Abstract
Purpose:
Minimal residual disease (MRD) presents a significant hurdle to curing metastatic neuroblastoma (NB). Biologic therapies directed against MRD can improve outcome. Evaluating treatment efficacy requires MRD measurement, which serves as surrogate endpoint. Because of tumor heterogeneity, no single marker will likely be adequate. Genome-wide expression profiling can uncover potential MRD markers differentially expressed in tumors over normal marrow/blood.
Experimental Design:
Gene expression array was carried out on 48 stage 4 tumors and 9 remission marrows using the Affymetrix U-95 gene chip. 34 genes with a tumor-to-marrow expression ratio higher than tyrosine hydroxylase were identified. Quantitative RT-PCR was performed on all 34 genes to study the dynamic range of tumor cell detection and the expression of these genes in normal marrow/blood samples and in stage 4 NB tumors. Top ranking markers were then tested for prognostic significance in the marrows of stage 4 patients collected from the same treatment protocol after 2 cycles of immunotherapy.
Results:
Based on sensitivity assays, 8 top-ranking markers were identified: CCND1, CRMP1, DDC, GABRB3, ISL1, KIF1A, PHOX2B, and TACC2. They were abundantly expressed in stage 4 NB tumors (n=20) and had low to no detection in normal marrow/blood samples (n=20). Moreover, expression of CCND1, DDC, GABRB3, ISL1, KIF1A, and PHOX2B in 116 marrows sampled after 2 treatment cycles was highly prognostic of progression-free and overall survival (p<0.001).
Conclusions:
Marker discovery based on differential gene expression profiling, stringent sensitivity and specificity assays, and well-annotated patient samples can rapidly prioritize and identify potential MRD markers of neuroblastoma.
INTRODUCTION
A major obstacle to curing metastatic neuroblastoma (NB) is the presence of minimal residual disease (MRD) in the bone marrow and peripheral blood even after the patient has achieved clinical remission. Before MRD can be targeted by either immunotherapy or myeloablative therapy, it needs to be detected and quantified. Even though there are only a few established MRD markers for NB, there is increasing evidence that MRD markers can be clinically useful (1-5). Residual occult tumor cells at the end of each phase of treatment can have an adverse impact on disease relapse and patient survival.
Despite their clinical utility in proof-of-principle studies, single markers are likely to be inadequate because tumor heterogeneity is a hallmark in cancers including NB. For example, even though GD2 synthase (GalNacT) is a highly sensitive MRD marker, it is generally believed that some tumors treated with GD2-directed therapy, be it antibody, immunocytokine, or scFv-modified T cells, can down-regulate the enzyme and the antigen GD2 as an escape mechanism. Similarly, tumors treated with another common modality (131I-MIBG) are expected to down-regulate its metabolic pathway, where tyrosine hydroxylase (TH) is a critical step. The rationale for using multiple markers is compelling (6). However, there is a paucity of MRD markers and previous efforts of tumor marker discovery have failed to identify candidates solely intended for MRD measurement because of suboptimal specificity and sensitivity (7).
For an orphan disease like NB with few known markers, genome-based expression screening is most likely to be useful. This is particularly true when marker discovery takes into account the context where tumor measurement is most relevant, informative, and feasible, i.e. the bone marrow and peripheral blood compartment. Using this approach our laboratory was able to identify new markers of subclinical disease for Ewing family of tumors (8). As to NB, this gene expression profiling strategy uncovered cyclin D1 as a novel molecular marker of MRD for patients with metastatic NB (9). In this report, an array analysis based on tumors from 48 stage 4 NB patients was used to rapidly filter 34 potential MRD markers from ∼16,000 unique genes. Sensitivity and specificity studies narrowed the list to 8 top-ranking markers. They were further evaluated for prognostic significance in the marrows of 116 stage 4 patients collected from the same treatment protocol after 2 cycles of immunotherapy.
MATERIALS AND METHODS
Identification of potential MRD markers of NB by genome-wide gene expression array analyses
Affymetrix human U-95 oligonucleotide array was carried out on 48 tumors (18/48 were MYCN amplified tumors), and 9 remission marrows from stage 4 NB patients diagnosed after 18 months of age and 12 NB cell lines (SH-SY5Y, SK-N-BE(1), SK-N-BE(2), SK-N-BE(2)M17, LAI-55N, SK-N-LP, SK-N-ER, SK-N-JD, BE(2)C, LAI-5S, SH-EP1, SK-N-BE(2)S). Absolute values of expression were calculated and normalized (scaling factor of 500) using Affymetrix Microarray Suite 5.0 (10).
Specimens for sensitivity and specificity studies
NB cell lines LAN1 and NMB7 were used for tumor cell seeding experiments. Buffy coat was obtained from New York Blood Center (New York, NY). Fresh-frozen stage 4 tumors were obtained at diagnosis and relapse at Memorial Sloan-Kettering Cancer Center in accordance to the guidelines of the institutional review board.
Patient characteristics of marrow samples
Bone marrows were collected from patients with stage 4 NB treated with an immunotherapy protocol using anti-GD2 monoclonal antibody 3F8 plus GM-CSF following chemotherapy (11). Archived marrows tested by qRT-PCR were all collected after 2 treatment cycles at a median time of 2.5 months from protocol entry. The median age at diagnosis was 3.3 years, with 108 of 116 patients diagnosed at 18 months or older, the highest age risk group. 27 patients had MYCN amplified tumors.
Molecular analysis
Mononuclear cells were isolated and total RNA isolated and quality assessed as previously described (12, 13). cDNA was synthesized from 1 ug of total RNA. One ul of cDNA was used for real-time quantitative PCR (qPCR) using Applied Biosystems (ABI) Sequence Detection System 7300 (Foster City, CA). All endogenous controls were purchased from ABI. For sensitivity assay, β2 microglobulin (β2M, 4326319E) was used for normalization. Tumor samples were normalized using the geometric mean of 2 endogenous controls: hypoxanthine phosphoribosyltransferase 1 (HPRT1, 4326321E) and succinate dehydrogenase complex, subunit A, flavoprotein (Fp) (SDHA, Hs00188166_ml) (14). Each sample was quantified using the comparative CT method (ABI) as a relative fold-difference to the positive control cell line NMB7. All genes selected from expression profiling data were tested using TaqMan Gene Expression Assays with fluorogenic probes from ABI; their assay ID shown in table 1.
Table 1.
34 genes identified from gene expression array data with tumor to bone marrow [T:BM] ratio higher than tyrosine hydroxylase
| U95 chip | Probe | Genbank | Gene Symbol | Gene Name | T:BM | ABI assay ID |
|---|---|---|---|---|---|---|
| A | 38800_at | D45352 | STMN2 | Stathmin-like 2 | 449 | Hs00199796_m1 |
| A | 33426_at | Y00064 | CHGB | Chromogranin B (secretogranin 1) | 287 | Hs00174956_m1 |
| A | 39297_at | U38810 | MAB21L1 | Mab-21-like 1 (C. elegans) | 263 | Hs00366575_s1 |
| A | 36149_at | D78014 | DPYSL3 | Dihydropyrimidinase-like 3 | 240 | Hs00181665_m1 |
| A | 36990_at | X04741 | UCHL1 | Ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 222 | Hs00188233_m1 |
| A | 35778_at | AB011103 | KIF5C | Kinesin family member 5C | 203 | Hs00189672_m1 |
| A | 37714_at | M25667 | GAP43 | Growth associated protein 43 | 182 | Hs00176645_m1 |
| B | 47939_at | AA102788 | ELAVL4 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | 175 | Hs00222634_m1 |
| A | 40272_at | D78012 | CRMP1 | Collapsin response mediator protein 1 | 140 | Hs00609714_m1 |
| A | 38551_at | U52112 | L1CAM | L1 cell adhesion molecule | 110 | Hs00544069_m1 |
| A | 36924_r_at | M25756 | SCG2 | Secretogranin II (chromogranin C) | 104 | Hs00185761_m1 |
| C | 64258_f_at | AW016235 | MEG3 | Maternally expressed 3 | 100 | Hs00292028_m1 |
| C | 61320_g_at | AL037611 | TACC2 | Transforming, acidic coiled-coil containing protein 2 | 99 | Hs00610617_m1 |
| A | 39990_at | U07559 | ISL1 | ISL LIM homeobox 1 | 93 | Hs00158126_m1 |
| A | 35020_at | D82344 | PHOX2B | Paired-like homeobox 2b | 85 | Hs00243679_m1 |
| C | 63848_s_at | AI199503 | PCSK1N | Proprotein convertase subtilisin/kexin type 1 inhibitor | 81 | Hs00560041_m1 |
| A | 39178_at | L10333 | RTN1 | Reticulon 1 | 71 | Hs00382515_m1 |
| A | 38418_at | X59798 | CCND1 | Cyclin D1 | 68 | Hs00277039_m1 |
| A | 32650_at | Z78388 | TAGLN3 | Transgelin 3 | 66 | Hs00203119_m1 |
| D | 73450_at | AI687064 | GRIA2 | Glutamate receptor, ionotropic, AMPA 2 | 59 | Hs00181331_m1 |
| A | 41771_g_at | AA420624 | MAOA | Monoamine oxidase A | 55 | Hs00165140_m1 |
| B | 43976_at | AI857856 | KIF21A | Kinesin family member 21A | 51 | Hs00286908_m1 |
| A | 36941_at | U16954 | MLLT11 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 | 50 | Hs00199111_m1 |
| A | 38604_at | AI198311 | NPY | Neuropeptide Y | 47 | Hs00173470_m1 |
| A | 38634_at | M11433 | RBP1 | Retinol binding protein 1, cellular | 47 | Hs00161252_m1 |
| E | 91882_at | AI573279 | SOX11 | SRY (sex determining region Y)-box 11 | 47 | Hs00846583_s1 |
| A | 40201_at | M76180 | DDC | Dopa decarboxylase (aromatic L-amino acid decarboxylase) | 47 | Hs00168031_m1 |
| E | 73596_at | AI377558 | CXXC4 | CXXC finger 4 | 46 | Hs00228693_m1 |
| E | 88926_at | AA029437 | CNTFR | Ciliary neurotrophic factor receptor | 45 | Hs00181798_m1 |
| B | 54897_at | AA167714 | MAP2 | Microtubule-associated protein 2 | 44 | Hs00159041_m1 |
| A | 33890_at | AB008109 | RGS5 | Regulator of G-protein signalling 5 | 43 | Hs00186212_m1 |
| B | 52176_at | W21875 | KIF1A | Kinesin family member 1A | 41 | Hs00188913_m1 |
| A | 38839_at | AL096719 | PFN2 | Profilin 2 | 40 | Hs00160050_m1 |
| C | 63823_at | AL120032 | GABRB3 | Gamma-aminobutyric acid (GABA) A receptor, beta 3 | 39 | Hs00241459_m1 |
| A | 32300_s_at | M17589 | TH | Tyrosine hydroxylase | 37 | Hs00165941_m1 |
Statistical analysis
Bone marrow was classified as marker-positive if the gene transcript level was greater than the upper limit of normal as defined as mean + 2SD of 20 normal marrow and blood samples. The clinical endpoint tested was progression-free survival (PFS) and overall survival (OS) from the beginning of immunotherapy using Kaplan-Meier method and compared by the log-rank test.
RESULTS
MRD marker discovery by gene expression profiling
For each probe in the U95 chip, the gene expression levels of 48 stage 4 NB tumors were compared to their levels in 9 remission stage 4 marrow samples and 12 NB cell lines. We employed three criteria for marker discovery: (1) statistical significance of gene expression in tumor over marrow, using Bonferroni correction for multiple comparisons, (2) the signal ratio of gene expression in tumor versus marrow for each gene for ranking, (3) superior ratio of tumor versus marrow when compared to TH, a widely used NB marker which served as the gold standard. Using student t-test, only genes with highly significant tumor expression were chosen (p < 6×10−7 using Bonferroni correction for multiple comparisons). TH was found to have a median tumor to marrow expression ratio of 37:1. After excluding genes of ubiquitous nature like collagen and pseudogenes, as well as genes with known expression in marrow, 34 genes with ratios >37 (i.e. superior to TH) and a median expression level of ≥2,500 units were filtered from ∼16,000 unique genes. The 34 genes identified were: CCND1, CHGB, CNTFR, CRMP1, CXXC4, DDC, DPYSL3, ELAVL4, GABRB3, GAP43, GRIA2, ISL1, KIF1A, KIF21A, KIF5C, L1CAM, MAB21L1, MAOA, MAP2, MEG3, MLLT11, NPY, PCSK1N, PFN2, PGP9.5, PHOX2B, RBP1, RGS5, RTN1, SCG2, SOX11, STMN2, TACC2, TAGLN3. They were ranked in descending tumor to marrow ratio (table 1).
Sensitivity of novel NB markers
Cells from NB cell lines LAN1 and NMB7 (defined as 100,000 transcript units) were seeded into 107 normal peripheral blood mononuclear cells, as well as peripheral blood stem cells, ranging from 1 to 1000 tumor cells. Sensitivity of all 34 genes was tested by quantitative RT-PCR. Detection by TH was used as a reference. In order to prioritize potential MRD markers, the acceptance criteria in these sensitivity assays were as follows: Detection limit must be at least 1 tumor cell in 106 normal cells in both cell lines tested. If normal hematopoietic cell alone had detectable expression, the gene expression signal of 10−6 tumor cells must be at least 2 times higher than normal cell expression. Moreover, the marker must be superior to the sensitivity of TH. The highest rank was 4, and the lowest was 0 (table 2). The rank of our reference TH was 2.
Table 2.
34 novel genes were ranked according to the sensitivity selection criteria+
| Gene Symbol | T:BM | Sensitivity Rank* |
|---|---|---|
| CCND1 | 68 | 4 |
| CRMP1 | 140 | 4 |
| DDC | 47 | 4 |
| GABRB3 | 39 | 4 |
| ISL1 | 93 | 4 |
| KIF1A | 41 | 4 |
| PHOX2B | 85 | 4 |
| TACC2 | 99 | 4 |
| CHGB | 287 | 3 |
| GAP43 | 182 | 3 |
| SOX11 | 47 | 3 |
| CNTFR | 45 | 2 |
| PFN2 | 40 | 2 |
| RBP1 | 47 | 2 |
| RGS5 | 43 | 2 |
| TH | 37 | 2 |
| CXXC4 | 46 | 1 |
| ELAVL4 | 175 | 1 |
| GRIA2 | 59 | 1 |
| MAB21L1 | 263 | 1 |
| MAOA | 55 | 1 |
| NPY | 47 | 1 |
| STMN2 | 449 | 1 |
| TAGLN3 | 66 | 1 |
| UCHL1 | 222 | 1 |
| DPYSL3 | 240 | 0 |
| KIF21A | 51 | 0 |
| KIF5C | 203 | 0 |
| L1CAM | 110 | 0 |
| MAP2 | 44 | 0 |
| MEG3 | 100 | 0 |
| MLLT11 | 50 | 0 |
| PCSK1N | 81 | 0 |
| RTN1 | 71 | 0 |
Selection criteria detailed in Results section.
Highest rank was 4.
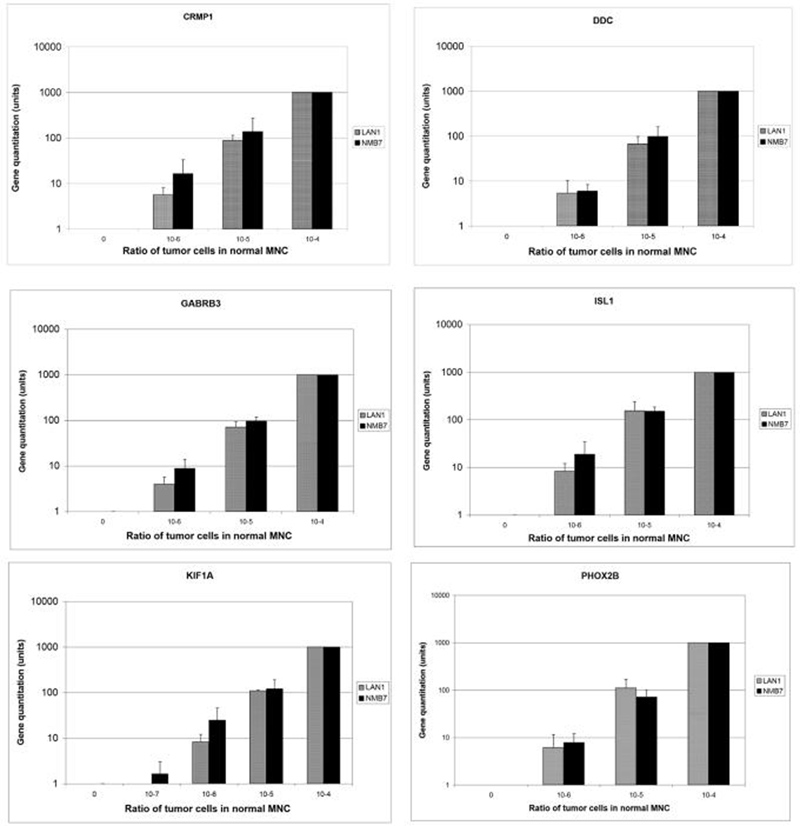
Of the 34 genes tested, 11 were superior to TH in these sensitivity assays. Among those that had the highest rank, CRMP1, DDC, GABRB3, ISL1, KIF1A, and PHOX2B also had no detectable expression in the normal cells (figure 1). As to CCND1 and TACC2, their sensitivity corrected for expression in normal mononuclear cells remained high (data not shown). In contrast, some genes which had high tumor to marrow ratios in the expression profiling lost their superiority when their sensitivity was tested by tumor cell spiking experiments. For example, as shown in table 1, STMN2, a gene with the highest tumor to bone marrow (BM) ratio (449:1), had a sensitivity rank of only 1. Another example was DPYSL3, with a tumor to BM ratio of 240:1, had an unacceptable profile with its high expression in normal mononuclear cells.
Figure 1.
Sensitivity of novel markers CRMP1, DDC, GABRB3, ISL1, KIF1A, and PHOX2B was evaluated by seeding NB cell line LAN1 and NMB7, ranging from 1 to 104 cells in 107 normal mononuclear cells (MNC). Gene expression level of 104 tumor cells in 107 normal MNC was defined as 1000 units. Quantitation was based on mean + S.D. of 3 experiments.
Marker specificity and expression in stage 4 NB tumors
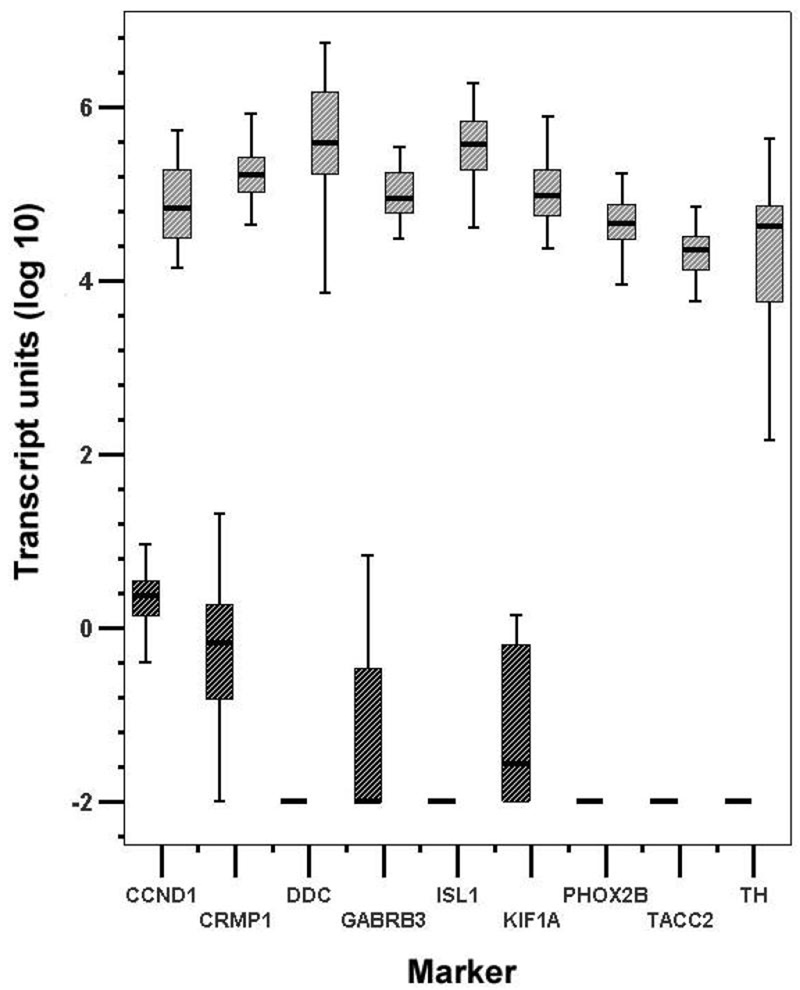
In order to further narrow down our selection of potential MRD markers of NB, only the top 8 genes, all ranked 4 in the sensitivity test, were studied using a panel of 20 normal peripheral blood and marrow samples to evaluate the specificity of these candidate markers in non-tumor bearing samples. In addition, these markers were tested by qRT-PCR in 20 stage 4 NB tumors. Transcript units in log scale of these 8 novel markers were all highly expressed in all stage 4 tumors tested with low to no detection in normal samples (figure 2). They compared favorably to TH.
Figure 2.
Differential expression of eight top ranking novel markers (CCND1, CRMP1, DDC, GABRB3, ISL1, KIF1A, PHOX2B, TACC2) and TH in 20 stage 4 NB tumors and 20 normal mononuclear cells. Tumors in gray, normal mononuclear cells in black, and transcript units in log (10) scale.
Detection of novel markers in bone marrows from 116 stage 4 NB patients treated after 2 cycles of immunotherapy
In order to determine the clinical relevance of these markers, their transcript levels were determined using marrows from 116 stage 4 patients after 2 cycles of immunotherapy on protocol IRB#9418. Since not all patients underwent full extent-of-disease workup until after the fourth cycle of treatment, their remission status at the time of marrow sampling could only be estimated: based on available tests, 50 patients were in complete remission (CR), the remaining 66 patients had clinical evidence of disease (18 very good partial response, 2 partial response, 24 stable disease, and 22 progressive disease). Disease status of patients according to marker positivity was analyzed (supplementary table 1). There was a general trend towards a higher percentage of patients with positive markers when they had clinical evidence of disease.
Marrow disease was evaluated histologically. Each marrow study consisted of 2 biopsies at 2 separate sites (usually posterior right iliac crest and posterior left iliac crest), plus 4 aspirates at 4 separate sites (usually posterior and anterior right and left iliac crests). Histology-negative marrows had complete negativity by biopsy (2/2 sites) and aspirates (4/4 sites). The frequency of multiple marker positivity according to histology status is summarized in supplementary table 2. By Kaplan-Meier analyses, both PFS (figure 3) and OS (data not shown) for 6/8 markers (CCND1, DDC, GABRB3, ISL1, KIF1A, and PHOX2B) were all highly prognostic (p<0.001). The median followup of survivors was 5.9 years.
Figure 3.
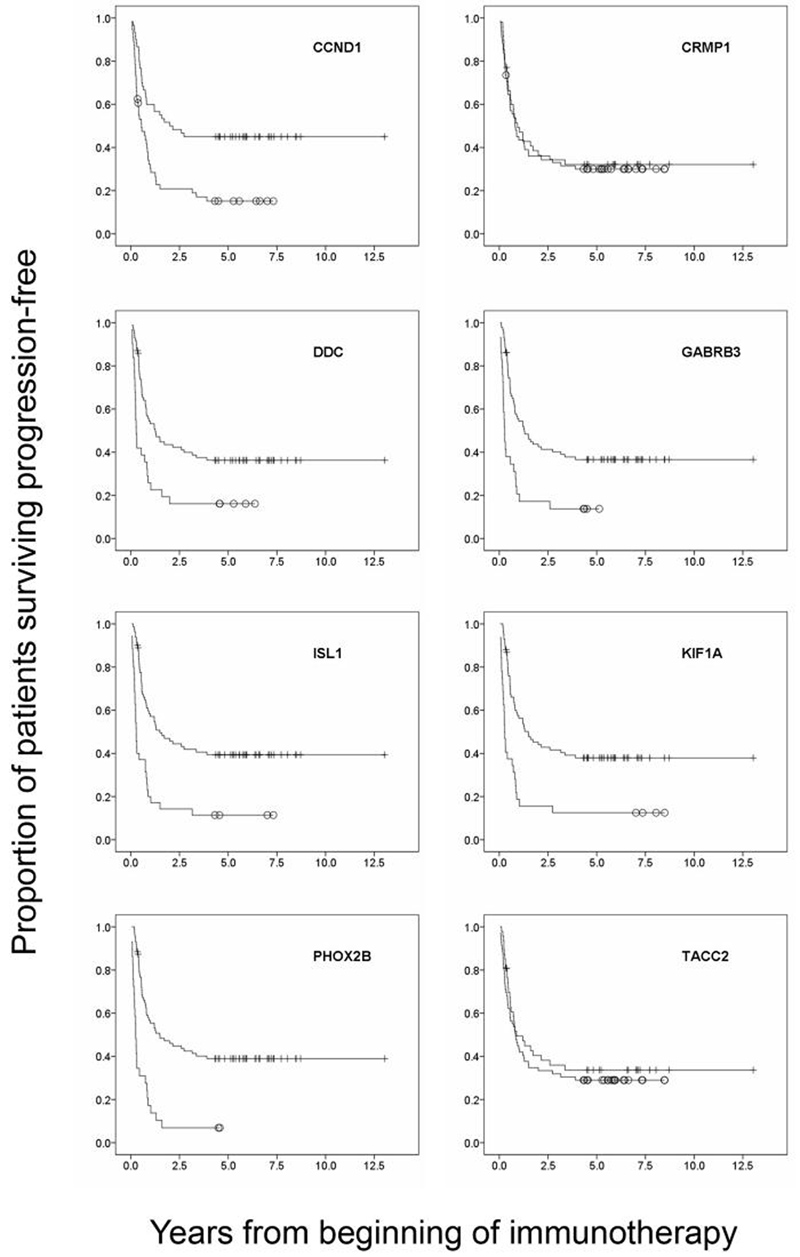
Kaplan-Meier plots of progression-free survival for the eight top ranking novel markers (CCND1, CRMP1, DDC, GABRB3, ISL1, KIF1A, PHOX2B, TACC2) with respect to marker status. Open circle: marker positive, vertical line: marker negative. Except for CRMP1 and TACC2, these markers were highly prognostic of outcome (p<0.001).
Among patients with MRD in the bone marrow, the prognostic impact of marker-positivity on PFS and OS is summarized in table 3. Three levels of marrow MRD were analyzed. For level 1, histology-negative MRD (2/2 negative biopsy plus 4/4 negative aspirates), only CCND1-positivity was found to correlate with PFS and OS with statistical significance. For level 2, aspirate-negative MRD (4/4 negative aspirates), both CCND1 and PHOX2B positivity was statistically associated with poorer PFS and OS. It should be noted that marrow samples for our marker study were all derived from pooled aspirates. Level 3 uses a broader definition of marrow MRD with ≥4/6 sampling sites being negative. CCND1, DDC, GABRB3, ISL1, KIF1A, and PHOX2B detection were all predictive of PFS, and all except GABRB3 of OS.
Table 3.
Univariate analyses of progression-free (PFS) and overall survival (OS) of stage 4 patients with minimal residual disease in the bone marrows after 2 cycles of immunotherapy
| Level 1:Histology-negative | Level 2: Aspirate-negative | Level 3: Isolated infiltration | ||||
|---|---|---|---|---|---|---|
| N=90 | N=94 | N=105 | ||||
| Marker | PFS p value | OS p value | PFS p value | OS p value | PFS p value | OS p value |
| CCND1 | 0.033 | 0.015 | 0.027 | 0.013 | 0.002 | 0.001 |
| DDC | NS | NS | NS | NS | 0.014 | 0.022 |
| GABRB3 | NS | NS | NS | NS | 0.009 | 0.067 |
| ISL1 | 0.08 | NS | 0.06 | 0.08 | <0.001 | <0.001 |
| KIF1A | NS | NS | 0.08 | 0.08 | 0.001 | 0.002 |
| PHOX2B | 0.07 | NS | 0.009 | 0.032 | <0.001 | <0.001 |
Histology-negative: negative in 2/2 biopsy and 4/4 aspirate sites
Aspirate-negative: only aspirates were considered, negative in 4/4 sites
Isolated infiltration: negative in ≥ 4/6 sampling sites
NS: p-value > 0.1
DISCUSSION
Historically, many tumor targets and markers were discovered by serology. With the advent of hybridoma technique, whole tumor cells were used to immunize lymphocytes, and clone selection was based on their differential expression of tumor over marrow/blood, or tumor over normal tissues. However, this marker discovery approach has major limitations. Lymphocytes, regardless of its being in vivo or in vitro, may be anergic or tolerized to certain antigens. Depending on the types of antigens studied, the immune repertoire of these lymphocytes can be limited, or skewed by dominant clones.
Tumor marker discovery using genome-wide expression array approach is an attractive alternative. Screening of expressed genes overcomes the issues of clonal frequency and lymphocyte restrictions. While the whole tumor approach identifies antibody before the antigen, the expression profiling approach pinpoints known genes, which are likely to have web-based tissue expression information available to filter out rapidly false leads. This marker discovery strategy was successfully applied to identify three novel markers of Ewing family tumors (8). The detection of STEAP1, CCND1, and/or NKX2-2 at diagnosis was informative and was predictive of whether patients had a higher likelihood to eventually develop new metastases and to die of this cancer. When applied to other tumor expression arrays, novel gene lists have recently been generated (data not shown). Besides genomics, proteomics and glycomics can take advantage of this approach.
The marker discovery outlined in this report was based on profiling a substantial number of stage 4 NB tumors, ensuring a representative spectrum of this metastatic cancer. The differential gene expression of NB tumors to remission marrow ratios as well as expression level was taken into account. Selection was based on tumor to marrow expression ratio superior to that of TH, a widely accepted NB marker, which serves as the gold standard. Based on these criteria, ∼16,000 genes were filtered to a list of 34 candidate genes as potential markers. However, there was no one-to-one corresponding correlation between gene expression array ranking and the ranking derived from the sensitivity experiments (table 2). This is likely because the expression level on the array depends on the target sequences of the specific gene chips. For example, even though 18/48 tumors in the array had genomic MYCN amplification, MYCN expression level did not meet the criteria we set, and was rejected in our final list of candidate genes. Additionally, glycolipid markers including beta-1,4-N-acetyl-galactosaminyl transferase (GD2 synthase) and sialyltransferase STX (ST8SiaII) were not detectable by the Affymetrix U95 Chip. This was most likely due to the substantial difference between the Affymetrix target sequence from the sequence deposited at GenBank. These findings reinforced our continual effort to discover novel molecular markers using a multipronged approach, be it genome-wide expression screen, or focusing on aberrations (e.g. MYCN) and pathways specific for tumor or metastases. We believe that both global screening and pathway-based approaches can complement the MRD marker discovery process.
Besides being highly sensitive with detection in 10−6 frequency, the 8 top ranking markers were also specific with high expression in stage 4 NB tumors (figure 2). CCND1 (Cyclin D1) has a pivotal role in controlling cyclin-dependent kinases during cell cycle progression (15), and it is over-expressed and has adverse prognostic impact in human cancers including NB (16). In addition to CCND1, DDC (dopa decarboxylase) and PHOX2B (paired-like homeobox 2b) are also associated with NB. DDC, an enzyme involved in the pathway of catecholamine synthesis, was shown to have utility as a tumor marker for NB (17). PHOX2B is a gene involved in the development of several major noradrenergic neuron populations. It is highly expressed in NB, and its germline mutation may be linked to hereditary NB (18).
The rest of our novel markers, namely CRMP1, GABRB3, KIF1A, ISL1, TACC2 have not been previously reported to be associated with NB. CRMP1 (collapsin response mediator protein 1) belonging to a family of cytosolic phosphoproteins expressed exclusively in the nervous system is involved in signal transduction pathway during neural development. GABRB3 (gamma-aminobutyric acid A receptor, beta 3), which encodes a member of the chloride ionic channel family, serves as the receptor for gamma-aminobutyric acid, the major inhibitory transmitter of the nervous system. KIF1A (kinesin family member 1A) belongs to the microtubule family involved with ATP binding. ISL1 (ISL LIM homeobox 1) is a zinc finger transcription factor, whereas TACC2 (transforming, acidic coiled-coil containing protein 2) belongs to a conserved family of proteins that are implicated in tumorigenesis by encoding a protein that concentrates at centrosomes throughout the cell cycle.
As part of our discovery algorithm, we tested these 8 candidate markers on the post-cycle 2 bone marrows of a cohort of stage 4 patients treated uniformly with an immunotherapy protocol. In addition to CCND1, a novel MRD response marker of NB reported previously (9), 5 additional markers, namely DDC, GABRB3, ISL1, KIF1A, and PHOX2B, were found to be useful in predicting patient outcome. These markers will need to be tested using an independent set of patient samples collected prospectively to confirm clinical utility. Moreover, a further fine-tuning will be required to determine which markers should be combined with GD2 synthase or TH in order to optimize the detection of marrow MRD in metastatic NB patients.
Supplementary Material
Acknowledgments
Sources of support: Supported in part by grants from the National Institutes of Health (grants CA106450 and CA118845), Robert Steel Foundation, and Hope Street Kids.
Footnotes
STATEMENT OF TRANSLATIONAL RELEVANCE
Despite achieving clinical remission, cancers like metastatic neuroblastoma often recur, due in part to the presence of subclinical minimal residual disease (MRD). Targeted therapy directed against MRD will improve outcome. The ability to measure MRD is critical for gauging the success of these targeting strategies, especially in the key metastatic compartments of marrow and blood. However, no single MRD marker will be adequate because of tumor heterogeneity. Tumor-selective mRNAs are sensitive and specific markers of active disease. In this report, using Affymetrix U95A-E gene expression array analysis on tumors from 48 stage 4 NB patients, 34 potential MRD markers were identified. Sensitivity and specificity studies narrowed the list to 8 top-ranking candidates. Using 116 well-annotated bone marrow samples collected from the same phase of an immunotherapy protocol, we further narrowed the list to 6 novel markers based on their prognostic impact on clinical outcome. We conclude that a genome-wide marker discovery approach could identify MRD molecular markers. This is particularly relevant for orphan diseases where known markers are generally scarce, and the ranking of potential markers by clinical significance can reduce false leads. We believe these novel markers will augment the precision in measuring MRD in metastatic neuroblastoma.
REFERENCES
- 1.Cheung IY, Lo Piccolo MS, Kushner BH, Kramer K, Cheung NK. Quantitation of GD2 synthase mRNA by real-time reverse transcriptase polymerase chain reaction: clinical utility in evaluating adjuvant therapy in neuroblastoma. J Clin Oncol. 2003;21:1087–93. doi: 10.1200/JCO.2003.02.055. [DOI] [PubMed] [Google Scholar]
- 2.Cheung IY, Lo Piccolo MS, Kushner BH, Cheung NK. Early molecular response of marrow disease to biologic therapy is highly prognostic in neuroblastoma. J Clin Oncol. 2003;21:3853–8. doi: 10.1200/JCO.2003.11.077. [DOI] [PubMed] [Google Scholar]
- 3.Burchill SA, Lewis IJ, Abrams KR, et al. Circulating neuroblastoma cells detected by reverse transcriptase polymerase chain reaction for tyrosine hydroxylase mRNA are an independent poor prognostic indicator in stage 4 neuroblastoma in children over 1 year. J Clin Oncol. 2001;19:1795–801. doi: 10.1200/JCO.2001.19.6.1795. [DOI] [PubMed] [Google Scholar]
- 4.Tchirkov A, Paillard C, Halle P, et al. Significance of molecular quantification of minimal residual disease in metastatic neuroblastoma. J Hematother Stem Cell Res. 2003;12:435–42. doi: 10.1089/152581603322286060. [DOI] [PubMed] [Google Scholar]
- 5.Cheung IY, Vickers A, Cheung NK. Sialyltransferase STX (ST8SiaII): a novel molecular marker of metastatic neuroblastoma. Int J Cancer. 2006;119:152–6. doi: 10.1002/ijc.21789. [DOI] [PubMed] [Google Scholar]
- 6.Cheung IY, Barber D, Cheung NK. Detection of microscopic neuroblastoma in marrow by histology, immunocytology, and reverse transcription-PCR of multiple molecular markers. Clin Cancer Res. 1998;4:2801–5. [PubMed] [Google Scholar]
- 7.Riley RD, Heney D, Jones DR, et al. A systematic review of molecular and biological tumor markers in neuroblastoma. Clin Cancer Res. 2004;10:4–12. doi: 10.1158/1078-0432.ccr-1051-2. [DOI] [PubMed] [Google Scholar]
- 8.Cheung I, Feng Y, Danis K, et al. Novel markers of subclinical disease for Ewing family tumors from gene expression profiling. Clin Cancer Res. 2007;13:6978–83. doi: 10.1158/1078-0432.CCR-07-1417. [DOI] [PubMed] [Google Scholar]
- 9.Cheung IY, Feng Y, Vickers A, Gerald W, Cheung NK. Cyclin D1, a novel molecular marker of minimal residual disease, in metastatic neuroblastoma. J Mol Diagn. 2007;9:237–41. doi: 10.2353/jmoldx.2007.060130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alaminos M, Mora J, Cheung NK, et al. Genome-wide analysis of gene expression associated with MYCN in human neuroblastoma. Cancer Res. 2003;63:4538–46. [PubMed] [Google Scholar]
- 11.Kushner BH, Kramer K, Cheung NKV. Phase II trial of the anti-G(D2) monoclonal antibody 3F8 and granulocyte-macrophage colony-stimulating factor for neuroblastoma. J Clin Oncol. 2001;19:4189–94. doi: 10.1200/JCO.2001.19.22.4189. [DOI] [PubMed] [Google Scholar]
- 12.Cheung IY, Cheung NKV. Molecular detection of GAGE expression in peripheral blood and bone marrow: utility as a tumor marker for neuroblastoma. Clin Cancer Res. 1997;3:821–6. [PubMed] [Google Scholar]
- 13.Cheung IY, Cheung NK. Quantitation of marrow disease in neuroblastoma by real-time reverse transcription-PCR. Clin Cancer Res. 2001;7:1698–705. [PubMed] [Google Scholar]
- 14.Fischer M, Skowron M, Berthold F. Reliable transcript quantification by real-time reverse transcriptase-polymerase chain reaction in primary neuroblastoma using normalization to averaged expression levels of the control genes HPRT1 and SDHA. J Mol Diagn. 2005;7:89–96. doi: 10.1016/S1525-1578(10)60013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnellan R, Chetty R. Cyclin D1 and human neoplasia. Mol Pathol. 1998;51:1–7. doi: 10.1136/mp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molenaar JJ, van Sluis P, Boon K, Versteeg R, Caron HN. Rearrangements and increased expression of cyclin D1 (CCND1) in neuroblastoma. Genes Chromosomes Cancer. 2003;36:242–9. doi: 10.1002/gcc.10166. [DOI] [PubMed] [Google Scholar]
- 17.Bozzi F, Luksch R, Collini P, et al. Molecular detection of dopamine decarboxylase expression by means of reverse transcriptase and polymerase chain reaction in bone marrow and peripheral blood: utility as a tumor marker for neuroblastoma. Diagn Mol Pathol. 2004;13:135–43. doi: 10.1097/01.pdm.0000128699.14504.06. [DOI] [PubMed] [Google Scholar]
- 18.Mosse YP, Laudenslager M, Khazi D, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75:727–30. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.