Summary
Two-hybrid screening is a standard methodology to identify and characterize protein-protein interactions that has become an integral component of many proteomic investigations. The two-hybrid system was initially developed using yeast as a host organism. However, bacterial two-hybrid systems have also become common laboratory tools and are preferred in some circumstances, although yeast and bacterial two-hybrid systems have never been directly compared. We describe here the development of a unified yeast and bacterial two-hybrid (YBTH) system in which a single bait expression plasmid is used in both organismal milieus. We use a series of leucine zipper fusion proteins of known affinities to compare interaction detection using both systems. While both two-hybrid systems detected interactions within a comparable range of interaction affinities, each demonstrated unique advantages. The yeast system produced quantitative readout over a greater dynamic range than that observed with bacteria. However, the phenomenon of “auto-activation” by baits was less problematic in the bacterial system than in yeast. Both systems identified physiological interactors for a library screen with a cI-Ras test bait; however, non-identical interactors were obtained in yeast and bacterial screens. The ability to rapidly shift between yeast and bacterial systems provided by these new reagents should provide a marked advantage for two-hybrid investigations. In addition, the modified expression vectors should be useful for any application requiring facile expression of a protein of interest in both yeast and bacteria.
Keywords: Two-hybrid, protein-protein interaction, leucine zipper, Ras, bacteria, yeast
Introduction
Yeast two-hybrid systems (1–4) are standard tools used to identify novel protein-protein interactions and to perform structure-function analysis on previously defined protein-protein interactions. Such systems are effective with a substantial fraction of eukaryotic proteins and have played an important role in high throughput proteomic analyses aimed at establishing sets of interacting proteins (e.g. (5–8). In order to increase the power of a two-hybrid approach to identify and analyze protein interactions in high throughput applications, one approach has been to translate the basic components of the yeast two-hybrid system to a bacterial host organism (9, 10). To date, the relative effectiveness of protein interaction detection in bacterial and yeast backgrounds has not been directly compared. However, there are a number of reasons to anticipate that differences might be observed. As yeast are eukaryotes, eukaryotic proteins used as “baits” in two-hybrid screens may be more likely to be appropriately folded and post-translationally modified in yeast than in bacteria, thereby increasing their chances of identifying physiological partners. However, certain proteins can be problematic as baits in the yeast two-hybrid system; for example, proteins that are normally excluded from the nucleus in eukaryotes, that are potentially sequestered via interaction with an abundant partner evolutionarily conserved in yeast, or that stimulate transcription in yeast (i.e. —that “autoactivate”). All of these potential issues would be expected to be less problematic in the bacterial two-hybrid system. To maximize chances of obtaining all relevant interactors for a protein of interest, it would be desirable to have the capability to rapidly test a given bait in both yeast and bacterial milieus.
In this current study, we have created and validated plasmids and strains that facilitate inter-conversion between yeast and bacterial protein interaction systems. We have designed a novel series of vectors in which a single plasmid containing a modified promoter drives the efficient expression of a bait protein in either yeast or bacteria, thereby permitting parallel studies in both organisms. In addition, we have constructed optimized supporting yeast and bacterial reporter strains. Using these reagents, we have generated constructs that permitted us to test a series of leucine zippers with interaction constants ranging between Kd ~10−4 and 10−15 M in both the yeast and bacterial systems using auxotrophic and quantitative reporters. We report that while both systems detect protein interactions within a comparable range of affinities, there are characteristic differences between the two systems: the yeast system possesses greater dynamic range for signal but the bacterial system appears to be less susceptible to the phenomenon of bait auto-activation. We discuss particular applications for our novel yeast/bacterial two-hybrid system.
Results and Discussion
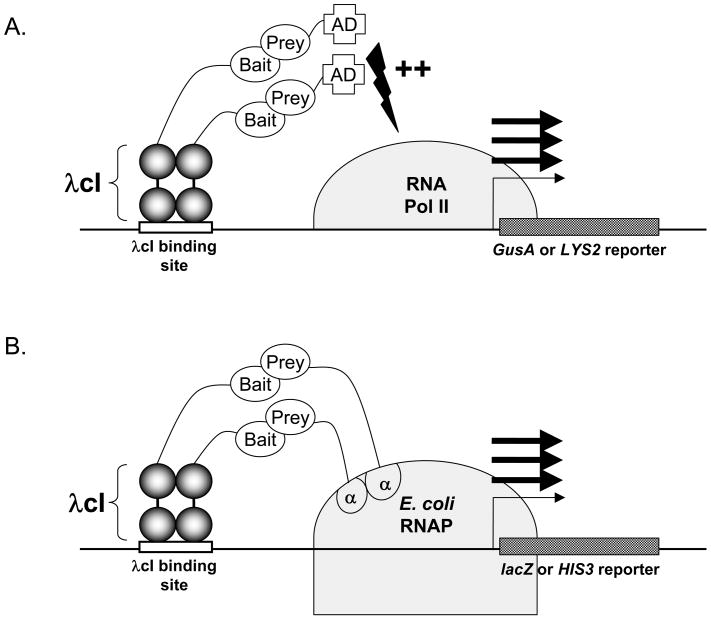
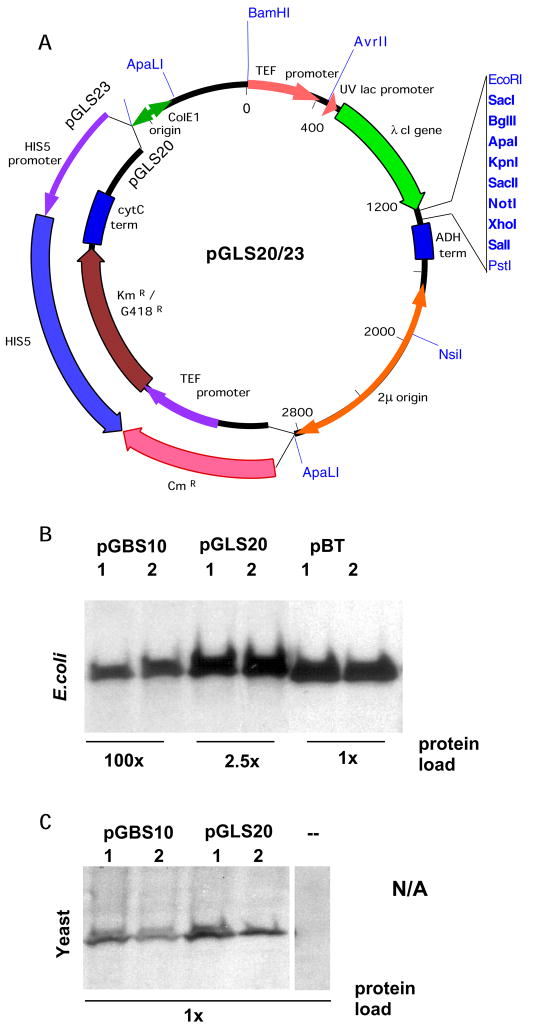
We have developed plasmids that allow the expression and parallel screening of a single bait protein in either a yeast or bacterial two-hybrid system using a single expression plasmid (Figure 1). As shown in Figure 1, bait proteins are expressed as fusions to the λ cI protein in both the yeast and bacterial two-hybrid systems. To enable this, we made several modifications to the plasmid pGBS9 (11), originally developed to express bait proteins as fusions to the λ cI repressor in a yeast two-hybrid system. The ADH1 promoter from this plasmid was replaced with a tandem promoter, in which the extremely powerful TEF1 promoter (12) from S. cerevisiae and the E. coli lpp/lacUV5 promoter both direct expression of a λ cI coding sequence and polylinker cloning site. The resulting plasmid, pGLS20, can be maintained in yeast or bacteria based on G418 or kanamycin resistance, respectively (Figure 2A). Other closely related plasmid derivatives (pGLS22, pGLS23) harbor the HIS5 gene to confer selection in yeast, and chloramphenicol resistance for selection in bacteria (Figure 2A). As shown in Figure 2B, expression of λ cI repressor using plasmid pGLS20 in bacteria is comparable to that obtained with plasmid pBT (a vector optimized for the bacterial two-hybrid system, Stratagene) and is more than 40-fold higher than that provided by the standard yeast two-hybrid expression plasmid pGBS10 (11). In yeast, expression of cI repressor fusions from pGLS20 and its derivatives is comparable to or exceeds that from pGBS10 (Figure 2C).
Figure 1. Schematics of the Yeast and Bacterial two hybrid systems.
A. In the yeast two-hybrid system shown, a dimeric λ cI-bait hybrid protein interacts with an activation domain (AD)-prey hybrid protein thereby stimulating transcription from an adjacent promoter that directs expression of a quantitative GusA or selectable LYS2 reporter gene. B. In the bacterial two-hybrid system shown, a dimeric λ cI-bait hybrid protein interacts with an E. coli RNA polymerase (RNAP) α-subunit-prey hybrid protein, thereby recruiting RNAP to an adjacent promoter that directs expression of a quantitative lacZ or selectable HIS3 reporter gene. Note that both systems utilize a λ cI-bait hybrid protein from a single plasmid effective in either organism.
Figure 2. Bait Expression from a combined bacterial/yeast expression plasmid.
A. Plasmid pGLS20, pGLS22 and pGLS23 (pGLS22 and pGLS23 only differ in the presence of an extra EcoRI site in the CmR gene of pGLS22) use a combined TEF1/uvLac promoter to express λ cI fused baits in yeast or bacteria. Plasmids are selected in yeast by selection for G418 resistance (pGLS20) or HIS5 complementation (pGLS23), and in bacteria by selection for kanamycin resistance (pGLS20) or chloramphenicol resistance (pGLS23). Relative expression of cI baits from these plasmids, versus the previously described pGBS10 (yeast two-hybrid, (11)) or pBT (bacterial two-hybrid, Stratagene) vectors is shown in bacteria (center panel). B. To demonstrate relative bait levels, equal total protein concentration was confirmed by Coomassie staining of a PAGE gel loaded with equivalent amounts of cell lysate for bacteria expressing each plasmid (not shown). Then, equal volumes of 1:40 (for pGLS20) or 1:100 (for pBT) dilutions of extracts in sample buffer were loaded in parallel with the same volume of undiluted extract from pGBS10-bearing cells. Western blots using anti-cI antibodies are shown. C. pGBS10 and pGLS20 express comparable levels of λ cI baits in yeast, based on Western analysis with antibodies to λ cI. 1, 2 denotes two independent transformants in bacteria or yeast; -, denotes yeast containing no bait plasmid.
We used these bi-functional pGLS plasmids to determine whether the yeast and bacterial two-hybrid systems exhibited any differences in their abilities to detect a series of interactions with differing affinities. To do this, we created a series of bait and prey fusion proteins using a set of previously characterized leucine zipper variants (13–15) with defined interaction affinities ranging from Kd >10−4 to 10−15 M as determined in vitro (Table 1). For analysis in the bacterial two-hybrid system, plasmid pAC-AMP-αLPL (Table 2) was used to express preys from the strong inducible lpp/lacUV5 tandem promoter as fusions to the amino-terminal domain of the RNA polymerase α subunit. For the yeast two-hybrid system, pJG4-5 (3) was used to express preys from the inducible GAL1 promoter as fusions to the synthetic transcriptional activation domain B42 (Figure 1). The ability of each zipper pair to activate transcription of a quantitative and an auxotrophic reporter was then assessed in bacteria and in yeast.
Table 1. Properties of leucine zippers used in this study.
pI calculations were made using the site at http://us.expasy.org/tools/pi_tool.html. Leucine zippers for many of the baits and their in vitro interaction properties were described previously (13–15).
| Combination | Bait | pI | Prey | Kd for bait-prey (in M) |
|---|---|---|---|---|
| 1 | EE12345L | 4.2 | EE12345L | Not detectable |
| 2 | RR12EE345L | 6.5 | RR12EE345L | |
| 3 | EE34 | 5.3 | EE34 | 8.1 × 10−4 |
| 4 | RR34 | 10.5 | RR34 | 3.9 × 10−5 |
| 5 | RR1234 L | 11.8 | RR1234 L | 2.5 × 10−7 |
| 6 | RR34 | 10.5 | EE34 | 1.0 × 10−8 |
| 7 | EE34 | 5.3 | RR34 | 1.0 × 10−8 |
| 8 | RR12EE345L | 6.5 | EE12RR345L | 1.3 × 10−11 |
| 9 | EE12RR345L | 10.4 | RR12EE345L | 1.3 × 10−11 |
| 10 | RR12345L | 12.2 | EE12345L | 1.1 × 10−11 |
| 11 | RR1234 L | 11.8 | EE1234 L | 1.0 × 10−15 |
| 12 | EE1234 L | 4.3 | RR1234 L | 1.0 × 10−15 |
Table 2.
Strains and plasmids used in this study.
| Plasmids | Selection in yeast/in E. coli | Comment/description | |
|---|---|---|---|
| Baits | |||
| pGLS20* | G418R | KmR | TEF1 promoter ensures expression of cI in yeast, while lpp/lacUV5 promoter provides for expression in E. coli |
| pGLS22/23* | HIS5 | CmR | Similar to pGLS20, see text for details |
| Reporters | |||
| pRG61 | URA3* | (KmR) | λcI operators direct transcription of the gusA gene; pRG61 is less sensitive and lower background reporter than pDR8. |
| pDR8 | |||
| pOR6 | (AmpR) | λcI operators direct transcription of the lacZ gene; | |
| Activation Domain Fusions | |||
| pJG4-5 | TRP1 | ApR | GAL1 promoter provides efficient expression in yeast of a gene fused to a cassette consisting of nuclear localization sequence, transcriptional activation domain, and HA epitope tag. |
| pAC-AMP-αLPL* pBR_UV5-αLP_* |
N/A | ApR | tandem lpp/lacUV5 promoters provides efficient expression in E. coli of a gene fused to E. coli RNAP alpha subunit residues 1-248. Plasmids differ in replication origins. |
| Strains | Relevant Genotype | Comment/description | |
| S. cerevisiae SKY191 | MATα trp1, his3, ura3, cIop-LYS2 | Reporter strains in which the expression of the LYS2 reporter gene is directed by a weak promoter bearing a λcI DNA binding site. | |
| S. cerevisiae PRT475* | MATα trp1, his3, his5, ura3, cIop-LYS2 | ||
| S. cerevisiae SKY54 | MATα trp1, his3, ura3, cIop-LYS2 | ||
| S. cerevisiae PRT50* | MATα trp1, his3, his5, ura3, cIop-LYS2 | ||
| E. coli KJ1567* | ΔhisB463, Δ (gpt-proAB-arg-lac)XIII zaj::Tn10 [F′ lacIq HIS3 aadA KanR] | Reporter strains in which the expression of the HIS3 and aadA reporter genes is directed by a weak promoter bearing a λcI DNA binding site | |
| Bacteriomatch I(Stratagene)I |
Δ (mcrA)183 Δ (mcrCB-hsdSMR-mrr)173endA1 hisB supE44 thi-1 recA1 gyrA96 relA1 lac [F′ laqIq HIS3 aadA Kanr] |
||
| E. coli AG58A(RP28)* | ΔhisB463, Δ (gpt-proAB-arg-lac)XIII zaj::Tn10 [F′ lacIq lacZ KanR] | Reporter strain in which the expression of the lacZ reporter gene is directed by a weak promoter bearing a λcI DNA binding site | |
| Libraries | Vector | Comment/description | |
| YTH Human HeLa Cell Library | pJG4-5 | Hela S-3 cells, Primary size: 9.6×106 Primer: UdT, average insert size: 1.5 kb | |
| BacterioMatch® II Human HeLa Cell Library | PTRG: see Stratagene Manual for details | Hela S-3 cells, Primary size: 4.5×106 Primer: UdT, average insert size: 1.3 kb | |
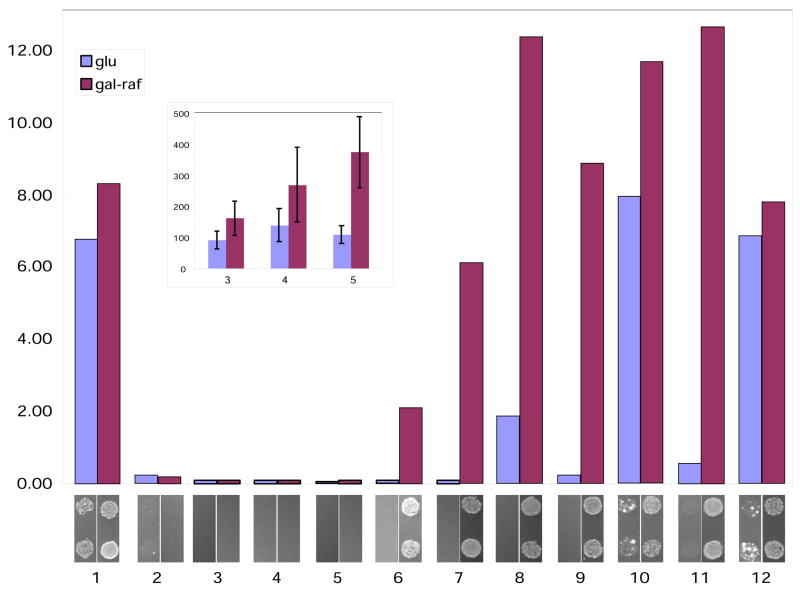
Our results in the yeast-based system demonstrate that zipper bait-prey combinations activate transcription of a quantifiable β-glucuronidase (GusA) reporter over a substantial range of affinities (Figure 3, bar graph). In this assay, zipper pairs with reported interaction dissociation constants of 1 × 10−8 M or lower (lanes 6–12) strongly activated reporter gene expression, as detected using a colorimetric substrate (PNP-gluc). Those with Kd values of 2.5 × 10−7 M or higher (with one exception—see below) did not strongly activate the reporter gene (Figure 3A, lanes 1–5). β-glucuronidase activity was generally induced ~30–180 fold over baseline values with the higher affinity leucine zipper pairs. Additional testing of the lower affinity interacting pairs using a more sensitive fluorescent substrate for beta-glucuronidase, MU-gluc (Figure 3A, inset), indicated that it was also possible to convincingly detect interactions in the range of 10−7 M, although the stimulation of GusA gene expression seen in these samples is markedly less strong than those obtained with interactions in the 10−8 M range. With the auxotrophic reporter strain (Figure 3, panels below bar graph), cells grew under selective conditions only if the interacting zippers possessed dissociation constants of 1 × 10−8 M or lower, paralleling the results obtained with the quantitative GusA reporter. The system did not have significant ability to discriminate between interactions with dissociation constants of 10−8 M or lower, suggesting the expression of the reporter gene was saturated. Importantly, for some of the baits examined, expression of the bait alone in the absence of the prey was sufficient to strongly activate transcription of the reporters, making it difficult to convincingly demonstrate protein interaction (see Figure 3, samples 1, 10, and 12).
Figure 3. Activation of colorimetric and auxotrophic reporters by zipper interaction in yeast.
Lane numbers below bar graph represent pairs of samples defined in Table 1. Bar graph reflects relative reporter activity measured by beta-glucuronidase assay using PNP-gluc as a substrate. Expression of the AD-fusion protein in yeast is inducible by galactose. Therefore, beta-glucuronidase activity revealed upon the growth on glucose (blue bars with “glu”) represents mainly contribution of bait alone, while activity upon the growth in the presence of galactose (dark-red with “gal-raf”) reflects the interaction between bait and prey. Results shown represent mean values for 3 independent experiments. Standard deviation (not shown) was variable but did not exceed 25% of the experimental value, as is typical for yeast two hybrid experiments. Inset, indicated samples re-analyzed using MU-gluc as a substrate. For context, values obtained for combination 6 (with a Kd of 1 × 10−8 M), were more than 10-fold higher than those with combination 5 with the MU-gluc substrate, indicating a significant discriminating function of the yeast two-hybrid system in this affinity range (not shown). Shown below bar graph is the growth of two representative spots of colonies 2 days after plating to selective medium.
Data shown are obtained using the SKY191 strain and pGLS20 as bait plasmids. Similar results were obtained using a combination of the PRT50 strain and pGLS22 bait plasmid (data not shown).
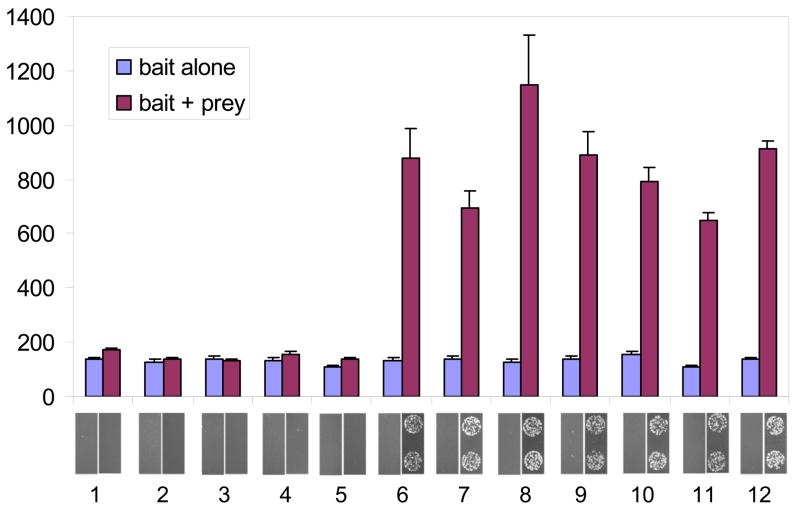
We next examined the abilities of the same zipper bait-prey combinations to activate transcription in the bacterial two-hybrid system (Figure 1) using the quantifiable lacZ reporter (Figure 4). Consistent with our results in the yeast-based system, leucine zipper pairs with reported dissociation constants lower than 10−8 M clearly stimulated expression of the lacZ reporter gene (Figure 3, samples 6–12) whereas interaction pairs with dissociation constants 2.5 × 10−7 M or higher failed to stimulate lacZ expression (Figure 3, samples 1–5). We also analyzed zipper-based activation of the auxotrophic reporter HIS3 (Figure 4, panels below bar graph). Results obtained using the auxotrophic HIS3 reporter gene closely paralleled those obtained with the lacZ reporter: only cells harboring zipper pairs with dissociation constants of 1 × 10−8 M or lower showed growth after 24 hours on selective plates. In contrast to the results obtained in the yeast-based system, none of the baits tested exhibited autoactivation in the absence of prey partners (compare samples 1, 10 and 12 in Figures 3 and 4).
Figure 4. Activation of colorimetric and auxotrophic reporters by zipper interaction in bacteria.
Lane numbers below bar graph represent pairs of samples defined in Table 1. Bar graph reflects relative reporter activity measured by β-galactosidase assay using ONPG as a substrate. β-galactosidase values are expressed in Miller units and represent the mean of three independent measurements with standard errors of the mean shown. The panel below the bar graph presents the growth of two representative spots of colonies 24 hours after plating to selective medium.
These results suggest differential advantages for detecting protein-protein interactions in the yeast and bacterial two-hybrid systems. First, our results using quantifiable reporters suggest that the yeast-based system possesses a broader dynamic range for detecting interactions (contrast Figures 3 and 4). In the yeast system, interactions characterized by dissociation constants as high as 10−8 M could be detected as an increase in GusA reporter gene expression (or as high as 10−7 M if a more sensitive substrate for GusA detection was used). In contrast, in the bacterial system, only interactions characterized by dissociation constants 10−8 M or lower could be detected as an increase in lacZ expression. Second, we note that the experiments performed using bacterial two-hybrid system yield colonies on selective medium somewhat more quickly than those done in the yeast system (one day versus two). Third, our results also suggest that autoactivation by bait proteins may be less problematic in bacteria than in yeast, as at least some proteins that are autoactivators in the yeast two hybrid system are not in the bacterial two-hybrid system (compare lanes 1, 10, 12 in Figures 3 and 4). This finding is not entirely surprising given the fundamental differences in mechanisms of gene activation and the evolutionary distance between prokaryotes and eukaryotes. The ability to use some baits that are auto-activating (unusable) in yeast in the bacterial two-hybrid system is a potentially significant advantage.
Our data also suggest that the threshold interaction strength required for robust transcriptional activation is similar in both organisms. In both the yeast and bacterial systems, full activation appears to require an interaction affinity between bait and prey fusion proteins defined by a dissociation constant in the 10−8 M range or lower. Although our results demonstrate a sharp transition between no activation and full activation of the reporter genes, previous studies in both systems have demonstrated that the magnitude of transcriptional activation observed can be correlated with the affinity of the bait-prey interaction (10, 16). While we do not know the precise reason for this difference in our results compared with previous studies, we note that Estojak and coworkers assessed interactions using a series of reporters of varying stringency (i.e. containing differing numbers of binding sites for the baits) to expand the detection range: there is no technical limitation to using a similar strategy with this new system. Overall, our results strongly suggest that use of the current system as a selection tool will work best for detecting interactions with dissociation constants in the mid-to-high nanomolar range.
The most demanding test of a protein interaction system is its ability to identify physiologically relevant interacting proteins from library screens. To test the YBTH system, we used a cI-Ras bait, as the interaction profile of Ras is well defined (17, 18). This bait was used to screen HeLa cDNA libraries in yeast and in bacteria, screening comparable numbers of primary transformants in each organism (see Methods). As shown (Table 3), both screens identified at least some clearly relevant interactors for Ras, with the yeast screen identifying clones for A-Raf (4) and Krit-1 (19), and the bacterial screen identifying RGL2/(20). Both screens also identified a number of clones with uncertain relevance towards Ras, that may or may not represent non-specific interactors; as well as a number of proteins frequently identified as false positives. Somewhat surprisingly, given that both libraries were prepared from HeLa cell mRNA, there were no overlapping isolates in the two screens, even though multiple isolates of some clones were obtained and the number of primary transformants was in excess of 3 × 106 in each case. This may indicate that specific Ras interactions are more readily detected in one or the other organism. Hypothetically, a protein requiring post-translational modification to interact with Ras may be more readily detected in yeast; while a protein that not only interacts with Ras, but with other eukaryotically conserved signaling partners, may be more available to interact with Ras in bacteria. Together, these results indicate that this system is robust for screening purposes: and the facile use of the pGLS vectors in both organisms in parallel may increase coverage and accuracy in screening.
Table 3. Comparative results of yeast and bacterial two-hybrid screening.
Accession number for clones are available on request.
| YTH clones total/indep | BTH clones total/indep | |
|---|---|---|
| Clear Ras Relevance | ||
| Krit-1 | 1/1 | N |
| ARaf | 5/1 | |
| RGL-2 | N | 7/2 |
| Ras relevance not known | ||
| Homo sapiens suppression of tumorigenicity 13 | 2/1 | N |
| zinc finger protein 616 | 3/1 | |
| hematopoietically | 1/1 | |
| expressed homeobox | ||
| Ras-related GTP binding | 1/1 | |
| dynein | 1/1 | |
| maspin | 1/1 | |
| clone CS0DM007YF21 | 1/1 | |
| chromosome 1 open reading frame 37 | 1/1 | |
| general transcrip-tion factor IIIC | N | 1/1 |
| glutathione peroxidase 4 | 3/1 | |
| guanine nucleotide binding protein | 1/1 | |
| gamma-interferon-inducible lysosomal thiol reductase | 1/1 | |
| latent transforming growth factor beta binding protein 4 | 1/1 | |
| Previously described as common false positive | ||
| CPEO mitochondrion | 2/1 | N |
| heat shock 90kDa protein 1 | 2/1 | |
| ribosomal protein L10a | 1/1 | |
| ribosomal protein S7 | 1/1 | |
| T108 mitochondrion | 1/1 | |
| ribosomal protein L26 | N | 1/1 |
| Ferritin | 1/1 | |
| NADH dehydrogenase | 1/1 | |
| ribosomal protein S2 | 1/1 | |
Finally, we note that (to our knowledge) this is the first description of a promoter combination that is potent in both yeast and bacterial milieus. In fact, we have found that our pGLS plasmids express sufficient levels of bait fusion proteins for activity in the bacterial two-hybrid system even without inducing the strong bacterial promoter. Lastly, while this article focuses on the use of the pGLS plasmids in a two-hybrid context, we anticipate that our general promoter design might also be useful in other functional characterization studies.
Methods
Molecular and Microbiological Manipulation
The cloning of novel constructs was performed using conventional protocols. Details of the sequences and cloning sites encompassed in the plasmids described in the Results section, as well as other basic characterizations of expression properties of these plasmids, are available at (21). Media and growth conditions used are described in (22).
Briefly, plasmid pGLS20 was constructed by replacing the ADH1 promoter of pGKS9 with a combination of the TEF1 promoter (from the pLexZeo plasmid, Invitrogen) and a lpp/lacUV5 promoter (from the pBT plasmid, Stratagene). To produce pGLS23, a HIS5CmR cassette was constructed in pCR2.1 vector by combining a HIS5 cassette from pJFK (R. Hopkins, unpublished) and a CmR cassette from pMW108. This cassette was then used to replace the G418R cassette in pGLS20. The bacterial two-hybrid prey plasmid pAC-AMP-αLPL was constructed by replacing the chloramphenicol resistance gene present in plasmid pKJ1267 (J.K.J, unpublished) with the ampicillin resistance gene from plasmid pACYC177. To fuse the various leucine zippers to the amino-terminal domain and inter-domain linker of the E. coli RNA polymerase-α subunit, DNA fragments encoding the zipper variants were inserted into the plasmid using unique NotI and XhoI restriction sites. To fuse leucine zippers to λ-cI and B42 moieties of the bait and yeast prey (pJG4-5) plasmids, DNA fragments encoding the zipper variants were inserted into the plasmid using unique EcoRI and XhoI restriction sites. Further information about cloning strategies used for plasmid construction, or details of yeast or bacterial strain construction and characterization, are available upon request.
Leucine Zippers
Leucine zipper sequences were chosen from among peptides described in (13–15). DNA was synthesized artificially to encode the described peptide sequences. All leucine zippers have the same length and differ only in the amino acids in positions g and e of the coiled coil (marked above sequence). Shown is the amino acid sequence of the zipper RR12EE345; helices 1, 3, 5 are underlined and helices 2 & 4 are double underlined, and the variable amino acids are shown in bold. Thus, in the example shown there are R’s in positions g and e of helices 1 &2, (hence the nomenclature of the molecule starts with RR12), while E’s in the corresponding positions of the helices 3,4 &5 cause nomenclature of the molecule to end with EE345. Complete details are available upon request.

Bait and prey expression
The expression of the bait and prey proteins (except for bacterial RNA polymerase-α fusions, for which no antibody was available) was confirmed by Western analysis, with primary antibody to cI for baits (1:5000), or hemagglutinin (1:1000) for preys expressed in yeast. To compare expression levels of cI protein in E. coli, corresponding plasmids were transformed into the DH5α strain and protein extracts prepared from exponentially growing cultures. Equal protein concentration was confirmed by Coomassie staining of a PAGE gel, then equal volumes of 1:40 (for pGLS20) or 1:100 (for pBT) dilutions of extracts in sample buffer were loaded in parallel with the same volume of undiluted extract from pGLS10-bearing cells. Proteins were resolved on a PAGE gel, and Western blot analysis was performed, using anti-cI antibodies. To compare expression levels of cI protein in yeast, corresponding plasmids were transformed in SKY191 strain and protein extracts prepared from the exponentially growing cultures. Equal protein loading was confirmed by Coomassie staining samples resolved on a PAGE gel (not shown). Then, equal volumes of extracts in sample buffer were loaded on the gel, and Western blot analysis was performed.
Reporter assays
For yeast, the activity of quantitative reporters was determined on a plate reader using a technique modified from Serebriiskii et al. 2000. Briefly, 50 μl of cultures exponentially growing in the wells of 96-well cells was added an equal volume 2 × Z-buffer containing 2mg/ml of PNPGluc and 50% Y-PER (Pierce). Activity was calculated as (OD420f-OD420i) divided by OD600, where the difference between OD420i and OD420f (initial and final readings) reflects the conversion of the colorless substrate into yellow product over a period of time from ~10–30 minutes, and OD600 is a measure of cell density in a given sample. For each data point for each yeast experiment, activities of 5 to 8 clones were measured and averaged. All readings were taken in a plate reader; it was previously shown (23) that plate reader measurements and derivative units are proportionally correlated with the OD units taken on a spectrophotometer.
For fluorescence detection, an equal volume of 2 × Z-buffer/50% Y-PER containing 0.8 mg/ml of MU-Gluc was added. Increase in fluorescence (excitation 355 nm, emission 460 nm) reflected the conversion of the colorless substrate into fluorescent product over a period of time from ~3–10 minutes, while OD600 was a measure of cell density in a given sample. The pRG61 plasmid was used as reporter in these experiments. For bacterial β-galactosidase reporter gene measurements, assays were performed essentially as described (24). Briefly, cultures inoculated from a fresh single colony were grown to mid-log phase and lysed by adding 1/10 volume PopCulture™ (Novagen). In a 96 well microtiter plate, 15 μl cell lysate was added to a mixture of 135 μl Z buffer and 30 μl 4 mg/ml ONPG to start the reaction. Kinetic assays were carried out by monitoring OD415 from 0–30 minutes using a plate reader. Additional details can be found at www.zincfingers.org. All bacterial β-galactosidase assays were done in triplicate. Auxotrophic reporters were assayed as described in (22). Bait and prey plasmids were transformed into corresponding selection strain, S. cerevisiae SKY191 or pRT50, or E. coli KJ1567. Growth on selection plates was measured over 5 days (yeast; note, all colonies that grew were prominent at 2 days) or 1 day (bacteria).
Targeted interactions using HRas as a bait
Full-length cRaf, BRaf, RalGDS and Rin1 genes were cloned in pJG4-5 plasmid. cRaf-RBD, RalGDS-RBD and full-length BRaf, wer cloned in pBR-UV5-αLP plasmid. To analyze interactions between defined pairs of proteins, bait and preys were co-transformed in the corresponding yeast or bacterial reporter strains. YTH interaction were assayed in SKY54, and BTH in Bacteriomatch II reporter strain.
Library screenings
Screening of the YTH library and analysis of primary isolates was done essentially as described in (22) using the pOR6 lacZ reporter. Briefly, about 3.5 × 106 cells carrying plasmids from the HeLa cDNA library were plated. 130 clones appearing on the auxotrophic selection plates were further examined. Off 28 positives (in which an initial positive phenotype was repeated), 24 were sequenced. The YBTH system developed here is fully compatible with strains and library reagents from Stratagene (the Bacteriomatch system). To emphasize compatibility, screening of the BTH library and analysis of primary isolates was done with this system (see Table II) essentially as recommended by the supplier. Briefly, about 5.5 × 106 cells carrying plasmids from the HeLa cDNA library (avg. insert size 1.3 kb) were plated. 96 clones appearing on the auxotrophic selection plates were characterized, and of 22 positives with a reproducible phenotype, 18 were sequenced.
Acknowledgments
We thank Dr. Paul Watt for critical review of the manuscript. We are grateful to Yijing Groeber for assistance in library screening. This work was supported by ACS pilot funding (to IGS); awards from NCI Translational Pilot Project funding, NIH RO1CA63366, and the Pennsylvania Tobacco Health Research Formula Fund (to EAG); core grant CA06927 (to Fox Chase Cancer Center); NIH K08 DK02883 and MGH Department of Pathology start-up funds (to JKJ); and support from the National Health and Medical Research Council of Australia (to RH).
References
- 1.Fields S, Song O. A novel genetic system to detect protein-protein interaction. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Nat Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gyuris J, Golemis EA, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 4.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 5.Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–7. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 7.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci U S A. 2000;97:7382–7. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 11.Serebriiskii IG, Mitina O, Pugacheva E, Benevolenskaya E, Kotova E, Toby GG, Khazak V, Kaelin WG, Chernoff J, Golemis EA. Detection of peptides, proteins, and drugs that selectively interact with protein targets. Genome Res. 2002;12:1785–91. doi: 10.1101/gr.450702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagashima K, Kasai M, Nagata S, Kaziro Y. Structure of the two genes coding for polypeptide chain elongation factor 1 alpha (EF-1 alpha) from Saccharomyces cerevisiae. Gene. 1986;45:265–73. doi: 10.1016/0378-1119(86)90024-7. [DOI] [PubMed] [Google Scholar]
- 13.Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. Embo J. 1994;13:2849–61. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krylov D, Barchi J, Vinson C. Inter-helical interactions in the leucine zipper coiled coil dimer: pH and salt dependence of coupling energy between charged amino acids. J Mol Biol. 1998;279:959–72. doi: 10.1006/jmbi.1998.1762. [DOI] [PubMed] [Google Scholar]
- 15.Moll JR, Ruvinov SB, Pastan I, Vinson C. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10(-15) M. Protein Sci. 2001;10:649–55. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbett KD, Alber T. The many faces of Ras: recognition of small GTP-binding proteins. Trends Biochem Sci. 2001;26:710–6. doi: 10.1016/s0968-0004(01)01974-0. [DOI] [PubMed] [Google Scholar]
- 18.Campbell PM, Der CJ. Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin Cancer Biol. 2004;14:105–14. doi: 10.1016/j.semcancer.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21-22. Oncogene. 1997;15:1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 20.Peterson SN, Trabalzini L, Brtva TR, Fischer T, Altschuler DL, Martelli P, Lapetina EG, Der CJ, White GC., 2nd Identification of a novel RalGDS-related protein as a candidate effector for Ras and Rap1. J Biol Chem. 1996;271:29903–8. doi: 10.1074/jbc.271.47.29903. [DOI] [PubMed] [Google Scholar]
- 21.Serebriiskii I, Golemis EA. ( http://www.fccc.edu/research/labs/golemis/InteractionTrapInWork.html)
- 22.Serebriiskii IG, Joung JK. Yeast and bacterial two-hybrid selection systems for studying protein-protein interactions. In: Golemis E, editor. Protein-Protein Interactions: A Molecular Cloning Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. pp. 93–142. [Google Scholar]
- 23.Serebriiskii IG, Toby GGGEA. Streamlined yeast colorimetric reporter assays, using scanners and plate readers. Biotechniques. 2000;29:278–9. 282–4, 286–8. doi: 10.2144/00292st03. [DOI] [PubMed] [Google Scholar]
- 24.Thibodeau SA, Fang R, Joung JK. High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. Biotechniques. 2004;36:410–5. doi: 10.2144/04363BM07. [DOI] [PubMed] [Google Scholar]
- 25.Serebriiskii I, Khazak V, Golemis EA. A two-hybrid dual bait system to discriminate specificity of protein interactions. J Biol Chem. 1999;274:17080–17087. doi: 10.1074/jbc.274.24.17080. [DOI] [PubMed] [Google Scholar]