes in all populations tested (Tables 1 and 3). The haplotypes of OCTN1 had no statistically significant differences in luciferase activity in either ACHN or HCT-116 cells compared with the reference (Fig. 2). The results suggest that the identified variants in the proximal promoter of OCTN1 do not affect the mRNA level of the transporter. SNPs in the coding region of OCTN1 have been reported to affect the expression or function of the transporter (Urban et al., 2007, 2008). For example, recent studies from our laboratory demonstrated that the nonsynonymous OCTN1 variant L503F was associated with a decrease in the renal tubular secretion and in the in vitro transport of the anticonvulsant gabapentin (Urban et al., 2008). In addition, an SNP in intron 1 of the gene affects the expression level of OCTN1 by decreasing the binding affinity for RUNX1, which is an essential hematopoietic transcription factor (Tokuhiro et al., 2003). A RUNX1 binding site is also located in the proximal promoter region of the OCTN1 gene but is not essential for promoter activity (Maeda et al., 2007). We did not identify any SNPs in the RUNX1 binding sites of the OCTN1 proximal promoter region among the four ethnic groups (Fig. 7).
In the OCTN2 promoter region, three of the seven SNPs had allele frequencies greater than 29.5% (-399G>C, -368T>G, and -207G>C), and three major haplotypes accounted for >98% of the total chromosomes in African-, European-, and Mexican-Americans. Two of the three haplotypes also occurred in more than 88% of total chromosomes in the Asian-Americans (Table 4). Although not observed in the Asian sample, -207C (Table 2) has a high allele frequency in the three other ethnic groups. Furthermore, the haplotypes containing the -207G with the exception of SLC22A5-P*6 showed significantly higher luciferase activities compared with the reference haplotype, SLC22A5-P*3 (Fig. 3). The -207G alone was associated with increased reporter activity (Fig. 4) and seems to have produced the increase in luciferase activity of the six haplotypes containing the SNP. The reason for the low luciferase activity of SLC22A5-P*6 haplotype may be explained by the presence of the -319A, which produced a lower reporter activity (Fig. 4). Thus, the increase in luciferase activity produced by the -207G in the SLC22A5-P*6 haplotype is likely to be cancelled out by the -319A. However, the MatInspector transcription factor prediction program and the TFSearch indicated that there is no transcriptional binding factor in the -319C>A region. Unknown factor would be involved in the transcriptional activity of OCTN2 promoter.
Our analyses also suggest that the -207G>C polymorphism may explain racial differences in OCTN2 expression level in lymphoblastoid cell lines. OCTN2 is expressed at significantly higher levels in lymphoblastoid cell lines from individuals of Chinese or Japanese ancestry in comparison with individual European ancestry. However, when grouped according to the OCTN2 G allele at position -207, the expression level of OCTN2 was not significantly different in lymphoblastoid cell lines derived from individuals of European ancestry compared with those of Chinese/Japanese ancestries, all of whom are homozygous for the -207G (Fig. 5). Because the OCTN1-1507 variant is not present in the YRI population, we were not able to identify the YRI samples that harbored the OCTN2-207G>C variant using HapMap data. However, it is noteworthy that the mRNA levels of OCTN2 in lymphoblastoid cell lines from the YRI population were significantly higher than the levels in the CEU sample (Fig. 6). The YRI population has a greater allele frequency of OCTN2-207G in comparison with the CEU population (62% compared with 50%; Table 2). Expression levels of OCTN1 mRNA in lymphoblastoid cell lines from the two GEO data sets were not different among the various ethnic groups (data not shown).
Previously, we have evaluated the contribution of genetic variability in the OCTN2 promoter to interindividual differences in gene expression and carnitine transport in the lymphoblastoid cell lines derived from individuals homozygous for either G/G or C/C at position -207. We found that lymphoblastoid cell lines from individuals homozygous for the G/G variant had significantly greater expression levels of OCTN2 mRNA and l-carnitine uptake than the cell lines from individuals homozygous for the -207C allele (Urban et al., 2006). The ex vivo result suggested that the -207G>C polymorphism affects function as well as basal expression of OCTN2 in lymphoblastoid cell lines from healthy volunteers. In this study, we analyzed the association between -207G>C and OCTN2 mRNA expression using data from the HapMap and GEO databases. A nonsynonymous SNP of OCTN1, 503Leu>Phe (1507C>T), and the promoter SNP of OCTN2, -207G>C, are in strong linkage disequilibrium and create a two-point haplotype (T/C*OCTNs or C/G*OCTNs haplotype) in individuals of European ancestry (Peltekova et al., 2004). Because the -207G>C SNP was not genotyped in the CEU cell lines, we associated expression level of OCTN2 with promoter genotype using the OCTN1-1507C>T SNP as a proxy for the -207G>C SNP in the OCTN2 promoter. As shown in Fig. 5, the mRNA level of OCTN2 was higher for the OCTN1-1507(C/C) homozygotes (reflecting the OCTN2-207G/G homozygotes) compared with that for the OCTN1-1507(T/T) homozygotes in the CEU population, in agreement with our previous results (Urban et al., 2006). Therefore, we investigated whether OCTN2 genotype (-207G/G or -207C/C) was predictive of carnitine plasma level or renal clearance. Despite the evidence from in vitro and ex vivo studies, however, we found that OCTNs haplotype, at least for the common haplotypes as defined here, did not predict carnitine plasma concentrations or carnitine CLR in vivo (Table 6). Although there does not seem to be a large effect of the C/G*OCTNs versus. T/C*OCTNs haplotype on carnitine disposition in vivo, there may be some heterogeneity within each haplotype group that offsets or masks the effects of the OCTN1 (1507C>T) and OCTN2 (-207G>C) SNPs. In addition, the fact that carnitine levels were positively correlated with carnitine renal clearance strongly suggests that dietary factors play an overwhelming role in carnitine disposition within the normal range, i.e., at normal levels of functional OCTN2 activity, renal tubular reabsorption of carnitine is sufficient to maintain adequate plasma carnitine levels, and variation in plasma carnitine levels are governed primarily by dietary intake. This is in contrast to the scenario at work in SCD, where complete loss of functional OCTN2 activity leads to extreme decreases in carnitine reabsorption and results in severe deficits in plasma carnitine levels. The data from the current study were in good agreement with recent data from Bene et al. (2007), who have reported that there were no statistically significant differences in esterified carnitine levels between C/G*OCNTs and T/G*OCTNs haplotype groups in both normal and Crohn disease patients. Others also have investigated the effect of the -207G>C polymorphism in human cardiac muscle and have found that OCTN2 expression in this tissue was not affected by genotype (Grube et al., 2006). Therefore, we speculate that the -207G>C polymorphism in the promoter may have variable effects on OCTN2 expression depending on cell system as well as tissue type.
In summary, this is the first extensive study of polymorphisms in the proximal promoters of OCTN1 and OCTN2. Our data suggest that one common polymorphism in OCTN2 (-207G>C) may produce significant differences in the transcription rate and expression of the gene and may explain ethnic differences in the mRNA expression levels of the gene in lymphoblastoid cell lines. Our data suggest that the OCTN2-207G>C does not contribute to the renal clearance of l-carnitine or the maintenance of l-carnitine levels in plasma in healthy individuals. Because the SNP is clearly functional and is associated with variation in expression levels of the transporter in lymphoblastoid cell lines, the data may suggest that OCTN2 expression levels in the kidney (and consequently carnitine clearance) are not regulated by this SNP. The data are consistent with tissue specific effects of the -207G>C on OCTN2 expression levels (Grube et al., 2006). Future studies of the influence of the -207G>C on other phenotypes that may be tissue specific, such as the disposition and response to various drugs including β-lactam antibiotics and cardiovascular drugs, may be fruitful (Ganapathy et al., 2000; Iwata et al., 2008; Ohnishi et al., 2008).
Acknowledgments
We acknowledge Dr. Richard M. Myers and Yuya Kobayashi for helpful discussions and technical advice on the promoter assay setup. Data in this article have been deposited in www.pharmgk-b.org. We acknowledge fellowship support for K.M.G. from the Japanese Society for the Promotion of Sciences.
This work was supported by the National Institutes of Health [Grant GM61390].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.108.146449.
ABBREVIATIONS: SNP, single-nucleotide polymorphism; OCTN, novel organic cation transporter; SCD, systemic carnitine deficiency; GEO, Gene Expression Omnibus; bp, base pairs(s); CEU CEPH, Utah residents with ancestry from northern and western Europe; YRI, Yoruba in Ibadan, Nigeria; CHB, Han Chinese; JPT, Japanese; CLr, renal clearance.
References
- Bene J, Komlósi K, Magyari L, Talián G, Horváth K, Gasztonyi B, Miheller P, Figler M, Mózsik G, Tulassay Z, et al. (2007) Plasma carnitine ester profiles in Crohn's disease patients characterized for SLC22A4 C1672T and SLC22A5 G-207C genotypes. Br J Nutr 98 345-350. [DOI] [PubMed] [Google Scholar]
- Bleasby K, Castle JC, Roberts CJ, Cheng C, Bailey WJ, Sina JF, Kulkarni AV, Hafey MJ, Evers R, Johnson JM, et al. (2006) Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: a resource for investigations into drug disposition. Xenobiotica 36 963-988. [DOI] [PubMed] [Google Scholar]
- Cooper SJ, Trinklein ND, Anton ED, Nguyen L, and Myers RM (2006) Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res 16 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropp CD, Yee SW, and Giacomini KM (2008) Genetic variation in drug transporters in ethnic populations. Clin Pharmacol Ther 84 412-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong FA, Kehrer DF, Mathijssen RH, Creemers GJ, de Bruijn P, van Schaik RH, Planting AS, van der Gaast A, Eskens FA, Janssen JT, et al. (2006) Prophylaxis of irinotecan-induced diarrhea with neomycin and potential role for UGT1A1*28 genotype screening: a double-blind, randomized, placebo-controlled study. Oncologist 11 944-954. [DOI] [PubMed] [Google Scholar]
- Eichelbaum M, Ingelman-Sundberg M, and Evans WE (2006) Pharmacogenomics and individualized drug therapy. Annu Rev Med 57 119-137. [DOI] [PubMed] [Google Scholar]
- Ganapathy ME, Huang W, Rajan DP, Carter AL, Sugawara M, Iseki K, Leibach FH, and Ganapathy V (2000) beta-Lactam antibiotics as substrates for OCTN2, an organic cation/carnitine transporter. J Biol Chem 275 1699-1707. [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, Johnson JA, Hayes DF, Klein T, Krauss RM, et al. (2007) The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther 81 328-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube M, Meyer zu Schwabedissen HE, Präger D, Haney J, Möritz KU, Meissner K, Rosskopf D, Eckel L, Böhm M, Jedlitschky G, et al. (2006) Uptake of cardiovascular drugs into the human heart: expression, regulation, and function of the carnitine transporter OCTN2 (SLC22A5). Circulation 113 1114-1122. [DOI] [PubMed] [Google Scholar]
- Ieiri I, Takane H, Hirota T, Otsubo K, and Higuchi S (2006) Genetic polymorphisms of drug transporters: pharmacokinetic and pharmacodynamic consequences in pharmacotherapy. Expert Opin Drug Metab Toxicol 2 651-674. [DOI] [PubMed] [Google Scholar]
- Iwata D, Kato Y, Wakayama T, Sai Y, Kubo Y, Iseki S, and Tsuji A (2008) Involvement of carnitine/organic cation transporter OCTN2 (SLC22A5) in distribution of its substrate carnitine to the heart. Drug Metab Pharmacokinet 23 207-215. [DOI] [PubMed] [Google Scholar]
- Koepsell H and Endou H (2004) The SLC22 drug transporter family. Pflugers Arch 447 666-676. [DOI] [PubMed] [Google Scholar]
- Kuwajima M, Lu K, Sei M, Ono A, Hayashi M, Ishiguro K, Ozaki K, Hotta K, Okita K, Murakami T, et al. (1998) Characteristics of cardiac hypertrophy in the juvenile visceral steatosis mouse with systemic carnitine deficiency. J Mol Cell Cardiol 30 773-781. [DOI] [PubMed] [Google Scholar]
- Lahjouji K, Mitchell GA, and Qureshi IA (2001) Carnitine transport by organic cation transporters and systemic carnitine deficiency. Mol Genet Metab 73 287-297. [DOI] [PubMed] [Google Scholar]
- Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, et al. (2003) Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci U S A 100 5896-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Hirayama M, Kobayashi D, Miyazawa K, and Tamai I (2007) Mechanism of the regulation of organic cation/carnitine transporter 1 (SLC22A4) by rheumatoid arthritis-associated transcriptional factor RUNX1 and inflammatory cytokines. Drug Metab Dispos 35 394-401. [DOI] [PubMed] [Google Scholar]
- Meier Y, Eloranta JJ, Darimont J, Ismair MG, Hiller C, Fried M, Kullak-Ublick GA, and Vavricka SR (2007) Regional distribution of solute carrier mRNA expression along the human intestinal tract. Drug Metab Dispos 35 590-594. [DOI] [PubMed] [Google Scholar]
- Nishimura M and Naito S (2005) Tissue-specific mRNA expression profiles of human ATP-binding cassette and solute carrier transporter superfamilies. Drug Metab Pharmacokinet 20 452-477. [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Terada T, Motohashi H, Asaka J, Aoki M, Katsura T, Kamba T, Ogawa O, and Inui K (2008) Analysis of regulatory polymorphisms in organic ion transporter genes (SLC22A) in the kidney. J Hum Genet 53 607-614. [DOI] [PubMed] [Google Scholar]
- Ohashi R, Tamai I, Nezu Ji J, Nikaido H, Hashimoto N, Oku A, Sai Y, Shimane M, and Tsuji A (2001) Molecular and physiological evidence for multifunctionality of carnitine/organic cation transporter OCTN2. Mol Pharmacol 59 358-366. [DOI] [PubMed] [Google Scholar]
- Ohnishi S, Okamura N, Sakamoto S, Hasegawa H, Norikura R, Kanaoka E, Takahashi K, Horie K, Sakamoto K, and Baba T (2008) Role of Na+l-carnitine transporter (OCTN2) in renal handling of pivaloylcarnitine and valproylcarnitine formed during pivalic acid-containing prodrugs and valproic acid treatment. Drug Metab Pharmacokinet 23 293-303. [DOI] [PubMed] [Google Scholar]
- Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, et al. (2004) Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet 36 471-475. [DOI] [PubMed] [Google Scholar]
- Poonkuzhali B, Lamba J, Strom S, Sparreboom A, Thummel K, Watkins P, and Schuetz E (2008) Association of breast cancer resistance protein/ABCG2 phenotypes and novel promoter and intron 1 single nucleotide polymorphisms. Drug Metab Dispos 36 780-795. [DOI] [PubMed] [Google Scholar]
- Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, Sheardown SA, Yue L, Burchard EG, Brett CM, et al. (2008) Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther 83 273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Duerr RH, Brant SR, Bromfield G, Datta LW, Jani N, Kane SV, Rotter JI, Philip Schumm L, Hillary Steinhart A, et al. (2007) Refined genomic localization and ethnic differences observed for the IBD5 association with Crohn's disease. Eur J Hum Genet 15 328-335. [DOI] [PubMed] [Google Scholar]
- Sissung TM, Gardner ER, Gao R, and Figg WD (2008) Pharmacogenetics of membrane transporters: a review of current approaches. Methods Mol Biol 448 41-62. [DOI] [PubMed] [Google Scholar]
- Stephens M and Donnelly P (2003) A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73 1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tein I (2003) Carnitine transport: pathophysiology and metabolism of known molecular defects. J Inherit Metab Dis 26 147-169. [DOI] [PubMed] [Google Scholar]
- Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, Suzuki M, Nagasaki M, Ohtsuki M, Ono M, et al. (2003) An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet 35 341-348. [DOI] [PubMed] [Google Scholar]
- Urban TJ, Brown C, Castro RA, Shah N, Mercer R, Huang Y, Brett CM, Burchard EG, and Giacomini KM (2008) Effects of genetic variation in the novel organic cation transporter, OCTN1, on the renal clearance of gabapentin. Clin Pharmacol Ther 83 416-421. [DOI] [PubMed] [Google Scholar]
- Urban TJ, Gallagher RC, Brown C, Castro RA, Lagpacan LL, Brett CM, Taylor TR, Carlson EJ, Ferrin TE, Burchard EG, et al. (2006) Functional genetic diversity in the high-affinity carnitine transporter OCTN2 (SLC22A5). Mol Pharmacol 70 1602-1611. [DOI] [PubMed] [Google Scholar]
- Urban TJ, Yang C, Lagpacan LL, Brown C, Castro RA, Taylor TR, Huang CC, Stryke D, Johns SJ, Kawamoto M, et al. (2007) Functional effects of protein sequence polymorphisms in the organic cation/ergothioneine transporter OCTN1 (SLC22A4). Pharmacogenet Genomics 17 773-782. [DOI] [PubMed] [Google Scholar]
- Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, and Sadée W (2008) Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood 112 1013-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]