Abstract
Background
Much effort has been devoted to development of cancer therapies targeting EGFR, based on its role in regulating cell growth. Small-molecule and antibody EGFR inhibitors have clinical roles based on their efficacy in a subset of cancers, generally as components of combination therapies. Many cancers are either initially resistant to EGFR inhibitors or become resistant during treatment, limiting the efficacy of these reagents.
Objective/Methods
To review cellular resistance mechanisms to EGFR-targeted therapies.
Results/Conclusions
The best validated of these mechanisms include activation of classic ATP-binding casette (ABC) multidrug transporters; activation or mutation of EGFR; and overexpression or activation of signaling proteins operating in relation to EGFR. We discuss current efforts and potential strategies to override these sources of resistance. We describe emerging systems-biology-based concepts of alternative resistance to EGFR-targeted therapies, and discuss their implications for use of EGFR-targeted and other targeted therapies.
Keywords: ABC transporter, antibody, cetuximab, EGFR, erlotinib, network, resistance, tyrosine kinase inhibitor
1. Introduction
Therapeutic targeting of biomolecules has become a dominant theme in modern day oncology. The success achieved with imatinib [1], bevacizumab [2] and a small number of other drugs [3–5] has been of enormous clinical benefit. However, these outcomes are in stark contrast with the no less spectacular failures of many other targeted agents (e.g. gefitinib [6], farnesyl transferase inhibitors [7]). Agents targeting EGFR fall into a middle ground: some of them are extremely successful for specific types of cancer, and in a subset of patients. Better understanding of why some tumor types respond and others do not, and why some patients respond to EGFR-targeting agents and others do not, would produce vast clinical benefits. Given the particular importance of the EGFR signaling pathway in cancer, we have here systematically analyzed potential factors affecting tumor susceptibility or resistance to EGFR inhibitors.
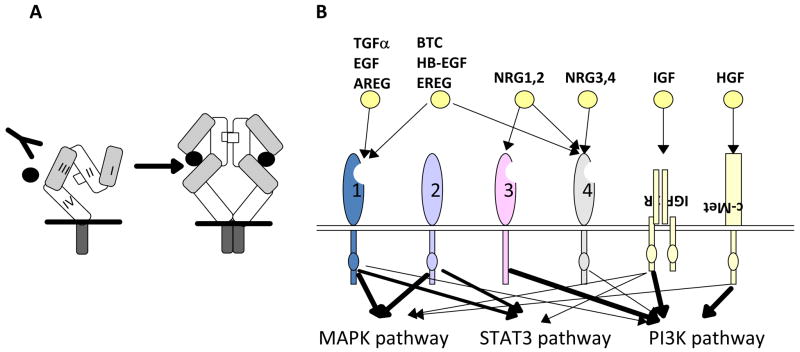
The reason EGFR has been viewed a critical target involves its role as a central regulator of cell proliferation, survival and migration in normal and cancerous cells, and EGFR has long been a target of intense interest and effort for development of targeted cancer therapeutic agents. The structure of EGFR is shown in Figure 1A. EGFR is a transmembrane receptor tyrosine kinase (RTK). The extracellular, cysteine-rich domain binds activating ligands. EGFR ligands are expressed on the cell surface as tethered precursors that require proteolytic cleavage to be released to the interstitial space. A family of such proteases (sheddases, also known as a disintegrin and metalloprotease, or ADAM, proteases) release the ligands EGF, betacellulin, epiregulin, TGF-α, amphiregulin, and heparin-binding EGF-like growth factor (HB-EGF) [8]. Once released from the membrane, the 49–85-amino acid mature growth factors are able to bind the EGF receptors. In 1997, Lemmon et al. first suggested a model in which one EGF monomer binds to one EGFR monomer, and receptor dimerization involves subsequent obligate association of two monomeric (1:1) EGF EGFR complexes, associated with activation [9]. The intracellular domain of EGFR includes a ligand- and dimerization-activated tyrosine kinase, which upon activation autophosphorylates EGFR at residues including Y992, Y1045, Y1068, Y1148 and Y1173 [10], and also phosphorylates other binding partners, as discussed below. Cooperative physical interaction between adjacent kinase domains of adjacent erythroblastic leukemia viral (v-erb-b) oncogene homolog (ErbB) family molecules is required for EGFR activation [11,12]. The normally auto-inhibited EGFR kinase undergoes conformational changes upon allosteric contact with another EGFR kinase domain, a mechanism strikingly similar to the activation of cyclin-dependent kinases by their cognate cyclins. The increased number of EGFR molecules on the cell surface associated with overexpression in cancer is predicted to increase the rate of kinase autoactivation [12].
Figure 1. EGFR/(v-erb-b) oncogene homolog 1 (ErbB1) and the ErbB protein family.
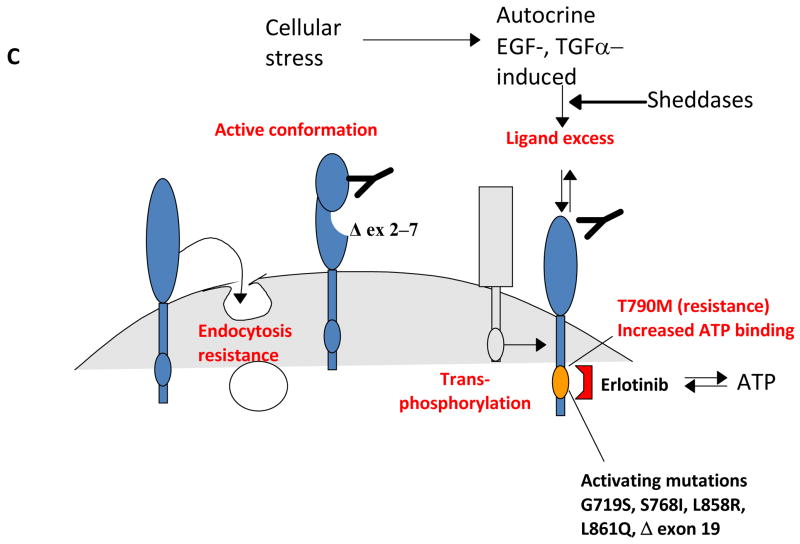
A, EGFR structure. Defined protein domains are numbered I‐IV [113]. Binding of ligand (black circles) to domains I and III of EGFR induces unfolding from a tethered to an ‘activated’ conformation. The resulting exposure of dimerization domain II allows it to pair with another EGFR molecule, or heterodimerize with another ErbB family member. Intracellular juxtaposition of the EGFR kinase domains induces trans-phosphorylation of essential tyrosine residues, activating downstream signaling. Cetuximab targets domain III, preventing ligand binding and subsequent activation steps. B. The ErbB receptor family, collaborating receptors, and receptor ligands [10]. EGFR homodimerizes, and heterodimerizes with all other ErbB family members [219]. All family members except ErbB3 contain a kinase domain (small ovals); ErbB3 is active in downstream signaling because of transphosphorylation. The profile of ligand binding to specific ErbB family members is indicated. AREG: amphiregulin; BTC: betacellulin; EREG: epiregulin; HB-EGF: heparin-binding EGF-like growth factor; HGF: hepatocyte growth factor; NRG: neuregulin; PI3K: phosphinositol 3 kinase; STAT :Signal transducer and activator of transcription. Signals emanating from c-Met, IGF-1R and ErbB3 modulate similar downstream outputs as ErbB family members: see text for details. C. Mechanisms of resistance to anti-EGFR agents. Deletion of EGFR exons 2-7 leads to constitutively active conformation of the EGFR [101,104]. EGFR kinase domain mutations, such as the common T790M mutation [86], may result in increased ATP binding and resistance to erlotinib. Receptor non-intrinsic mechanisms of resistance to anti-EGFR agents include ligand excess [115], resistance to antibody-induced endocytosis [16], or transphosphorylation [129,130].
The intracellular domain of EGFR also encompasses a motif regulating internalization [13]. Oligomerization of the ligand-activated EGFR triggers a rapid process of receptor internalization in parallel with the autophosphorylation/activation process, which allows regulated removal of the protein from the cell surface into intracellular compartments, limiting the duration of signaling [14,15].
EGFR (also known as ErbB1) is the initially characterized member of a four-member group of proteins that also includes ErbB2/human EGFR 2 (HER2), ErbB3, and ErbB4 (Figure 1B). As part of its activation process, EGFR homodimerizes, but also in some cases heterodimerizes with ErbB2 and other family members to create an active signaling unit [16]. The four ErbB proteins have overall structural similarity, with some key differences. For example, ErbB2 is not ligand-activated, but activated based on its heterodimerization with other family members [17]; in normal cells, EGFR/ErbB2 heterodimers are more active than EGFR homodimers due to constitutive kinase activity of ErbB2, but this is regulated by the relative low expression level of ErbB2 in normal cells [18]. As another example, ErbB3 is ligand-activated, but by a different set of ligands than EGFR, and lacks an active kinase domain, so that its main transmission of signals downstream is through its heterodimeric partners [19,20].
There are three very well described and physiologically important signaling outputs of activated EGFR homo- or heterodimers:
EGFR activates RAS viral oncogene homolog (Ras) signaling. The adaptor protein growth factor receptor-bound protein 2 (GRB2) either directly or with assistance of another scaffold protein, v-src sarcoma viral oncogene homology 2 domain-containing protein (Shc), binds activated EGFR at auto-phosphorylated phosphotyrosines, and activates son of sebvenless (SOS), a GTP-exchange factor for Ras, initiating signaling cascades that proceed through v-raf murine sarcoma viral oncogene homolog (Raf)/MAP kinse-ERK kinase (MEK)/extracellular signal-regulated kinase (ERK), influencing proliferation and cell cycle; v-ral simian leukemia viral oncogene homolog guanine nucleotide dissociation stimulator (RalGDS), influencing proliferation and migration; and phospho-inositol 3 kinase (PI3K), and thence to the important PI3K effector v-akt murine thymoma viral oncogene homolog (AKT), governing survival responses.
EGFR activates signal transducer and activator of transcription (Stat)3 and 5 leading to their phosphorylation, nuclear translocation, and transcriptional activation of genes involved in proliferative response.
EGFR directly binds and activates the p85 subunit of PI3K, thus providing a second stimulus to activate AKT. Although these are some of the most studied interactions, EGFR also interacts with a number of other signaling proteins; a recent peptide library screen identified multiple overlapping sites for over 40 interactions between EGFR and cytosolic proteins [10,21]. Soon after its initial discovery in 1980 [22], abnormal function of EGFR was identified as cancer-promoting, based on the discovery that the v-erbB oncogene, encoded by the erythroblastosis virus, represented a truncated form of EGFR [22–24]. The v-erbB truncation constitutively activated the tyrosine kinase activity of the protein, driving proliferation and apoptosis resistance in tumors. After three decades of study, several distinct categories of oncogenic lesions influencing endogenous EGFR activity in human tumors have been documented as clinically significant (Figure 1C). One of the best studied is activation of the EGFR signaling arising either from increased gene copy number (induced by amplification or polysomy [25,26]) or from feedback upregulation in response to cellular stresses [27–29]. Second, in some cancers, the EGFR ligands are overexpressed [30]. Third, increased activity of the EGFR signaling is mediated through activating mutations in its kinase domain, or via structural alterations of the extracellular domain such as in variant III (EGFRvIII) discussed below. These direct mechanisms of EGFR activation are observed in many different types of cancer, at significant frequencies. For example, increase in EGFR gene copy number is seen in approximately one fifth of non-small cell lung cancers (NSCLC) [31] and is found in populations partially overlapping with those possessing EGFR kinase mutations (most commonly seen in female non-smoking lung adenocarcinoma or bronchoalveolar carcinoma patients) [32]. Finally, in addition to cancers with lesions directly related to EGFR hyperactivity, EGFR and its family members are also central components of critical autocrine feedback circuits that promote growth (Figure 1C). Hence, even in cancers in which EGFR itself is not oncogenically activated, inhibition of EGFR would be predicted to have therapeutic value.
For these reasons, enormous effort has gone into the development of targeted therapeutic agents that inhibit EGFR. The most useful clinical agents fall into two classes: small-molecule tyrosine kinase inhibitors (TKIs), and EGFR-directed antibodies (Table 1). The small-molecule inhibitors of EGFR include reversible inhibitors such as erlotinib and gefitinib, and irreversible ones, such as HKI-272, EKB-569 and CI-1033. These TKIs target the intracellular kinase domain of EGFR by blocking the ATP-binding pocket thus preventing autophosphorylation on cytoplasmic tyrosines, and subsequent assembly of downstream macromolecular signaling complexes mediated through v-src sarcoma viral oncogene homology 2 domain (SH2)- or polypyrimidine tract binding protein (PTB)-domain interactions with EGFR phospho-tyrosines.
Table 1.
Current use of anti-EGFR agents in clinical settings
| Class | Synergy in clinical use |
|---|---|
| Antibodies (cetuximab panitumumab) | Cetuximab plus radiation: improved cure rate and survival in squamous cell carcinomas of the head and neck (SCCHN) [161]. Cetuximab plus platinum-based chemotherapy: improved response rate and progression free survival (PFS) in metastatic/recurrent SCCHN [162,222]; improved survival in metastatic non-small-cell lung cancer (NSCLC) [223]. Cetuximab plus irinotecan or oxaliplatin: improved PFS in patients with metastatic colorectal cancer (mCRC) [3,224,225]. |
| Small molecule kinase inhibitors (Erlotinib Gefitinib Lapatinib ) | With gemcitabine, mild survival gains (0.33 months) in metastatic pancreatic cancer. Erlotinib is approved by the FDA for treatment of pancreatic adenocarcinoma in combination with gemcitabine [226]. So far, limited benefit has been seen in combinations of tyrosine kinase inhibitors (TKIs) with radiation in early phase clinical trials marked by increased toxicity. Both gefitinib and erlotinib are antagonistic with some forms of chemotherapy in Phase III trials [97,170]. Lapatinib improves time to progression of human EGF recepter 2 (HER-2)-positive metastatic breast cancer patients [119]. |
The first of the EGFR-targeting antibodies, m225, was generated in John Mendelsohn’s laboratory in the early 1990’s and was subsequently transformed into an acclaimed anti-cancer targeting agents, cetuximab [33]. There are presently at least five EGFR-targeting antibodies in clinical use or under development, including cetuximab, panitumumab, matuzumab (EMD 72000), nimotuzumab (hR3), and zalutumumab [34]. Upon receptor activation, EGFR domains I and III are brought within proximity of each other, and create a binding site for the ligand molecule [35,36] (Figure 1A). Structural analyses have shown that most of the EGFR-targeting antibodies, including cetuximab, sterically hinder the interactions between ligand and binding site on EGFR [25]. Alternatively, several recently developed antibodies (e.g., 806 [37] and zalutumumab [34]) act by blocking the conformational rearrangements that are required during EGFR dimerization, while some target EGFR dimerization with partners (e.g., pertuzumab, which disrupts EGFR ErbB2 interactions [38]). Besides directly inhibiting EGFR-dependent signaling, a second action of these antibodies is to promote receptor removal from the cell surface, reducing the active pool of the protein available to signal [14]. A third, in vivo, activity of therapeutic antibodies is to induce antibody-dependent, cell-mediated cytotoxicity (ADCC) [39]. In this process, the Fc domain of the antibody engages the Fc receptors on macrophages and natural killer (NK) cells, driving their activation and promoting cancer cell destruction.
In rare cancers, in which there is absolute dependence of the malignant phenotype on a unique oncogenic lesion, the use of a targeted agent as a monotherapy is appropriate and yields a dramatic clinical response. This is the underlying basis for efficacy of imatinib and other drugs of its class, which block the highly potent oncogenic driver breakpoint cluster region-Abelson murine leukemia viral (v-abl) oncogene homolog (BCR-ABL), in chronic myeloid leukaemia CML [1]. For the many reasons discussed below, EGFR-targeting inhibitors are very effective as single agents only in a small subset of patients. However, because of the central role of EGFR signaling in apoptosis resistance, neo-angiogenesis, cell proliferation and damage repair, inactivation of EGFR was immediately predicted as being likely to enhance the action of cytotoxic chemotherapies, radiation or other targeted agents. EGFR inhibitors are commonly used in combination therapies with these other treatments in the clinic, and hence, in studying resistance to EGFR-targeting therapies, it is also necessary to consider interactions between EGFR and its co-administered agents.
In this review, we summarize current and emerging concepts of tumor resistance to pharmacologic signaling inhibitors, particularly bearing on resistance to EGFR. These topics include (1) pharmacokinetic resistance mechanisms, in which drugs fail to reach their targets; (2) resistance due to changes in the drug target; (3) compensatory mechanisms and lateral rescue pathways in cancer cells, that render targeting ineffective; (4) the potential for intelligent development of combinatorial strategies, that utilize understanding of biological networks to devise synergistic therapies that override resistance. Although EGFR is the main focus of this review, this discussion emphasizes commonalities that can be applied to many other pathway-targeted inhibitors.
2. Interaction of EGFR and the classic cellular machinery for drug resistance
In thinking about drug resistance in relation to inhibitors targeting EGFR, a natural tendency is to focus primarily on EGFR-centric signaling processes. However, because the primary clinical use of EGFR inhibitors is in combination with classic chemotherapeutic agents, it is important to also consider the interactions between EGFR and the ‘classic’ drug resistance machinery when evaluating mechanisms of resistance to therapies involving EGFR inhibitors.
2.1 Drug transporters
ATP Binding Cassette (ABC) transporters, contribute to resistance against chemotherapeutic agents by pumping drugs out of the cancer cell. ABC transporter group members, classified in subfamilies ABCA ABCG based on the homology in their ATP binding domains and other criteria [40], are widely expressed in tissues. ABC transporters efflux a wide range of agents, including the EGFR-targeted TKIs gefitinib, erlotinib, and lapatinib [41–43], with considerable but incomplete overlap in drug specificity among the different ABC family members [44] (Figure 2A). These transporters limit the amount of active drug entering into some tissues, and therefore may limit in vivo availability of a specific signaling inhibitor.
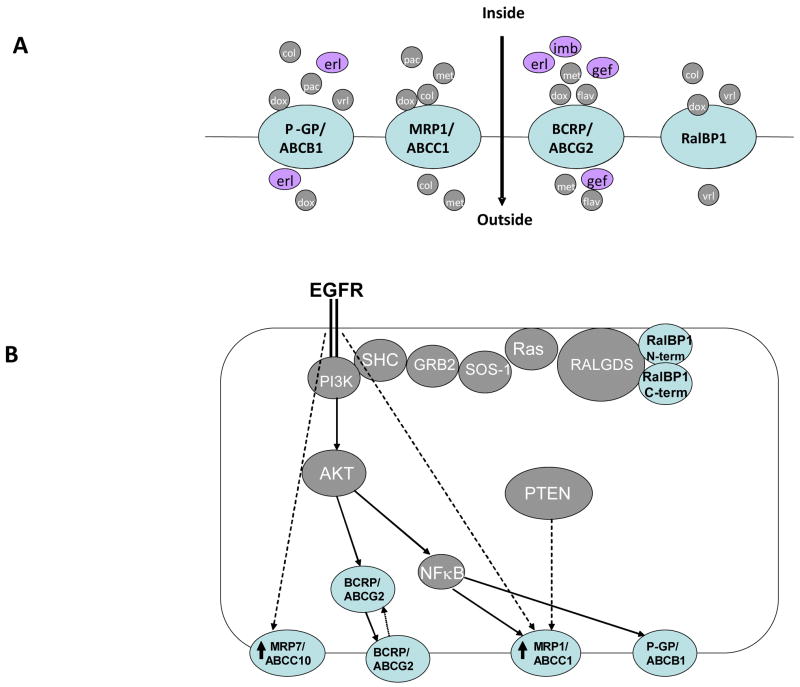
Figure 2.
A. Transported substrates of P-glycoprotein (P-GP), also known as multidrug resistance gene 1 (MDR1), and as ATP binding casette B1 (ABCB1), multidrug resistance protein 1 (MRP1)/ABCC1, breast cancer resistance protein (BCRP)/ABCG2 and Ral binding protein 1 (RalBP1)/Ral interacting protein, 76 kDa (RLIP76). ABC transporters efflux a wide range of xenobiotics from the cell. Among these, erlotinib (erl), gefitinib (gef), and imatinib (imb) target EGFR; colchicine (col), doxorubicin (dox), flavopyridol (flav), methotrexate (met), paclitaxel (pac) and vinorelbine (vrl) are cytotoxic agents commonly used in conjunction with EGFR-targeted therapies. B. EGFR signaling mechanisms that regulate efflux pumps. EGFR signaling pathways regulate the expression of the P-GP/MDR1, MRP1, BCRP/ABCG2 and RalBP1/RLIP76 transporters. At least three ABC transporters are regulated by EGFR via the phosphinositol 3 kinase (PI3K)–v-akt murine thymoma viral oncogene homolog (AKT) arm of the EGFR signaling pathway; phosphatase and tensin homolog (PTEN) and NF-κB contribute to this regulation. GRB2: growth factor receptor-bound protein 2; RALGDS: v-ral simian leukemia viral oncogene homolog guanine nucleotide dissociation stimulator; Ras: RAS viral oncogene homolog; SHC: v-src sarcoma viral oncogene homology 2 domain-containing protein; SOS-1: son of sevenless homolog 1.
Significantly, a growing body of experimental evidence links the activity of the EGFR signal transduction pathway to regulation of ABC transporters. A number of recent studies indicate that changes in the activity of EGFR and its effectors in cancer cells regulate the expression and activity of many transport proteins (Figure 2B). EGF-induced transient activation of EGFR transcriptionally upregulates members of the multidrug resistance protein (MRP, also known as ABCC) transporter subfamily, including MRP1 (also known as ABCC1) and MRP7 (ABCC10), in the breast adenocarcinoma MCF-7 cell line [45], compatible with the idea that active EGFR signaling may result in drug resistance [45]. Exogenous overexpression of constitutively active Ras increases expression of the important ABC transporter P-glycoprotein (P-GP, also known as multidrug resistance gene, or MDR1, and as ABCB1), and induces colchicine resistance in human and other mammalian cell lines [46,47]. Conversely, Schaich et al. reported an inverse correlation between activating Ras mutations and the mRNA expression of the P-GP/MDR1 transporter in acute myeloid leukemia (AML) [48]. Taken together these studies suggest a cell-type-dependent relationship between Ras and MRP1 activity.
The EGFR effector PI3K, and PI3K-activated effectors, regulate cell survival and protect against a wide range of apoptotic inducers. PI3K activation selectively upregulates transcription of MRP1 but not P-GP/MDR1/ABCB1, and selects for chemoresistant cells, in a prostatic carcinoma model [49]. A corroborating report indicates that phosphatase and tensin homolog (PTEN) phosphatase activity, which inhibits the PI3K pathway, correlates with the mRNA and protein expression levels of MRP1 and another transporter, breast cancer resistance protein (BCRP, also known as ABCG2), but does not correlate with P-GP/MDR1/ABCB1 status, in prostate cell lines [47,50]. BCRP/ABCG2 a somewhat divergent ‘half transporter’ has only one ATP binding cassette domain [51], and one transmembrane domain [52]. This is in contrast to the two ATP-binding cassette domains and two transmembrane domains contained in MDR and MRP subfamily members. Interestingly, the BCRP/ABCG2 transporter is expressed at different levels in leukemia and solid tumors samples [53] and five independent studies have correlated BCRP/ABCG2 expression to AML therapeutic response. Higher levels of BCRP/ABCG2 are found in patients that do not go into post-treatment remission, and have been linked to lower survival rates [54]. The anti-carcinogenic agent curcumin has been shown to inhibit the PI3K/Akt/NF-κB pathway, and thus downregulate the ability of P-GP/MDR1/ABCB1 to confer resistance to adriamycin [55]. Choi et al. suggested that this work provides direct evidence that inhibition of an EGFR effector pathway can counter the efflux capabilities of P-GP/MDR1/ABCB1, possibly by suppressing its expression [55].
EGFR signaling through Ras activates RalGDS, which subsequently triggers Ral which then recruits the Ral binding GTPase activating protein (GAP) Ral binding protein 1 (RalBP1) (also known as (Ral interacting protein, 76 kDa (RLIP76)) [56–58]. RLIP76 mediates a rather unusual connection between the Ras signaling pathway and transport activity for xenobiotics. Besides acting as a RalGAP, the ubiquitously expressed RLIP76 has the features of an unusual ABC transporter: it contains two nucleotide-binding domains, but does not contain clearly defined transmembrane domains, although it has integral membrane associations. RLIP76 confers drug resistance to anthracyclines, vinca alkaloids, and 4-hydroxy-2-nonenal (4-HNE). Hence, EGFR-initiated activation of RLIP76 can induce transport of a variety of cationic and anionic xenobiotics, including doxorubicin and vinorelbine, which have been tested successfully in combination therapy with the EGFR targeting agents lapatinib, cetuximab and gefitinib. [59–62] Separately, RalBP1 is involved in the clathrin-coated-vesicle-mediated endocytosis of important growth factor receptors, including not only EGFR, but also the insulin and TGF-β receptors [63], connecting its action to feedback loops, as discussed in following sections.
Based on the above connections, EGFR-targeting TKIs are hence well positioned to be inhibitors of the ABC transporters. Indeed, gefitinib has been shown to function as a potent ABC inhibitor, reversing resistance to and enhancing cytotoxicity of the well known BCRP/ABCG2 and P-GP/MDR1/ABCB1 substrates, topotecan, irinotecan and mitoxantrone, in BCRP/ABCG2 or P-GP/MDR1/ABCB1 overexpressing cells [42]. As another example of its anti-pump activity, gefitinib inhibits P-GP/MDR1/ABCB1-mediated transport of its substrate calcein AM [64]. A similar range of clinically relevant anti-BCRP/ABCG2 effects was demonstrated for lapatinib, a dual inhibitor of EGFR and HER2 [43]. Furthermore, Shi et al. have reported that erlotinib inhibits P-GP/MDR1/ABCB1-mediated resistance to paclitaxel, colchicine and vinblastine [65]. These data all suggest that one action of EGFR pathway TKIs in the clinic involves increasing the activity of co-administered cytotoxic agents.
As noted above, EGFR-targeting TKIs are simultaneously substrates of the same transporters they inhibit [42]. For example, experiments performed in BCRP/ABCG2- or P-GP/MDR1/ABCB1-transfected cells show that both of these proteins transport erlotinib [42]. Deletions of the P-gp/Mdr1/AbcB1 or Bcrp/AbcG2 genes in mice result in higher levels of systemic exposure after erlotinib administration [42]. These results suggest that for therapeutic gain, erlotinib efflux could be suppressed by co-administration of P-GP/BCRP inhibitors. Such inhibitors of P-GP/MDR1/ABCB1 include verapamil, quinidine, cyclosporine A and tariquidar [66] and for BCRP/ABCG2 include tariquidar, cyclosporine A and fumitremorgin C [44]. To date, no work has addressed this possibility. Further studies are needed to better establish the role of transporters in EGFR resistance mechanisms.
As a side note, one of the most intriguing connections between specific ABC transporters and drug resistance is their recently described association with normal and cancer stem cells [67]. A so-called ‘side population’ (SP) enriched for stem cells is easily identifiable by flow cytometry in clinical samples and cell lines [67,68]. SP cells are characterized by their accelerated efflux of Hoechst 33342 and Rhodamine 123 (Rh123) fluorescent dyes. Both P-GP/MDR1/ABCB1 and BCRP/ABCG2, are highly expressed by these SP cells. One recent study has shown that BCRP/ABCG2 is directly required for the maintenance of the SP in the breast cancer cell line, MCF7 [69]. Furthermore, both BCRP/ABCG2 expression and SP presence was suppressed during TGF-β-induced epithelial mesenchymal transition. Whether signaling pathways essential for survival of majority of cancer cells are similarly active in stem cells remains be documented; however, certainly one current concept of drug resistance suggests it is dependent on the protected status of stem cells [67]. Acquired resistance to EGFR-targeted agents due to changes in cancer stem cells may be a rational avenue for future investigations, since novel stem-cell-directed agents are emerging. One of these drugs, the BCRP/ABCG2 inhibitor fumitremorgin C, selectively affects the cancer stem cell population in an NSCLC cell line [68]. Furthermore, erlotinib inhibits BCRP/ABCG2-mediated resistance to flavopyridol, SN38 and mitoxantrone, raising the possibility of a potential role of EGFR signaling in regulating BCRP/ABCG2 activities related to drug resistance in stem cells [65]. It is likely that this will be an area of intense investigation in coming years.
2.2 Survival machinery
For EGFR-targeting agents as for other targeted and non-targeted agents in cancer the execution steps of cellular apoptosis may be yet another source of cancer cell resistance. A detailed discussion of the cell death machinery in cancer therapy is beyond the scope of this review, but has recently been reviewed thoroughly by Lessene et al. [70]. Notably, inhibition of the EGFR pathway with erlotinib in sensitive cell lines leads to upregulation of the BH3-containing apoptosis activator proteins B-cell lymphoma protein 2 (BCL2)-interacting mediator of cell death (BIM) and p53 upregulated modulator of apoptosis (PUMA, also known as BCL2 binding component 3 (BBC3)) [71], leading to subsequent oligomerization of BCL2-associated X protein (BAX) and BCL2-associated agonist of cell death (BAD) and formation of mitochondrial pores [72]. This pro-apoptotic effect of erlotinib, operating via the intrinsic mitochondrial pathway, can be counteracted by overexpression of BCL-2 even in the presence of effective suppression of Erk and Akt phosphorylation [71]. Gefitinib killing of NSCLC cells has also been shown to require a contribution from BIM [73]. Additional studies have tied resistance to EGFR-pathway-targeting agents such as lapatinib to increased expression of the anti-apototic myeloid cell leukemia sequence 1 (MCL-1) and reduced expression and activity of BCL2-antagonist/killer 1 (BAK) and BAX [74]. Intriguingly, one study has suggested that the intracellular domain of ErbB4 itself has properties of a Bcl-2 homology 3 (BH3)-domain protein, and communicates directly with the proteins BCL-2 and BAK to regulate apoptosis [75]. These studies clearly indicate the potential of manipulating the core cellular survival to increase potency of EGFR-targeted and other cancer therapies.
3. EGFR pathway-specific resistance mechanisms
3.1 EGFR-intrinsic factors
A major determinant of whether tumors will or will not respond to EGFR inhibitors often involves changes directly affecting the EGFR protein (Figure 1C). A minority of human cancers with hyperactive EGFR signaling are exquisitely sensitive to withdrawal of signaling inputs mediated by such specific inhibitors, because they have become pathway ‘addicted’ [76]. Certainly, the increased EGFR gene copy number observed in some tumors provides genetic evidence for the positive selection of potentially EGFR-addicted cancer clones, and in NSCLC and in colon cancer has been associated with higher clinical efficacy of EGFR antagonists, erlotinib and cetuximab [25,26,77].
Oncogene addiction, or at least increased sensitivity to EGFR-targeting TKIs, has in some cases been associated with specific mutations affecting the EGFR protein. For example, Lynch et al reported that somatic mutations in the kinase domain of EGFR in NSCLC impart susceptibility to the TKI gefinitib [78]. Mutations in the activation loop of the EGFR kinase (exon 21) or deletion in exon 19 constitute 40% and 50%, respectively, of all kinase domain alterations associated with sensitivity to gefitinib and erlotinib in NSCLC, and are associated with longer patient survival following treatment with specific inhibitors [32,79,80]. These mutated receptors exhibit less dependency on ligand binding for their kinase activity, and are constitutively active.
The ability of this mutant EGFR to act as an oncogene was demonstrated in a mouse model of lung cancer using ΔE746-A750 and L858R transgenic EGFR constructs, which simulate the naturally occurring lesions found in human tumors [81]. The ‘addictive’ mutations unite cancers of different histological and organ site origin and similarly confer sensitivity to relevant signaling inhibitors. For example, EGFR gene amplifications or mutations are rarely seen in the upper gastrointestinal tract malignancies however, when present, they make these cancer types similarly susceptible to the effects of the EGFR inhibitors [82–85].
Balancing the identification of mutations that sensitize cells to EGFR TKIs was the observation that tumors treated with these inhibitors eventually acquire secondary mutations within the EGFR kinase domain that induce TKI resistance. The most common resistance mutation, T790M, is associated with about 50% of cases of acquired resistance to the EGFR kinase inhibitors gefitinib and erlotinib. T790M introduces a methionine in place of a ‘gatekeeper’ threonine residue. EGFR mutants that retain sensitivity to EGFR have an approximately 10-fold greater affinity for gefitinib compared with wild type l EGFR kinase domains [86]. The acquired resistance mutation T790M only slightly (twofold) reduces the affinity of the double-mutant receptor to gefitinib, but leads to a greater increase in competitive ATP binding and more dramatic resistance to gefitinib.
In contrast to the reversible TKIs gefitinib or erlotinib, irreversible TKIs or anti-EGFR antibodies can overcome this source of resistance, as has been shown in preclinical models [87–90]. The activating effect of kinase-mutant T790M EGFR on other ErbB-family receptors and STAT3 can be counteracted by blockade of ErbB2 with lapatinib [91], and further enhanced by the anti-EGFR antibody, cetuximab [91]. This dual EGFR inhibition strategy is now being pursued in clinical settings [92].
In addition to marked sensitivity of EGFR mutant cancers to TKIs, experience in clinical practice with gefitinib and erlotinib indicates that a significant percentage of tumors with non-mutant EGFR also respond to these compounds (about 10% of all squamous cell carcinomas of the head and neck (SCCHN) [93] and 7% of NSCLC [94]), indicating that other factors are involved. One potential contributing factor to the responsiveness of wild type cancer to EGFR inhibitors may be increased production of the EGFR ligands, which has been demonstrated in clinical samples of colorectal cancer [30] and in cell lines [95]. Increased copy number of the wild type or mutant EGFR (fluorescenece in situ hybridization (FISH) positivity) has been also associated with sensitivity to EGFR antagonists [31,77]. Tailoring therapy according to specific EGFR mutational status can dramatically improve the response rates in lung cancer patients treated with EGFR TKI, gefitinib. Sequist et al. [96] reported a 55% response rate in a small Phase II study of gefitinib in 31 patients with NSCLC selected on the premise of activating mutations in the EGFR kinase domain. The frequency of EGFR kinase mutations in large randomized studies without genotype pre-selection within the American patient population is in the vicinity of 15% (13% in the Tarceva responses in conjunction with paclitaxel and carboplatin (TRIBUTE) trial [97], 17% in the BR.21 trial [94]), overlapping with FISH positivity in ~38% of patients. The BR.21 trial showed median survival improvement of nearly 2 months (10 months versus 8.3 months) in EGFR-mutant patients treated with second line erlotinib versus placebo, respectively [94,98]. In this large study, ad hoc analysis showed a 27% response rate to erlotinib in patients harboring activating mutations of the EGFR versus 7% in a subgroup of patients with wild type EGFR. The efficacy of EGFR antagonists in lung cancer is further complicated by the response-favorable contribution of FISH positivity, or negative contributions of activating mutations in Kirsten rat sarcoma viral oncogene homolog (KRas) or PI3K, or loss of PTEN, to the responsiveness to EGFR antagonists. These issues are discussed in the following sections of this review.
One of the first described mutations in EGFR, producing EGFR variant III (EGFRvIII), is an in-frame deletion from exons 2 through 7 (amino acids 6–273) in the extracellular domain that results in ligand-independent constitutive activation of EGFR [99]. Although not present in normal adult tissues, this mutated form of EGFR is clonally expressed on the surface of up to 40% of glioblastomas [100], and has also been recently reported as being common in SCCHN [101]. In glioblastoma cell lines, the presence of EGFRvIII confers resistance to gefitinib that is correlating with sustained signaling activity of the EGFR and persistence of phospho-Akt [102]. In contrast to the other deletions and mutations of EGFR discussed above, the role of EGFRvIII expression in response of cancer cells to EGFR-targeting treatments is more complicated. Significantly, although tumors bearing EGFRvIII have decreased sensitivity to the reversible TKIs gefitinib and erlotinib, they are more sensitive to treatment with irreversible EGFR TKIs such as HKI-272 [103]. Likewise, cancer cells expressing EGFRvIII are less sensitive to EGFR-targeting antibodies such as cetuximab [101,104], even though the in vitro binding of the antibody to its site in domain III of the EGFR remains preserved [105]. One recently described monoclonal antibody, mAb-h806, is capable of neutralizing this deletion variant of EGFR, and is effective in vitro and in vivo [37]. It is plausible that mAb-h806 operates through the same mechanism as the dual ErbB inhibitors [37,106,107], by preventing ErbB2-dependent transphosphorylation of normal or vIII EGFR. It has been suggested that structural differences associated with the vIII deletion create an open conformation of EGFR that is more available for interactions with its dimerization partners, including ErbB2. The new generation of irreversible ErbB inhibitors targeting both EGFR and ErbB2 (e.g. HKI-272 and EKB-569 [108,109]) may prevent cross-activation of the EGFR from its co-receptor, ErbB2/HER2.
3.2 Factors upstream of EGFR; ligand excess
In cancers in which EGFR is non-mutated, and maintains ligand dependence, an abnormal excess of the EGFR ligands may be a potent oncogenic driver and resistance factor for reversible EGFR-targeting TKIs. Amphiregulin overexpression contributes to the malignant phenotype of many epithelial tumors, including hepatomas [110]. mRNA analysis of a series of NSCLC tumors from patients treated with gefitinib identified increased amphiregulin expression as a resistance marker [111]. In some colorectal cancers (CRCs), excess production of the EGFR ligands epiregulin and amphiregulin correlates with longer disease control by cetuximab [30]. This may be due to a greater dependency on upstream regulation of EGFR signaling in these CRCs: in contrast to lung cancers, CRCs and gliomas do not typically harbor EGFR kinase domain mutations [112], as screening of 293 colorectal tumors revealed only one such mutation. Depletion of the EGFR receptor from the cell surface and blocking ligand binding by cetuximab [113] may be particularly effective in cancers bearing no receptor alterations.
Outcomes in ovarian cancer negatively correlate with expression of HB-EGF and a sheddase, ADAM17, which releases the ligand from its cell surface bound precursor [114]. Specific ADAM inhibitors, such as INCB3619, can be synergistic with EGFR TKIs by suppressing ligand-mediated activation of both EGFR and its co-receptor, ErbB3 [115]. Suggestively, the upregulation of upstream activators of EGFR (both sheddases and ligands) has been observed in response to cellular stresses induced by cytotoxic chemotherapies sometimes used in conjunction with EGFR inhibitors: Wang et al. [116] discovered HB-EGF induction and activation of NF-κB in colon and pancreatic cancer cell lines as an early response element to multiple cytotoxic chemotherapy agents. A gefitinib-resistant subclone of the highly EGFR-dependent A431 cell line was shown to upregulate IGF-R1 signaling via loss of IGF binding proteins, a mechanism that effectively increases the level of autocrine IGF [117].
Together with the studies discussed above addressing EGFR and ABC transporters, these results suggest further close connections between regulation of EGFR signaling and resistance to commonly used chemotherapies.
3.3 Lateral signaling through EGFR co-receptors
EGFR signaling activity, and susceptibility to inhibition, is governed in part by its dimerization partners. Hence, the dimerization of EGFR with ErbB2 and ErbB3 has attracted attention as a potential source for therapeutic resistance, or as a potential source of novel therapeutic targets. For example, a subset of cancers co-expressing ErbB2 and EGFR are refractory to inhibitors of a single co-receptor [118]. Lapatinib, a selective TKI designed to target both EGFR and ErbB2, has shown promise in early clinical trials in patients with EGFR/ErbB2-expressing metastatic breast cancer [119], although it is not yet clear whether the biological activities of lapatinib arise from its inhibition of both receptors, or predominantly from its activity against ErbB2 [120]. Additional agents targeting EGFR and ErbB2 are under development (e.g. HKI-272 and EKB-569 [108,109]), as are agents such as pertuzumab, that interrupt pairing between EGFR and ErbB2 [121], and pan-ErbB family inhibitors [122].
EGFR heterodimerization with ErbB3 [123], coupled with upregulation of ErbB3 [11,124] also provides cellular resistance to EGFR kinase blockade. For example, Rothenberg et al inactivated endogenous EGFR mRNA in the highly EGFR-dependent cell line PC9, and tested a panel of mutant EGFR constructs for ability to rescue cell survival [125]. Rescue did not require retention of autophosphorylated tyrosines on EGFR, but was strictly dependent on retention of sequences allowing heterodimerization with ErbB3. Although ErbB3 lacks a kinase domain, its activation by dimerization with EGFR promoted further activating association with the p85 of PI3K, which was sufficient to compensate for disabled EGFR-dependent signaling. ErbB3 signaling is biased towards PI3K-Akt signaling, with six discrete PI3K binding sites present on this receptor [10]; Erbb3 also signals to activate the Src protein, providing separate survival signals [126,127].
Besides dimerizing with ErbB family members, several other EGFR-related heterodimerizations are of particular note for therapeutic resistance. In cases of acquired resistance to erlotinib, dimerization between the hepatocyte growth factor (HGF) receptor c-Met [128] and ErbB3 activated survival signaling pathways involving PI3K and Src, and thus completely replaced signals from the inhibited EGFR [20]. It has become apparent that in trans signaling is a key feature of ErbB family. Specifically, activation of the PI3K/Akt pathway is driven predominantly through phosphorylation in trans of the kinase-inactive member ErbB3 [129]. This phenomenon, initially described in cell lines, was subsequently validated as occurring in vivo in clinical samples from lung cancer patients who acquired resistance to erlotinib over time while on therapy [130] via genomic amplification of the c-Met locus and upregulation of phospho-ErbB3. These alterations are a source of acquired resistance to EGFR antagonists although this distinction may be blurred by discoveries of a high incidence of Met amplifications (sometimes coincidental with EGFR amplifications) in the untreated NSCLC tumor tissues [128,131]. The issue may be that the tumors are heterogeneous at their origin and selective pressure of a particular drug or treatment modality enriches for a predominant phenotype/genotype [132].
Finally, interactions between EGFR and the insulin growth factor receptor (IGF-1R) can also provide resistance to TKIs targeting EGFR [133]. This interaction is specifically induced when IGF-1R-expressing cancer cells are treated with gefitinib [134]. IGF-IR activates the PI3-K/Akt pathway by signaling through insulin receptor substrate-1 (IRS-1) and activates Ras and downstream signaling proteins via direct interactions with the adaptor Shc. A negative feedback loop mechanism operating through IGF-PI3K-Akt signaling is responsible for derepression of IGF-R1 and increased Akt activity, leading to resistance to mammalian target of rapamycin (mTOR) inhibitors in clinical settings [135,136]. Cross-talk between EGFR and VEGF signaling has also been well documented in vitro and in situ, where induction of angiogenesis requires EGFR activity [137] for upregulation of both proangiogenic ligands (e.g., VEGF) and the VEGF receptors. Clinically, this relationship provides an explanation for the striking parallelism between the high degree of vascularization observed in resistant tumors and the vasculature collapse when tumors are sensitive to EGFR antagonists. Combined treatment modalities with VEGFR/EGFR dual kinase inhibitors [138] are aimed at overcoming acquired resistance to EGFR inhibitors via upregulation of alternative signaling via VEGFR1, a scenario reminiscent of that seen with Met/ErbB3 and IGF-R1.
3.4 Downstream ‘core’ EGFR effectors
Critical EGFR-dependent signaling proceeds through its proximal downstream effectors, including notably the Ras group of paralogs (K-, H-, and N-Ras). Ras protein mutations have been identified as causative lesions in more than a quarter of human cancers, with K-Ras most commonly affected [139]. At the cell membrane, Ras is activated not only by ErbB family proteins, but additional growth factor receptors including IGF-1R [133], Met [20,140], and ErbB3 [123,124]. Ras mutations, typically affecting codons 12 and 13, result in predominantly GTP-bound, activated Ras, which constitutively activates downstream pathways of cell proliferation (e.g. RAF/MEK/ERK), motility and metastasis (e.g. RalGDS/RalA), and survival (PI3K/Akt) [141]. Because of this central signaling role, it is not surprising that K-Ras mutations have recently emerged as a major predictive marker of resistance to anti-EGFR therapies.
K-Ras mutation is associated with resistance to EGFR-targeting antibody therapy in approximately 40% of CRC [30,142,143]. Lievre et al. reported that among 30 participants with metastatic CRC (mCRC) treated with cetuximab, a K-Ras mutation was found in 68% of non-responding participants and none of the responding participants [144]. Analysis of a large randomized study of patients with metastatic CRC treated with panitumumab showed that those with tumors harboring K-Ras mutations had a 0% response rate and 7.4-week median time to progression (TTP) compared with a 17% response rate and 12.3-week TTP for participants with tumors without K-Ras mutations [145]. Similar results have been seen in several randomized studies to date (Table 3). In fact, the use of cetuximab in patients with K-Ras mutations may be harmful, as demonstrated by the retrospective analysis of the capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO)-2 study. In this analysis, the addition of cetuximab to capecitabine, oxaliplatin, and bevacizumab appeared worse than capecitabine, oxaliplatin, and bevacizumab in the K-Ras mutation group (12.5 versus 8.6 months of progression-free survival (PFS), p = 0.043). The trend toward worse outcome with cetuximab (against Ras mutant tumors) in combination with folinic acid, fluorouracil and oxiplatin (FOLFOX) was also seen in the smaller OPUS study, in which PFS was 5.5 months with FOLFOX/cetuximab and 8.6 months with FOLFOX alone (hazard ratio 1.83 p = 0.02). Thus, analysis of K-Ras mutation status appears to predict those metastatic CRC patients who will not benefit from treatment, and perhaps have a worse outcome, with the use of cetuximab or panitumumab as either single agents or in combination with chemotherapy. The K-Ras-mediated primary resistance to EGFR TKI has also been reported in lung cancer [146].
Table 3.
Rational synergies under exploration with EGFR inhibitors.
| Preclinical co-targets | Trials | |
|---|---|---|
| Target | Rationale | Phase |
| VEGF/VEGFR | VEGF signaling confers resistance by activating PI3K-Akt [137,138] | II |
| ADAM17 | Inhibition of sheddases prevents release of ligand [8,115] | I |
| ErbB3 | Antibody to ErbB3 blocks survival signals to PI3K-Akt [20] | I |
| c-Met, IGF-1R | Antibodies and TKIs prevent transphosphorylation and PI3K-Akt signaling [20,220] | I |
| STAT3 | Decoy oligonucleotide blocks STAT3-mediated activation of transcription, resistance to apoptosis [169] | II |
| mTOR | Synergistic effects in preclinical models [227], tolerated combination in mCRC | I |
| PI3K | Synergistic with EGFR inhibition, PI3K activation via other surface receptors (ErbB3, IGF-R1, Met) protects from apoptosis [55,228] | I |
| Src | Dasatinib, a multitargeted kinase inhibitor, blocks Src activity and increases efficacy of Cetuximab. Src activates signaling of ErbB family receptors [127] |
ADAM: a disintegrin and metalloprotease; Akt: v-akt murine thymoma viral oncogene homolog; ERB3: v-erb-b oncogene homolog 3; mCRC: metastatic colorectal cancer; mTOR: mammalian target of rapamycin; PI3K: phosphinositol 3 kinase; PTEN: phosphatase and tensin homolog; Src: v-src sarcoma viral oncogene homolog; STAT3 :Signal transducer and activator of transcription 3; TKI: tyrosine kinase inhibitors.
The fact that EGFR-inhibited cancers can switch to dependence on alternative receptors such as ErbB3, IGF-1R, VEGFR1, and Met (which are biased to the PI3K/AKT signaling cascade) suggests the potential importance of mutations targeting effectors in this pathway in promoting resistance to EGFR-directed therapies. The PI3K axis is often hyperactivated in tumors: PI3K gene amplification has been reported in ovarian cancer [147], while the mutational activation of the catalytic p110 subunit of PI3K have been identified in ~10–15% of all cancers [148]. Overexpression of AKT, activating mutations in the regulatory p85 and p110 subunits of PI3K, or inactivation of PTEN have been shown to confer resistance to growth factor withdrawal and promote metastatic behavior in model systems [149,150]. PTEN is itself mutationally inactivated in many tumors [151], while direct binding of Ras to the catalytic p110 subunit of PI3K is required for tumorigenesis in animal models [141]. Finally, a recent high throughput sequencing study by Wood et al [139] sought to define the genomic ‘mutational landscape’ of breast and colon cancers. The authors concluded that the Ras-PI3K-Akt pathway was enriched for mutations in both breast and colon cancers, while for a given type, the specific genes targeted varied between individuals.
Ongoing efforts are beginning to assess the role of these additional-pathway-directed lesions in influencing response to EGFR-targeted inhibitors. For example, the genetic deletion of PTEN in the MDA-468 breast cancer cell line was shown to be responsible for resistance to gefitinib, while reconstitution of PTEN in this cell line restored sensitivity to gefitinib and resulted in gefitinib-induced inhibition of Akt [152] (as Akt localization to the cell membrane, required for its activity, involves interaction between the Akt PH domain and the third phosphate group of phosphatidylinositol-3,4,5 trisphosphate; an interaction regulated by PTEN). For the most part, clinical studies exploring the role of these genetic changes in resistance to EGFR-targeted therapies have not yet progressed in clinical settings to the degree that mutation of Ras has, but it is highly likely that a similar correlation with treatment resistance will be found. Other EGFR/RAS proximal proteins such as RAF and its paralog B-RAF, which is mutated in 10–15% of spontaneously arising cancers [148], or RalGDS and its effectors, should also be scrutinized as likely modulators of treatment response in clinic al settings. For example, a recent study of a panel of CRC cell lines demonstrated that cell lines that are dually PI3K catalytically active mutants or PTEN null, and Ras or BRAF catalytic mutants, are highly resistant to cetuximab, in contrast to singly mutated or non-mutated cells [153]. Extending such studies to clinical use could have rapid and powerful benefits in stratification of patients for therapy.
3.5 Alternative EGFR-centered resistance mechanisms
Although most of the functions of EGFR that have been discussed above bear on its kinase activity, EGFR also takes part in complex set of interactions in the cytosol or even in the nucleus [154,155]. Upon receptor engagement by a ligand and oligomerization, rapid changes in the cytosol ensue which involve EGFR ubitiquination and endocytosis. Within minutes of physiological ligand stimulation of EGFR in normal cells, or and in cancer cell lines susceptible to EGFR antagonists, the ligand receptor complexes are removed from cell surface [156]. Two distinct pathways regulate subsequent movement of the receptor from the endosomal compartment: clathrin-mediated trafficking leads to sustained signaling and receptor recycling, while non-clathrin-coated endosomes direct the internalized EGFR to the lysosome for degradation [156].
Hence, activation of EGFR in oncogenesis may also involve genetic or epigenetic changes affecting the rates of receptor endocytosis, lysosomal trafficking and membrane recycling [157]. Similarly, cellular changes that lead to resistance to therapies that target EGFR may influence the same processes. Such resistance mechanisms have not been investigated as intensively as the mechanisms discussed in the preceding sections. However, emerging data suggest that resistance to the anti-EGFR antibody cetuximab is linked to receptor internalization process, which is significantly impaired in resistant clones. Cetuximab-resistant SCCHN clones show abnormal EGFR persistence on plasma membranes after EGF stimulation [16]. This observation may be of more general significance, as the gefitinib-sensitive lung cancer cell line PC9 is characterized by rapid EGFR endocytosis. The endocytic trafficking is impaired in the resistant QG56 line [158]. However, contrasting data from Kwak et al. [159] showed increased internalization of EGFR in a treatment-resistant subclone of NCI-H1650. Together, these studies suggest that proteins that control EGFR internalization may emerge as important modulators of therapeutic efficacy, and that this mechanism requires further exploration.
4. Current targeted combination therapies and resistance
Consistent with the role of EGFR as survival factor for cancer cells, combinatorial strategies of EGFR targeting antibodies and chemotherapy or radiotherapy have been developed. In such contexts, blockade of the receptor removes the on-demand EGFR activation and prevents apoptosis-based resistance and sublethal damage repair from radiation. After the initial observation by Balaban et al. [160] that treatment with EGF-targeting antibodies during and after irradiation augmented cell survival, even in cells with non-mutated EGFR, the use of monoclonal EGFR antibodies to sensitize cancer cells to radiation has become one of the major breakthroughs in the treatment of SCCHN [161].
Development of combinatorial strategies (Tables 1 and 3) with EGFR-targeting antibodies such as cetuximab and chemotherapy drugs identified a number of combined approaches that are currently in broad use for the treatment of colorectal [3], head and neck [162] and lung [163] cancers Chemotherapy agents introducing strand breaks (topoisomerase I inhibitors [164]) or DNA adducts (platinum compounds [165]) interact synergistically with cetuximab, preventing DNA repair processes regulated by EGFR [27]. Cetuximab, by blocking the ligand-to-receptor access, prevents compensatory EGFR activation and phosphorylation of STAT3 [166] in response to radiation induced damage. Work by Grandis et al. [167] identified STAT3 as a critical element of EGFR-induced proliferation. Strategies simultaneously targeting EGFR and STAT3 have been effective in the preclinical models of SCCHN, both increasing apoptosis and reducing B cell lymphoma like X (Bcl-xL) expression [168]. One novel pharmacological strategy that inactivates STAT3 using decoy oligonucleotides mimicking its cognate binding sequence of the DNA [169] is now entering a phase I clinical trial (ClinicalTrials.gov, NCT00696176).
It is still poorly understood why combinations of an EGFR-TKI and chemotherapy have failed to improve survival of NSCLC patients in several large randomized clinical trials, regardless of whether the drugs were given concurrently [97,170] or sequentially [171]. Enthusiasm for the adjuvant use of EGFR inhibitors has been tempered by the results of Southwest Oncology Group (SWOG) trial 0023 which treated patients with inoperable stage III NSCLC with cisplatin, etoposide, and radiation, followed by consolidation docetaxel. Patients were then randomized to maintenance gefinitib or placebo. This study unexpectedly showed a decrement in survival in the gefitinib arm of 23 months (n = 118) versus 35 months for placebo (n = 125; two-sided p = .013) [171]. At the same time, the initial report from the FLEX trial [172] (presented at the 2008 American Society of Clinical Oncology Annual Meeting, abstract #3) showed significant improvement of survival in the cetuximab plus cisplatin and vinorelbine chemotherapy arm compared with chemotherapy alone (11.3 versus 10.1 months, p = 0.0441). Preclinical models attempting to address possible antagonistic pharmacological interactions between the EGFR-TKI causing G1 arrest, and cell-cycle-active anti-microtubular agents [173] do not correlate with the obvious clinical efficacy of cetuximab dosed to attain a steady state blood level. The efficacy differences between the two classes of EGFR inhibitors may stem from the differences in modulation of inducible EGFR signaling activity via ligand upregulation (a process which is most effectively blocked with cetuximab, which competes for the ligand binding site), versus modulation of constitutive ‘addictive’ signaling of hyperactive EGFR kinase by TKIs in the minority of NSCLC patients in which both drugs are active. Alternatively, they may also reflect additional antibody-specific tumor targeting contributions, such as activation of ADCC responses. More mechanistic investigations are required.
5. Conclusion
As illustrated above and in other recent reviews [174,175], an increasingly detailed knowledge of EGFR signaling relationships has begun to allow the development of rationally designed combination therapies that have begun to improve effective use of EGFR inhibitors in clinical practice. Because of the expense and length of time required to establish new trials, a number of promising combinations predicted based on preclinical work have not yet been assessed in humans, raising the potential for significant clinical gains as these combinations move through the Phase I/Phase II process. However, incorporation of new models of cell signaling now emerging from proteomic- and systems biology-based analyses suggests that these combinations are more likely to yield incremental rather than transformative therapeutic gains. The reasons why more work is required are discussed below.
6. Expert opinion: network analysis of resistance mechanisms, to develop new combination therapies
Clearly, exploiting available information about well-defined, EGFR-associated signaling pathways provides better clinical outcomes with available EGFR-targeted inhibitors, but many tumors remain resistant to these therapies. Suggestively, studies such as those of Kassouf et al note that in some cancers treated with gefitinib, an ‘uncoupling’ is observed, in which the upstream signaling proteins (e.g. EGFR) are inhibited, but downstream outputs (e.g. ERK1/2, cyclin D1) remain active because additional, undefined proteins have probably compensated for the deficiency in EGFR signals [176,177]. Efficient application of EGFR-targeting agents would clearly be enhanced if it were possible to readily identify those proteins capable of providing compensatory signaling, and blocking their activity. Recent conceptual and technical advances bearing on cellular signaling networks provide powerful new tools to achieve this goal.
6.1 Defining cell signaling networks
Since the early 1990s, new methods for detecting protein protein interactions, including the yeast and other two-hybrid systems [178], tandem affinity purification (TAP) [179,180], protein complementation [181,182], and mass spectrometry [183] have yielded complementary [184] large data sets regarding protein associations. These data have in turn been merged with large genetic datasets derived from studies of functional interactions in model organisms [185], as well as bioinformatics analyses that address issues of protein co-expression, or potential conserved interactions between protein ortholog groups from different organisms [186]. It is becoming appreciated that the classic view of linear or slightly branching signaling pathways is very far removed from the reality pertaining in cells, and Friedman and Perrimon have addressed at length the need to transition from canonical and simple pathway-centric models to instead think of signaling network models [187,188]. By some estimates, many signaling proteins interact with more than 20–30 partners, providing a dense degree of connection between ‘pathways’ that were predominantly thought to be independent. Using currently freely available tools for network modeling [189], it is possible to begin to think about alternative sources of resistance to EGFR-targeted therapies intrinsic in a network structure.
To introduce some key network concepts, biological networks have been constructed and topologically analyzed for a variety of species. These networks provide robust resistance to perturbations such as environmental conditions and mutations [190]. For example, a large-scale gene knockout study in yeast indicated that 50% of the total genes not only were not essential for viability, but also could be eliminated without a significant consequence for organism fitness, because other genes could functionally compensate for their loss [191]. Evolutionarily, this characteristic of functional redundancy poses a great survival advantage [192,193], as single mutations are unlikely to be lethal, but can instead provide the basis for the selection of new functions [194].
Redundancy can arise in cellular signaling networks in discrete ways [195]. Homogeneous redundancy is exemplified by the existence of numerous copies of key genes existing as backups in case one copy is disabled or lost. For example, protein paralogs may share a similar structure and at least a partially congruent function (e.g. the paralogs p130 v-crk sarcoma virus CT10 oncogene homolog-associated substrate (p130Cas) and neural precursor cell expressed, developmentally down-regulated 9 (Nedd9) both bind and activate overlapping substrates in integrin signaling networks [196]). In heterogeneous redundancy, distinctly different noninteracting proteins regulate similar functional pathways with convergent endpoints: for example, protein kinase C (PKC)-ζ provides collateral activation of the MAPK signaling pathway downstream of EGFR [197].
The concept of redundancy can be further qualified by considering network structure. Nodes and edges, representing molecules (i.e. genes, proteins, small molecules) and the interactions between them, respectively, define the constituents of biological networks. Barbabasi and Albert have examined the architectural properties of biological networks and found the degree distribution (representing the probability that a node has exactly k links or interactions) of nodes in these networks is ‘scale-free’, differing from that found in random networks based on a power law distribution as compared with a Poisson distribution [198]. In a random network, the degree of each node remains close to the average degree of the network: if cells were random networks, all proteins would have approximately the same numbers of ‘partners’. However, biological networks contain many nodes with few links to other nodes and a few nodes, referred to as hubs, which are highly connected. Hubs can be broken down into two categories: date and party hubs [199]. Party hubs tend to be constitutively coexpressed with their interacting partners, while date hubs only transiently overlap in expression with partner proteins. Biologically, an example of a party hub might be a core scaffolding subunit of the ribosome [200], while a date hub might be a dynamically regulated scaffolding protein that assembles protein complexes only in specific regulated circumstances (e.g. kinase suppressor of Ras (KSR) [201]). Finally, some analyses have suggested that because of the dense connectivity of biological networks, any given protein node in the cellular network is separated by no more than about five or six edge/node ‘hops’ from any other [195].
In general, the biological networks found in normal cells do not experience systemic failure upon ‘random attack’. The odds of a spontaneous mutation hitting a hub, versus a minimally connected inessential node, are low: moreover, inactivation of an individual non-hub node will not have any general consequences for the stability of the overall network connectivity. However, if a targeted node is highly connected, not supported by a redundant paralog, and thus essential for cellular function, the result is fatal (see discussions of centrality lethality theory [202,203]. These network properties, with some further qualifications [190], have critical implications for both cancer progression and targeted cancer therapies.
6.2 Defining and targeting networks in cancer cells
Early stage cancer cells have a limited number of oncogenic lesions promoting proliferation. These lesions are likely to involve proteins within the EGFR signaling cascade, as these drive cell division and increase the pool size of tumor cells. Proteins within this cascade are highly connected [187], and at least some are likely to be hubs (although we note, at the present time, the state of biological databases does not allow for a totally reliable estimate of the number of edges for each EGFR-related node, nor whether this number is similar to or higher than those seen for other signaling systems. Although unbiased large-scale datasets are accumulating rapidly [204], much information about protein interactions is curated from the published literature [205,206], and the high degree of interest in EGFR signaling ensures some degree of sample bias). Targeted therapies (especially therapeutic monoclonal antibodies) are typically designed with specificity to only one or at most two proteins. In a cancer at a relatively early stage, a reagent targeting a hub such as EGFR or an EGFR effector, particularly supported by a cytotoxic agent, is perhaps most likely to be effective.
As tumors grow beyond an initial small size, accumulating a large population of individual cancer cells, other properties affecting network structure and robustness come into play: anti-redundancy and feedback signaling. Krakauer and Plotkin have discussed the specific selection for ‘anti-redundancy’ in larger populations (whether the populations are species, pathogens, or cells within an organism [192]). In cancers, anti-redundancy arises when checkpoints that induce apoptosis of mutationally damaged cells are removed, with the classic example being the loss of tumor suppressors such as p53 [207]. The value of such a lesion, which becomes prevalent in the population, is that it allows the rapid emergence of small clonal populations with one, two or a larger number of additional genetic changes. These multiple mutations may be slightly deleterious to the overall survival of an unselected tumor, and extremely difficult to detect within a larger tumor mass, but be highly protective of tumor growth under the selective conditions posed by targeted chemotherapy or irradiation. Certainly, many aggressive tumors are well-known to be clonally heterogenous [208,209]; in fact, even prior to treatment, the growth of a solid tumor beyond an initial small size results in imposition of stressors such as high acidity, hypoxia and nutrient deprivation, that provide selection pressures for ability to maintain anti-apoptotic and proliferative functions in a hostile environment [210].
In this context, it is perhaps instructive that large-scale efforts to sequence ‘the cancer genome’ for various cancer types have identified a limited number of mutations, which target many genes already known to have pro-oncogenic function, that are found in the majority of tumors [135,136,211]. These studies also identify a much larger pool of mutations occurring rarely in tumors [139,212]. It is likely that the focus on mutations found commonly in the bulk of a tumor mass may miss the presence of multiple mutations affecting genes not clearly known to be pro-oncogenic, and existing only in a small tumor cell compartment. In this view, a p53-mutated tumor, or any tumor that is able to survive and proliferate while simultaneously manifesting aneuploidy or other signs of genomic instability would be intrinsically likely to possess a medium- to high- number of sub-populations colonizing different microenvironmental niches, analogous to the diversity of Darwins’ finches exploiting different macroenvironments [213].
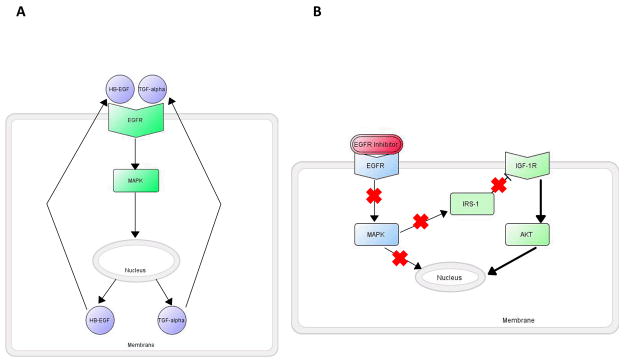
In addition to potential genetic modification of networks, feedback loops are epigenetic mechanisms for signaling control that vary signaling input to control output levels, based on signaling sensors that gauge system output in reference to benchmark levels. Both negative and positive feedback loops exist in biological networks. Negative-feedback loops autoregulate the system, promoting homeostasis and providing stability. In contrast, positive-feedback loops are highly sensitive and have the ability to promote an active signal independent of perturbations. However, these mechanisms that maintain normal cellular function can also introduce fragilities into the network that can be exploited during cancer. For example, in normal cells, the EGFR signaling pathway is an example of a regulated positive-feedback loop. Activated EGFR signaling through the MAPK cascade to the nucleus induces transcription of EGFR ligands, TGF-α and HB-EGF, which bind EGFR and re-initiate EGFR signaling until negative signals from other sources interrupt the loop [214] (Figure 3A). Positive feedback loops can be hijacked by tumor cells and promote uncontrolled cell growth through their self-sustaining attribute, or via loss of the negative ‘stop’ signals due to mutation. As another example, drug resistance to erlotinib can be thought of as a product of a negative feedback loop activating the AKT pathway, also controlled by EGFR, through IGF-IR signaling [215] (Figure 3B). Phosphorylation of IRS-1 at selective serine sites causes a decrease in IGF-IR signaling [216]. Inhibition of MAPK pathway activity via blockade of EGFR signaling decreases IRS-1 serine phosphorylation, and hence increases IGF-IR-mediated AKT signaling. This circuit is robust to both upstream EGFR inhibition and downstream (MEK) inhibition, demonstrating resilience to multiple perturbations through its ability to regulate the IGF-IR IRS-1 negative feedback loop.
Figure 3. Feedback signaling loops modulate EGFR signaling.
A. Positive feedback. In this example, EGFR-dependent activation of transcription of the genes for the EGFR ligands heparin-binding EGF-like growth factor (HB-EGF) and TGF-α (output) augments the signaling input. B. Negative feedback. Ligand activation of EGFR activates MAPK downstream signaling. Activated MAPK activates insulin receptor substrate 1 (IRS1), which provides negative feedback that inhibits signaling from the IGF-1 receptor. Blockade of EGFR cancels this negative feedback, and induces compensatory activation of IGF-1R. IGF-1R activates PI3K and v-akt murine thymoma viral oncogene homolog (Akt) leading to apoptosis resistance [220].
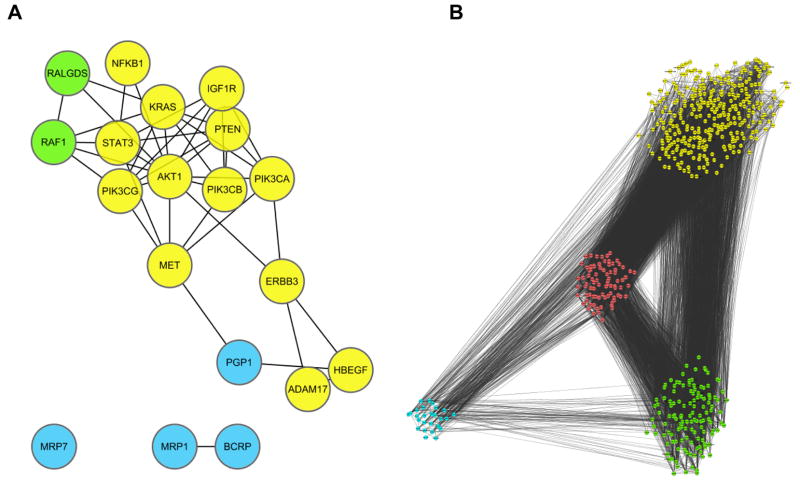
If an advanced stage tumor is a product of a combination of functional redundancy of specific signaling nodes, selected anti-redundancy to identify multiple discrete clonal sub-populations, overall degree of the complexity of network structure, and auto-regulatory feedback mechanisms [190], what are the consequences for an EGFR-targeted therapy? Inhibition of EGFR, typically in combination with a cytotoxic agent, will provide feedback signals that upregulate MDRs/MRPs, limiting available drug concentrations, and will also select for any available anti-redundant (mutationally tolerant) pre-existing cell populations, to help avoid the apoptosis induced by the cytotoxic agent. The shutdown of an important hub, EGFR, will provide feedback/feedforward signals that help maintain the activation of key EGFR effectors. One elegant RNAi study has demonstrated the existence of an extensive network of previously unsuspected proteins regulating ERK signaling [187]. Following EGFR and cytotoxic treatment, these (and other) ‘alternative’ regulators are likely to experience epigenetic upregulation and activation, or alternatively, if the genomic instability in the tumor had fortuitously hit one of these alternative regulators, such clones would gain a survival advantage. As discussed above, the tumor genome studies (e.g. [139]) demonstrate that multiple members of a pathway become mutationally active in tumors: the degree to this phenomenon is related to specific treatment history remains to be determined (also see discussion in [217]). Figure 4A shows interactions, found using the protein protein interaction database STRING, between proteins discussed in above sections as modulators of the cellular response to EGFR targeted inhibitors. Figure 4B is a collection of these proteins (seeds) and their ‘first neighbor’ interactors (proteins binding directly to members of the seed group) displayed as a graphical respresentation to highlight the dense connectivity among the seeds and their proximal networks and the complexity of the EGFR network. Potentially, the mutational or epigenetic targeting of proteins within this expanded group would also help induce recurrence of a drug-resistant tumor.
Figure 4. Networks of proteins confer resistance to EGFR-targeted inhibitors.
A. Known (yellow) and potential (green) signaling proteins that regulate resistance to EGFR inhibitors. Blue, canonical transporter proteins involved in drug resistance. Analysis of these proteins in Cytoscape [221] reveals numerous direct physical interactions (lines) involving many of these proteins. ADAM: a disintegrin and metalloprotease; AKT: v-akt murine thymoma viral oncogene homolog; BCRP: breast cancer resistance protein; ERB3: v-erb-b oncogene homolog 3; HB-EGF: heparin-binding EGF-like growth factor; KRAS: Kirsten rat sarcoma viral oncogene homolog; MRP: multidrug resistance protein; P-GP: P glycoprotein; PI3K: phosphinositol 3 kinase; PTEN: phosphatase and tensin homolog; RAF: v-raf murine sarcoma viral oncogene homolog; RALGDS: v-ral simian leukemia viral oncogene homolog guanine nucleotide dissociation stimulator; STAT :Signal transducer and activator of transcription. B. Cytoscape was used to retrieve the set of direct binding partners for the 19 EGFR resistance modulator proteins shown in A (a total of 549 proteins), then map physical interactions between these direct interactors (black lines). Yellow, green, and blue represent either the initial ‘seed’ groups shown in A, or their direct interactors; proteins indicated in red interact with two or more of the ‘seeds’. The network is highly connected, involving 8537 interactions.
What are alternative approaches to improve resistance? In considering the EGFR network structure, it is provocative to consider the date hubs, particularly those corresponding to nodes/proteins with an expression profile demonstrating they can only be readily upregulated or expressed in cell lineages corresponding to the tumor. Among these, simultaneous blockade of two or three different hubs may so significantly limit tumor growth that it is very difficult to route signals around the blockade. Nevertheless, development of an arsenal of antibody and small molecule inhibitors to target different hubs that can be used to substitute in combination therapies as specific hub-centered blocks are circumvented would be invaluable, as would the development and facile application of robust biomarkers that report critical downstream endpoints. Although there are some provocative recent reports that the tumor stroma and other associated cells may derive from tumors, and/or contain anti-redundancy-associated mutations in proteins such as p53 [218], nevertheless the tumor-associated stroma is much less likely to be genomically unstable than the primary tumor, and hence likely to have a less flexible capacity to reprogram its network. Hence, application of therapeutic agents targeting either endothelial or stromal cells, in combination with agents targeting tumor cell signaling hubs, would be predicted to further reduce the ability of tumors to override signaling blockades.
In summary, therapeutic interventions may best be improved by thinking of the War on Cancer not as a conventional war against a limited number of well-defined foes, but as a guerrilla action against an inventive foe operating while embedded in a host population that is essentially friendly to its cause. In this view, systems biology provides the information matrix that maps out key resistance hubs, while robust networks of biomarkers provide real-time tracking of how these hubs are progressively utilized over time in response to therapeutic assaults. In this situation, development of a deep armamentarium of targeted drugs will allow a rapid response to changing conditions, and flexible use of three and four agents in combination will prevent tumors from easily evading assaults. Notably, the pre-requisites for such a scenario would be a radical change in the structure and practice of clinical trials, as well as the investment of a very large amount of capital. These are challenges for the next decade.
Table 2.
Consequences of activating Kirsten rat sarcoma viral oncogene homolog (KRas) mutations on treatment outcomes in metastatic colorectal cancer (mCRC).
| Trial | Size | Treatment | PFS* (months)/Responses(%) | |
|---|---|---|---|---|
| KRasWT | KRasMut | |||
| CAIRO2 [225] | 755 | CapOxBev | −C : 10.7 months† | −C:12.5months |
| ±Cetuximab(C) | +C: 10.5 months | +C:8.6months, | ||
| p = 0.1 | p = 0.044 | |||
| CRYSTAL [224] | 540 | 1st line | −C: 8.1 months/43% | −C: 8.7 months/40% |
| FOLFIRI | +C:9.9 months/59% | +C:7.6 months/36% | ||
| ±Cetuximab(C) | p = 0.017/0.002 | p = 0.47 | ||
| OPUS.[229] | 337 | 1st line | −C: 7.7 months/37% | −C:8.6 months/49% |
| FOLFOX | +C:7.2 months/61% | +C:5.5mo/33% | ||
| ±Cetuximab(C) | p = 0.01 | p = 1 | ||
PFS (progression free survival) here defines time during which no tumor growth was detected radiographically; responses here are the sum of complete and partial responses.
The CAIRO2 study demonstrated no difference in PFS (shown above), response (40.6% versus 43.9%, p = 0.44 ) or median OS (20.4 versus 20.3 months, p = 0.21) upon adding cetuximab to capecitabine, oxaliplatin and bevacizumab. In KRas mutant colon cancer patients, the detrimental effect of the two-antibody combination reached statistical significance. A similar trend was reported in the OPUS study.
Acknowledgments
The authors were supported by NIH R01s CA63366 and CA113342, and DOD W81XWH-07-1-0676 from the Army Materiel Command (to EAG); K01 CA120091 (to EAH); by the Fox Chase-Genentech Career Development Award (to IA); by the Fox Chase Cancer Center (FCCC) Head and Neck Cancer Keystone Program; and by NIH core grant CA06927 and support from the Pew Charitable Fund (to FCCC).
Bibliography
Papers of special note have been highlighted as either of interest (*) or of considerable interest (**) to readers.
- 1.*.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–42. doi: 10.1056/NEJM200104053441402. The first trial in patients, to the authors’ knowledge, demonstrating the efficacy of imatinib; a breakthrough in the treatment of CML, and an optimal example of a targeted therapy. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 3.*.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. A proof-of-concept clinical trial showing synergy between an anti-EGFR antibody and chemotherapy. [DOI] [PubMed] [Google Scholar]
- 4.*.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. A demonstration of synergy between chemotherapy and trastuzumab in a subset of breast cancers selected on the basis of ErbB2/HER2. [DOI] [PubMed] [Google Scholar]
- 5.Pegram M, Hsu S, Lewis G, et al. Inhibitory effects of combinations of HER-2/neu antibody and chemotherapeutic agents used for treatment of human breast cancers. Oncogene. 1999;18:2241–51. doi: 10.1038/sj.onc.1202526. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson E. Surprise Phase III failure for ZD1839. Lancet Oncol. 2002;3:583. doi: 10.1016/s1470-2045(02)00883-5. [DOI] [PubMed] [Google Scholar]
- 7.Rao S, Cunningham D, de Gramont A, et al. Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J Clin Oncol. 2004;22:3950–7. doi: 10.1200/JCO.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 8.Sahin U, Weskamp G, Kelly K, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769–79. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.*.Lemmon MA, Bu Z, Ladbury JE, et al. Two EGF molecules contribute additively to stabilization of the EGFR dimer. Embo J. 1997;16:281–94. doi: 10.1093/emboj/16.2.281. The origin of the ‘one ligand-one receptor’ model for EGFR dimerization is described. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.*.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–74. doi: 10.1038/nature04177. This massive protein-array-based study analyzes the relative signaling properties of EGFR, ErbB2, and ErbB3 in regard to phospho-tyrosine-based interactions with numerous partner proteins. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Pickin KA, Bose R, et al. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–4. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Gureasko J, Shen K, et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Zhu F, Wang Z. Identification of EGF receptor C-terminal sequences 1005–1017 and di-leucine motif 1010LL1011 as essential in EGF receptor endocytosis. Exp Cell Res. 2007;313:3349–63. doi: 10.1016/j.yexcr.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Sigismund S, Argenzio E, Tosoni D, et al. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–19. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell. 2001;12:1897–910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.*.Wheeler DL, Huang S, Kruser TJ, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–56. doi: 10.1038/onc.2008.19. This paper describes deficient endocytosis as mechanism of resistance to cetuximab in SCCHN cell lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klapper LN, Vaisman N, Hurwitz E, et al. A subclass of tumor-inhibitory monoclonal antibodies to ErbB-2/HER2 blocks crosstalk with growth factor receptors. Oncogene. 1997;14:2099–109. doi: 10.1038/sj.onc.1201029. [DOI] [PubMed] [Google Scholar]
- 18.Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D. Quantitative analysis of HER2-mediated effects on HER2 and epidermal growth factor receptor endocytosis: distribution of homo- and heterodimers depends on relative HER2 levels. J Biol Chem. 2003;278:23343–51. doi: 10.1074/jbc.M300477200. [DOI] [PubMed] [Google Scholar]
- 19.Menendez JA, Mehmi I, Lupu R. Trastuzumab in combination with heregulin-activated Her-2 (erbB-2) triggers a receptor-enhanced chemosensitivity effect in the absence of Her-2 overexpression. J Clin Oncol. 2006;24:3735–46. doi: 10.1200/JCO.2005.04.3489. [DOI] [PubMed] [Google Scholar]
- 20.**.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. The discovery of Met amplification as a mechanism for erlotinib resistance, and illustrates the roles for transphosphorylation and ErbB3 in EGFR signaling is described. [DOI] [PubMed] [Google Scholar]
- 21.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100012. 2005 0008. Published online 25 May 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ushiro H, Cohen S. Identification of phosphotyrosine as a product of epidermal growth factor-activated protein kinase in A-431 cell membranes. J Biol Chem. 1980;255:8363–5. [PubMed] [Google Scholar]
- 23.Cohen S, Carpenter G, King L., Jr Epidermal growth factor-receptor-protein kinase interactions. Co-purification of receptor and epidermal growth factor-enhanced phosphorylation activity. J Biol Chem. 1980;255:4834–42. [PubMed] [Google Scholar]
- 24.*.Downward J, Yarden Y, Mayes E, et al. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–7. doi: 10.1038/307521a0. The initial discovery of sequence similarity between EGFR and v-erb, suggesting a role for EGFR in cancer is described. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–45. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 26.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 27.Vigneron A, Gamelin E, Coqueret O. The EGFR STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res. 2008;68:815–25. doi: 10.1158/0008-5472.CAN-07-5115. [DOI] [PubMed] [Google Scholar]
- 28.Benhar M, Engelberg D, Levitzki A. Cisplatin-induced activation of the EGF receptor. Oncogene. 2002;21:8723–31. doi: 10.1038/sj.onc.1205980. [DOI] [PubMed] [Google Scholar]
- 29.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. Published online 12 September 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch FR, Varella-Garcia M, Cappuzzo F, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18:752–60. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 32.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naramura M, Gillies SD, Mendelsohn J, et al. Therapeutic potential of chimeric and murine anti-(epidermal growth factor receptor) antibodies in a metastasis model for human melanoma. Cancer Immunol Immunother. 1993;37:343–9. doi: 10.1007/BF01518458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivera F, Vega-Villegas ME, Lopez-Brea MF, Marquez R. Current situation of Panitumumab, Matuzumab, Nimotuzumab and Zalutumumab. Acta Oncol. 2008;47:9–19. doi: 10.1080/02841860701704724. [DOI] [PubMed] [Google Scholar]
- 35.Lax I, Fischer R, Ng C, et al. Noncontiguous regions in the extracellular domain of EGF receptor define ligand-binding specificity. Cell Regul. 1991;2:337–45. doi: 10.1091/mbc.2.5.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.*.Ferguson KM, Berger MB, Mendrola JM, et al. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–17. doi: 10.1016/s1097-2765(03)00047-9. This paper describes the crystal structure of the EGF:EGFR complex, supporting a mechanistic understanding of the role for dimerization in the activation process. [DOI] [PubMed] [Google Scholar]
- 37.Mishima K, Johns TG, Luwor RB, et al. Growth suppression of intracranial xenografted glioblastomas overexpressing mutant epidermal growth factor receptors by systemic administration of monoclonal antibody (mAb) 806, a novel monoclonal antibody directed to the receptor. Cancer Res. 2001;61:5349–54. [PubMed] [Google Scholar]
- 38.Agus DB, Akita RW, Fox WD, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. 2002;2:127–37. doi: 10.1016/s1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 39.Weiner LM. Cancer immunotherapy the endgame begins. N Engl J Med. 2008;358:2664–5. doi: 10.1056/NEJMp0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003;8:411–24. doi: 10.1634/theoncologist.8-5-411. [DOI] [PubMed] [Google Scholar]
- 41.Lemos C, Jansen G, Peters GJ. Drug transporters: recent advances concerning BCRP and tyrosine kinase inhibitors. Br J Cancer. 2008;98:857–62. doi: 10.1038/sj.bjc.6604213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchetti S, de Vries NA, Buckle T, et al. Effect of the ATP-binding cassette drug transporters ABCB1, ABCG2, and ABCC2 on erlotinib hydrochloride (Tarceva) disposition in in vitro and in vivo pharmacokinetic studies employing Bcrp1−/−/Mdr1a/1b−/− (triple-knockout) and wild-type mice. Mol Cancer Ther. 2008;7:2280–7. doi: 10.1158/1535-7163.MCT-07-2250. [DOI] [PubMed] [Google Scholar]
- 43.Polli JW, Humphreys JE, Harmon KA, et al. The role of efflux and uptake transporters in N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethy l]amino}methyl)-2-furyl]-4-quinazolinamine ( GW572016, lapatinib) disposition and drug interactions. Drug Metab Dispos. 2008;36:695–701. doi: 10.1124/dmd.107.018374. [DOI] [PubMed] [Google Scholar]
- 44.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Garcia R, Franklin RA, McCubrey JA. EGF induces cell motility and multi-drug resistance gene expression in breast cancer cells. Cell Cycle. 2006;5:2820–6. doi: 10.4161/cc.5.23.3535. [DOI] [PubMed] [Google Scholar]
- 46.Stromskaya TP, Grigorian IA, Ossovskaya VS, et al. Cell-specific effects of RAS oncogene and protein kinase C agonist TPA on P-glycoprotein function. FEBS Lett. 1995;368:373–6. doi: 10.1016/0014-5793(95)00662-s. [DOI] [PubMed] [Google Scholar]
- 47.Stavrovskaya AA, Stromskaya TP. Transport proteins of the ABC family and multidrug resistance of tumor cells. Biochemistry (Moscow) 2008;73:592–604. doi: 10.1134/s0006297908050118. [DOI] [PubMed] [Google Scholar]
- 48.Schaich M, Ritter M, Illmer T, et al. Mutations in ras proto-oncogenes are associated with lower mdr1 gene expression in adult acute myeloid leukaemia. Br J Haematol. 2001;112:300–7. doi: 10.1046/j.1365-2141.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 49.Lee JT, Jr, Steelman LS, McCubrey JA. Phosphatidylinositol 3′-kinase activation leads to multidrug resistance protein-1 expression and subsequent chemoresistance in advanced prostate cancer cells. Cancer Res. 2004;64:8397–404. doi: 10.1158/0008-5472.CAN-04-1612. [DOI] [PubMed] [Google Scholar]
- 50.Scherbakova EA, Stromskaya TP, Rybalkina EY, et al. Role of the PTEN protein in the multidrug resistance of prostate cancer cells. Mol Biol (Moscow) 2008;42:430–435. [PubMed] [Google Scholar]
- 51.*.Yague E, Arance A, Kubitza L, et al. Ability to acquire drug resistance arises early during the tumorigenesis process. Cancer Res. 2007;67:1130–7. doi: 10.1158/0008-5472.CAN-06-2574. A provocative study demonstrating early appearance of elevated P-GP/MDR/ABCB1 and drug resistance in pre-malignant cells. [DOI] [PubMed] [Google Scholar]
- 52.Miyake K, Mickley L, Litman T, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: demonstration of homology to ABC transport genes. Cancer Res. 1999;59:8–13. [PubMed] [Google Scholar]
- 53.Robey RW, Polgar O, Deeken J, To KW, Bates SE. ABCG2: determining its relevance in clinical drug resistance. Cancer Metastasis Rev. 2007;26:39–57. doi: 10.1007/s10555-007-9042-6. [DOI] [PubMed] [Google Scholar]
- 54.Benderra Z, Faussat AM, Sayada L, et al. MRP3, BCRP, and P-glycoprotein activities are prognostic factors in adult acute myeloid leukemia. Clin Cancer Res. 2005;11:7764–72. doi: 10.1158/1078-0432.CCR-04-1895. [DOI] [PubMed] [Google Scholar]
- 55.Choi BH, Kim CG, Lim Y, Shin SY, Lee YH. Curcumin down-regulates the multidrug- resistance mdr1b gene by inhibiting the PI3K/Akt/NFκB pathway. Cancer Lett. 2008;259:111–8. doi: 10.1016/j.canlet.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Jullien-Flores V, Mahe Y, Mirey G, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113( Pt 16):2837–44. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 57.Awasthi S, Singhal SS, Sharma R, et al. Transport of glutathione conjugates and chemotherapeutic drugs by RLIP76 (RALBP1): a novel link between G-protein and tyrosine kinase signaling and drug resistance. Int J Cancer. 2003;106:635–46. doi: 10.1002/ijc.11260. [DOI] [PubMed] [Google Scholar]
- 58.Jullien-Flores V, Dorseuil O, Romero F, et al. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–7. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 59.Konecny GE, Venkatesan N, Yang G, et al. Activity of lapatinib a novel HER2 and EGFR dual kinase inhibitor in human endometrial cancer cells. Br J Cancer. 2008;98:1076–84. doi: 10.1038/sj.bjc.6604278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren W, Korchin B, Zhu QS, et al. Epidermal growth factor receptor blockade in combination with conventional chemotherapy inhibits soft tissue sarcoma cell growth in vitro and in vivo. Clin Cancer Res. 2008;14:2785–95. doi: 10.1158/1078-0432.CCR-07-4471. [DOI] [PubMed] [Google Scholar]
- 61.Rosell R, Robinet G, Szczesna A, et al. Randomized phase II study of cetuximab plus cisplatin/vinorelbine compared with cisplatin/vinorelbine alone as first-line therapy in EGFR-expressing advanced non-small-cell lung cancer. Ann Oncol. 2008;19:362–9. doi: 10.1093/annonc/mdm474. [DOI] [PubMed] [Google Scholar]
- 62.Lopez JP, Wang-Rodriguez J, Chang C, et al. Gefitinib inhibition of drug resistance to doxorubicin by inactivating ABCG2 in thyroid cancer cell lines. Arch Otolaryngol Head Neck Surg. 2007;133:1022–7. doi: 10.1001/archotol.133.10.1022. [DOI] [PubMed] [Google Scholar]
- 63.Awasthi S, Singhal SS, Singhal J, Cheng J, Zimniak P, Awasthi YC. Role of RLIP76 in lung cancer doxorubicin resistance: II. Doxorubicin transport in lung cancer by RLIP76. Int J Oncol. 2003;22:713–20. [PubMed] [Google Scholar]
- 64.Ozvegy-Laczka C, Hegedus T, Varady G, et al. High-affinity interaction of tyrosine kinase inhibitors with the ABCG2 multidrug transporter. Mol Pharmacol. 2004;65:1485–95. doi: 10.1124/mol.65.6.1485. [DOI] [PubMed] [Google Scholar]
- 65.Shi Z, Peng XX, Kim IW, et al. Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 2007;67:11012–20. doi: 10.1158/0008-5472.CAN-07-2686. [DOI] [PubMed] [Google Scholar]
- 66.Steinbach D, Legrand O. ABC transporters and drug resistance in leukemia: was P-gp nothing but the first head of the Hydra? Leukemia. 2007;21:1172–6. doi: 10.1038/sj.leu.2404692. [DOI] [PubMed] [Google Scholar]
- 67.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 68.Sung JM, Cho HJ, Yi H, et al. Characterization of a stem cell population in lung cancer A549 cells. Biochem Biophys Res Commun. 2008;371:163–7. doi: 10.1016/j.bbrc.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 69.Yin L, Castagnino P, Assoian RK. ABCG2 expression and side population abundance regulated by a transforming growth factor β-directed epithelial-mesenchymal transition. Cancer Res. 2008;68:800–7. doi: 10.1158/0008-5472.CAN-07-2545. [DOI] [PubMed] [Google Scholar]
- 70.Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov. 2008;7:989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- 71.Deng J, Shimamura T, Perera S, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–75. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- 72.Ling YH, Lin R, Perez-Soler R. Erlotinib induces mitochondrial-mediated apoptosis in human H3255 non-small-cell lung cancer cells with epidermal growth factor receptorL858R mutation through mitochondrial oxidative phosphorylation-dependent activation of BAX and BAK. Mol Pharmacol. 2008;74:793–806. doi: 10.1124/mol.107.044396. [DOI] [PubMed] [Google Scholar]
- 73.Cragg MS, Kuroda J, Puthalakath H, et al. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:1681–89. doi: 10.1371/journal.pmed.0040316. discussion 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin AP, Miller A, Emad L, et al. Lapatinib resistance in HCT116 cells is mediated by elevated MCL-1 expression and decreased BAK activation and not by ERBB receptor kinase mutation. Mol Pharmacol. 2008;74:807–22. doi: 10.1124/mol.108.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naresh A, Long W, Vidal GA, et al. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66:6412–20. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- 76.*.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–80. doi: 10.1158/0008-5472.CAN-07-3293. discussion 80. This paper thoroughly discusses the oncogene addiction view of cancer. [DOI] [PubMed] [Google Scholar]
- 77.Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008;26:3351–7. doi: 10.1200/JCO.2007.14.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.*.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. In this paper clinical evidence for the increased efficacy of erlotinib in patients with hyperactive EGFR arising from mutations in the EGFR kinase domain is presented. [DOI] [PubMed] [Google Scholar]
- 79.Taron M, Ichinose Y, Rosell R, et al. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5878–85. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 80.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 81.Li D, Shimamura T, Ji H, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 82.Leone F, Cavalloni G, Pignochino Y, et al. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clin Cancer Res. 2006;12:1680–5. doi: 10.1158/1078-0432.CCR-05-1692. [DOI] [PubMed] [Google Scholar]
- 83.Nakazawa K, Dobashi Y, Suzuki S, et al. Amplification and overexpression of c-erbB-2, epidermal growth factor receptor, and c-met in biliary tract cancers. J Pathol. 2005;206:356–65. doi: 10.1002/path.1779. [DOI] [PubMed] [Google Scholar]
- 84.Kwak EL, Jankowski J, Thayer SP, et al. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res. 2006;12:4283–7. doi: 10.1158/1078-0432.CCR-06-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Janmaat ML, Gallegos-Ruiz MI, Rodriguez JA, et al. Predictive factors for outcome in a Phase II study of gefitinib in second-line treatment of advanced esophageal cancer patients. J Clin Oncol. 2006;24:1612–9. doi: 10.1200/JCO.2005.03.4900. [DOI] [PubMed] [Google Scholar]
- 86.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.*.Kim HP, Han SW, Kim SH, et al. Combined lapatinib and cetuximab enhance cytotoxicity against gefitinib-resistant lung cancer cells. Mol Cancer Ther. 2008;7:607–15. doi: 10.1158/1535-7163.MCT-07-2068. This paper describes a novel mechanism of resistance to erlotinib arising from increased binding affinity of EGFR to ATP. [DOI] [PubMed] [Google Scholar]
- 88.Doody JF, Wang Y, Patel SN, et al. Inhibitory activity of cetuximab on epidermal growth factor receptor mutations in non small cell lung cancers. Mol Cancer Ther. 2007;6:2642–51. doi: 10.1158/1535-7163.MCT-06-0506. [DOI] [PubMed] [Google Scholar]
- 89.Perez-Torres M, Guix M, Gonzalez A, Arteaga CL. Epidermal growth factor receptor (EGFR) antibody down-regulates mutant receptors and inhibits tumors expressing EGFR mutations. J Biol Chem. 2006;281:40183–92. doi: 10.1074/jbc.M607958200. [DOI] [PubMed] [Google Scholar]
- 90.Steiner P, Joynes C, Bassi R, et al. Tumor growth inhibition with cetuximab and chemotherapy in non-small cell lung cancer xenografts expressing wild-type and mutated epidermal growth factor receptor. Clin Cancer Res. 2007;13:1540–51. doi: 10.1158/1078-0432.CCR-06-1887. [DOI] [PubMed] [Google Scholar]
- 91.Kim HP, Han SW, Kim SH, et al. Combined lapatinib and cetuximab enhance cytotoxicity against gefitinib-resistant lung cancer cells. Mol Cancer Ther. 2008;7:607–15. doi: 10.1158/1535-7163.MCT-07-2068. [DOI] [PubMed] [Google Scholar]
- 92.Ramalingam S, Forster J, Naret C, et al. Dual inhibition of the epidermal growth factor receptor with cetuximab, an IgG1 monoclonal antibody, and gefitinib, a tyrosine kinase inhibitor, in patients with refractory non-small cell lung cancer (NSCLC): a Phase I study. J Thorac Oncol. 2008;3:258–64. doi: 10.1097/JTO.0b013e3181653d1b. [DOI] [PubMed] [Google Scholar]
- 93.Karamouzis MV, Grandis JR, Argiris A. Therapies directed against epidermal growth factor receptor in aerodigestive carcinomas. JAMA. 2007;298:70–82. doi: 10.1001/jama.298.1.70. [DOI] [PubMed] [Google Scholar]
- 94.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–75. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 95.Yonesaka K, Zejnullahu K, Lindeman N, et al. Autocrine production of amphiregulin predicts sensitivity to both gefitinib and cetuximab in EGFR wild-type cancers. Clin Cancer Res. 2008;14:6963–73. doi: 10.1158/1078-0432.CCR-08-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 97.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a Phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:5892–9. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 98.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non- small-cell lung cancer. N Engl J Med. 2005;353:123–32. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 99.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–7. [PubMed] [Google Scholar]
- 100.Humphrey PA, Gangarosa LM, Wong AJ, et al. Deletion-mutant epidermal growth factor receptor in human gliomas: effects of type II mutation on receptor function. Biochem Biophys Res Commun. 1991;178:1413–20. doi: 10.1016/0006-291x(91)91051-d. [DOI] [PubMed] [Google Scholar]
- 101.Sok JC, Coppelli FM, Thomas SM, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–73. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 102.Learn CA, Hartzell TL, Wikstrand CJ, et al. Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Clin Cancer Res. 2004;10:3216–24. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- 103.Ji H, Zhao X, Yuza Y, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang W, Wu G, Barth RF, et al. Molecular targeting and treatment of composite EGFR and EGFRvIII-positive gliomas using boronated monoclonal antibodies. Clin Cancer Res. 2008;14:883–91. doi: 10.1158/1078-0432.CCR-07-1968. [DOI] [PubMed] [Google Scholar]
- 105.Patel D, Lahiji A, Patel S, et al. Monoclonal antibody cetuximab binds to and down-regulates constitutively activated epidermal growth factor receptor vIII on the cell surface. Anticancer Res. 2007;27:3355–66. [PubMed] [Google Scholar]
- 106.Johns TG, Perera RM, Vernes SC, et al. The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res. 2007;13:1911–25. doi: 10.1158/1078-0432.CCR-06-1453. [DOI] [PubMed] [Google Scholar]
- 107.Luwor RB, Zhu HJ, Walker F, et al. The tumor-specific de2-7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene. 2004;23:6095–104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- 108.Laheru D, Croghan G, Bukowski R, et al. A Phase I study of EKB-569 in combination with capecitabine in patients with advanced colorectal cancer. Clin Cancer Res. 2008;14:5602–9. doi: 10.1158/1078-0432.CCR-08-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wong KK. HKI-272 in non small cell lung cancer. Clin Cancer Res. 2007;13:s4593–6. doi: 10.1158/1078-0432.CCR-07-0369. [DOI] [PubMed] [Google Scholar]
- 110.Castillo J, Erroba E, Perugorria MJ, et al. Amphiregulin contributes to the transformed phenotype of human hepatocellular carcinoma cells. Cancer Res. 2006;66:6129–38. doi: 10.1158/0008-5472.CAN-06-0404. [DOI] [PubMed] [Google Scholar]
- 111.Kakiuchi S, Daigo Y, Ishikawa N, et al. Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839) Hum Mol Genet. 2004;13:3029–43. doi: 10.1093/hmg/ddh331. [DOI] [PubMed] [Google Scholar]
- 112.Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 113.**.Li S, Schmitz KR, Jeffrey PD, et al. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–11. doi: 10.1016/j.ccr.2005.03.003. This paper describes the structural basis for cetuximab action, based on interference with ligand access to EGFR. [DOI] [PubMed] [Google Scholar]
- 114.Tanaka Y, Miyamoto S, Suzuki SO, et al. Clinical significance of heparin-binding epidermal growth factor-like growth factor and a disintegrin and metalloprotease 17 expression in human ovarian cancer. Clin Cancer Res. 2005;11:4783–92. doi: 10.1158/1078-0432.CCR-04-1426. [DOI] [PubMed] [Google Scholar]
- 115.Zhou BB, Peyton M, He B, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang F, Liu R, Lee SW, et al. Heparin-binding EGF-like growth factor is an early response gene to chemotherapy and contributes to chemotherapy resistance. Oncogene. 2007;26:2006–16. doi: 10.1038/sj.onc.1209999. [DOI] [PubMed] [Google Scholar]
- 117.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–19. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang SE, Narasanna A, Perez-Torres M, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 119.Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–43. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 120.Arteaga CL, O’Neill A, Moulder SL, et al. A Phase I II study of combined blockade of the ErbB receptor network with trastuzumab and gefitinib in patients with HER2 (ErbB2)-overexpressing metastatic breast cancer. Clin Cancer Res. 2008;14:6277–83. doi: 10.1158/1078-0432.CCR-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gordon MS, Matei D, Aghajanian C, et al. Clinical activity of pertuzumab (rhuMAb 2C4), a HER dimerization inhibitor, in advanced ovarian cancer: potential predictive relationship with tumor HER2 activation status. J Clin Oncol. 2006;24:4324–32. doi: 10.1200/JCO.2005.05.4221. [DOI] [PubMed] [Google Scholar]
- 122.Wong TW, Lee FY, Yu C, et al. Preclinical antitumor activity of BMS-599626, a pan-HER kinase inhibitor that inhibits HER1/HER2 homodimer and heterodimer signaling. Clin Cancer Res. 2006;12:6186–93. doi: 10.1158/1078-0432.CCR-06-0642. [DOI] [PubMed] [Google Scholar]
- 123.Chen X, Yeung TK, Wang Z. Enhanced drug resistance in cells coexpressing ErbB2 with EGF receptor or ErbB3. Biochem Biophys Res Commun. 2000;277:757–63. doi: 10.1006/bbrc.2000.3731. [DOI] [PubMed] [Google Scholar]
- 124.Frolov A, Schuller K, Tzeng CW, et al. ErbB3 expression and dimerization with EGFR influence pancreatic cancer cell sensitivity to erlotinib. Cancer Biol Ther. 2007;6:548–54. doi: 10.4161/cbt.6.4.3849. [DOI] [PubMed] [Google Scholar]
- 125.Rothenberg SM, Engelman JA, Le S, et al. Modeling oncogene addiction using RNA interference. Proc Natl Acad Sci U S A. 2008;105:12480–4. doi: 10.1073/pnas.0803217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang J, Kalyankrishna S, Wislez M, et al. SRC-family kinases are activated in non-small cell lung cancer and promote the survival of epidermal growth factor receptor-dependent cell lines. Am J Pathol. 2007;170:366–76. doi: 10.2353/ajpath.2007.060706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–10. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- 128.Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–87. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 129.**.Sergina NV, Rausch M, Wang D, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–41. doi: 10.1038/nature05474. This paper demonstrates the redundancy of the ErbB family network as a mechanism for resistance to the ErbB2/HER2-inhibiting antibody trastuzumab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J Thorac Oncol. 2008;3:331–9. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- 132.Jiang SX, Yamashita K, Yamamoto M, et al. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. Int J Cancer. 2008;123:2480–6. doi: 10.1002/ijc.23868. [DOI] [PubMed] [Google Scholar]
- 133.Morgillo F, Kim WY, Kim ES, et al. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 134.Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res. 2002;62:200–7. [PubMed] [Google Scholar]
- 135.O’Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wan X, Harkavy B, Shen N, et al. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26:1932–40. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
- 137.Viloria-Petit A, Crombet T, Jothy S, et al. Acquired resistance to the antitumor effect of epidermal growth factor receptor-blocking antibodies in vivo: a role for altered tumor angiogenesis. Cancer Res. 2001;61:5090–101. [PubMed] [Google Scholar]
- 138.Bianco R, Rosa R, Damiano V, et al. Vascular endothelial growth factor receptor-1 contributes to resistance to anti-epidermal growth factor receptor drugs in human cancer cells. Clin Cancer Res. 2008;14:5069–80. doi: 10.1158/1078-0432.CCR-07-4905. [DOI] [PubMed] [Google Scholar]
- 139.*.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. A landmark study establishing the profile of genes mutationally targeted in 11 breast and 11 colorectal cancers. [DOI] [PubMed] [Google Scholar]
- 140.Guo A, Villen J, Kornhauser J, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–7. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gupta S, Ramjaun AR, Haiko P, et al. Binding of ras to phosphoinositide 3-kinase p110α is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 142.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 143.Freeman DJ, Juan T, Reiner M, et al. Association of K-ras mutational status and clinical outcomes in patients with metastatic colorectal cancer receiving panitumumab alone. Clin Colorectal Cancer. 2008;7:184–90. doi: 10.3816/CCC.2008.n.024. [DOI] [PubMed] [Google Scholar]
- 144.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer research. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 145.Amado RGWM, Freeman D, et al. Analysis of KRAS mutations in patients with metastatic colorectal cancer receiving panitumumab monotherapy. Euro J Cancer. 2007;5(6S):8. [Google Scholar]
- 146.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. published online 25 January, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 148.Weir B, Zhao X, Meyerson M. Somatic alterations in the human cancer genome. Cancer Cell. 2004;6:433–8. doi: 10.1016/j.ccr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 149.Guo XN, Rajput A, Rose R, et al. Mutant PIK3CA-bearing colon cancer cells display increased metastasis in an orthotopic model. Cancer Res. 2007;67:5851–8. doi: 10.1158/0008-5472.CAN-07-0049. [DOI] [PubMed] [Google Scholar]
- 150.Wang J, Kuropatwinski K, Hauser J, et al. Colon carcinoma cells harboring PIK3CA mutations display resistance to growth factor deprivation induced apoptosis. Mol Cancer Ther. 2007;6:1143–50. doi: 10.1158/1535-7163.MCT-06-0555. [DOI] [PubMed] [Google Scholar]
- 151.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 152.Bianco R, Shin I, Ritter CA, et al. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene. 2003;22:2812–22. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- 153.Jhawer M, Goel S, Wilson AJ, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–61. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dittmann K, Mayer C, Fehrenbacher B, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–9. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 155.Lin SY, Makino K, Xia W, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 156.*.Sigismund S, Woelk T, Puri C, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A. 2005;102:2760–5. doi: 10.1073/pnas.0409817102. This paper demonstrates that EGFR endocytosis may lead to receptor recycling versus receptor degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314:3093–106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nishimura Y, Bereczky B, Ono M. The EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis of EGFR via the early/late endocytic pathway in non-small cell lung cancer cell lines. Histochem Cell Biol. 2007;127:541–53. doi: 10.1007/s00418-007-0281-y. [DOI] [PubMed] [Google Scholar]
- 159.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Balaban N, Moni J, Shannon M, et al. The effect of ionizing radiation on signal transduction: antibodies to EGF receptor sensitize A431 cells to radiation. Biochim Biophys Acta. 1996;1314:147–56. doi: 10.1016/s0167-4889(96)00068-7. [DOI] [PubMed] [Google Scholar]
- 161.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 162.Burtness B, Goldwasser MA, Flood W, et al. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol. 2005;23:8646–54. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- 163.Rossi A, Bria E, Maione P, et al. The role of cetuximab and other epidermal growth factor receptor monoclonal antibodies in the treatment of advanced non-small cell lung cancer. Rev Recent Clin Trials. 2008;3:217–27. doi: 10.2174/157488708785700276. [DOI] [PubMed] [Google Scholar]
- 164.Kim S, Prichard CN, Younes MN, et al. Cetuximab and irinotecan interact synergistically to inhibit the growth of orthotopic anaplastic thyroid carcinoma xenografts in nude mice. Clin Cancer Res. 2006;12:600–7. doi: 10.1158/1078-0432.CCR-05-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Prewett M, Deevi DS, Bassi R, et al. Tumors established with cell lines selected for oxaliplatin resistance respond to oxaliplatin if combined with cetuximab. Clin Cancer Res. 2007;13:7432–40. doi: 10.1158/1078-0432.CCR-07-1768. [DOI] [PubMed] [Google Scholar]
- 166.Bonner JA, Raisch KP, Trummell HQ, et al. Enhanced apoptosis with combination C225/radiation treatment serves as the impetus for clinical investigation in head and neck cancers. J Clin Oncol. 2000;18:47S–53S. [PubMed] [Google Scholar]
- 167.Grandis JR, Drenning SD, Chakraborty A, et al. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102:1385–92. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Song JI, Grandis JR. STAT signaling in head and neck cancer. Oncogene. 2000;19:2489–95. doi: 10.1038/sj.onc.1203483. [DOI] [PubMed] [Google Scholar]
- 169.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–9. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 170.Gatzemeier U, Pluzanska A, Szczesna A, et al. Phase III study of erlotinib in combination with cisplatin and gemcitabine in advanced non-small-cell lung cancer: the Tarceva Lung Cancer Investigation Trial. J Clin Oncol. 2007;25:1545–52. doi: 10.1200/JCO.2005.05.1474. [DOI] [PubMed] [Google Scholar]
- 171.Kelly K, Chansky K, Gaspar LE, et al. Phase III trial of maintenance gefitinib or placebo after concurrent chemoradiotherapy and docetaxel consolidation in inoperable stage III non-small-cell lung cancer: SWOG S0023. J Clin Oncol. 2008;26:2450–6. doi: 10.1200/JCO.2007.14.4824. [DOI] [PubMed] [Google Scholar]
- 172.Stebbing J. FLEX data, KRAS and ERCC1 testing in oncology. Future Oncol. 2008;4:471–3. doi: 10.2217/14796694.4.4.471. [DOI] [PubMed] [Google Scholar]
- 173.Solit DB, She Y, Lobo J, et al. Pulsatile administration of the epidermal growth factor receptor inhibitor gefitinib is significantly more effective than continuous dosing for sensitizing tumors to paclitaxel. Clin Cancer Res. 2005;11:1983–9. doi: 10.1158/1078-0432.CCR-04-1347. [DOI] [PubMed] [Google Scholar]
- 174.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 175.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 176.Kassouf W, Dinney CP, Brown G, et al. Uncoupling between epidermal growth factor receptor and downstream signals defines resistance to the antiproliferative effect of Gefitinib in bladder cancer cells. Cancer Res. 2005;65:10524–35. doi: 10.1158/0008-5472.CAN-05-1536. [DOI] [PubMed] [Google Scholar]
- 177.Golemis EA, Serebriiskii I, Finley RL, et al. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Mol Biol. 2008 doi: 10.1002/0471142727.mb2001s82. Chapter 20:Unit 20 1. publishhed online April 2008. [DOI] [PubMed] [Google Scholar]
- 178.Ratushny V, Golemis E. Resolving the network of cell signaling pathways using the evolving yeast two-hybrid system. Biotechniques. 2008;44:655–62. doi: 10.2144/000112797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wissing J, Jansch L, Nimtz M, et al. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol Cell Proteomics. 2007;6:537–47. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- 180.Wissing J, Jansch L, Dickinson WJ. Marginal fitness contributions of nonessential genes in yeast. Mol Cell Proteomics. 1998;6:253–7. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hiatt SM, Shyu YJ, Duren HM, Hu CD. Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in Caenorhabditis elegans. Methods. 2008;45:185–91. doi: 10.1016/j.ymeth.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hiatt SM, Shyu YJ, Duren HM, Hu CD. Perspective: Evolution and detection of genetic robustness. Methods. 2003;57:185–91. [Google Scholar]
- 183.Kruger M, Moser M, Ussar S, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–64. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 184.Yu H, Braun P, Yildirim MA, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–10. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.**.Pujana MA, Han JD, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–49. doi: 10.1038/ng.2007.2. In this paper network modeling was used to identify novel regulators of centrosome function and their role in breast cancer. [DOI] [PubMed] [Google Scholar]
- 186.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.**.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–4. doi: 10.1038/nature05280. A functional genomics approach to identify complex modulators of RTK/ERK pathways is described. [DOI] [PubMed] [Google Scholar]
- 188.**.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–31. doi: 10.1016/j.cell.2007.01.007. An outstanding perspective on the emerging literature relevant to biological networks. [DOI] [PubMed] [Google Scholar]
- 189.Handcock MS, Hunter DR, Butts CT, et al. Statnet: Software tools for the representation, visualization, analysis and simulation of network data. J Stat Softw. 2008;24:1548–7660. doi: 10.18637/jss.v024.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Stelling J, Sauer U, Szallasi Z, et al. Robustness of cellular functions. Cell. 2004;118:675–85. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 191.Thatcher JW, Shaw JM, Dickinson WJ. Marginal fitness contributions of nonessential genes in yeast. Proc Natl Acad Sci U S A. 1998;95:253–7. doi: 10.1073/pnas.95.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Krakauer DC, Plotkin JB. Redundancy, antiredundancy, and the robustness of genomes. Proc Natl Acad Sci U S A. 2002;99:1405–9. doi: 10.1073/pnas.032668599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kitano H. Cancer as a robust system: implications for anticancer therapy. Nat Rev Cancer. 2004;4:227–35. doi: 10.1038/nrc1300. [DOI] [PubMed] [Google Scholar]
- 194.de Visser JA, Hermisson J, Wagner GP, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–72. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 195.*.Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. The use of high throughput two-hybrid screening to identify physical interactions between many human proteins, including many associated with disease is described. [DOI] [PubMed] [Google Scholar]
- 196.Singh M, Cowell L, Seo S, et al. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48:54–72. doi: 10.1007/s12013-007-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cohen EE, Lingen MW, Zhu B, et al. Protein kinase Cζ mediates epidermal growth factor-induced growth of head and neck tumor cells by regulating mitogen-activated protein kinase. Cancer Res. 2006;66:6296–303. doi: 10.1158/0008-5472.CAN-05-3139. [DOI] [PubMed] [Google Scholar]
- 198.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–12. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 199.Han JD, Bertin N, Hao T, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 200.Hinton TM, Coldwell MJ, Carpenter GA, et al. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem. 2007;282:1695–708. doi: 10.1074/jbc.M602780200. [DOI] [PubMed] [Google Scholar]
- 201.Therrien M, Wong AM, Rubin GM. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell. 1998;95:343–53. doi: 10.1016/s0092-8674(00)81766-3. [DOI] [PubMed] [Google Scholar]
- 202.**.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. An extremely thorough review establishing important principles necessary to understand and therapeutically target biological networks. [DOI] [PubMed] [Google Scholar]
- 203.Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 204.Yu YK, Gertz EM, Agarwala R, et al. Retrieval accuracy, statistical significance and compositional similarity in protein sequence database searches. Nucleic Acids Res. 2006;34:5966–73. doi: 10.1093/nar/gkl731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Peri S, Navarro JD, Amanchy R, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–71. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mishra GR, Suresh M, Kumaran K, et al. Human protein reference database 2006 update. Nucleic Acids Res. 2006;34:D411–4. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 208.Dexter DL, Leith JT. Tumor heterogeneity and drug resistance. J Clin Oncol. 1986;4:244–57. doi: 10.1200/JCO.1986.4.2.244. [DOI] [PubMed] [Google Scholar]
- 209.Gisselsson D, Pettersson L, Hoglund M, et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci U S A. 2000;97:5357–62. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gillies RJ, Gatenby RA. Hypoxia and adaptive landscapes in the evolution of carcinogenesis. Cancer Metastasis Rev. 2007;26:311–7. doi: 10.1007/s10555-007-9065-z. [DOI] [PubMed] [Google Scholar]
- 211.Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.*.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–15. doi: 10.1016/j.cell.2006.09.042. In this paper mathematical modeling was used tosimulate the role of harsh versus mild microenvironments in conditioning selection of mutations that promote invasion. [DOI] [PubMed] [Google Scholar]
- 214.Schulze A, Lehmann K, Jefferies HB, et al. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 2001;15:981–94. doi: 10.1101/gad.191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Buck E, Eyzaguirre A, Rosenfeld-Franklin M, et al. Feedback mechanisms promote cooperativity for small molecule inhibitors of epidermal and insulin-like growth factor receptors. Cancer Res. 2008;68:8322–32. doi: 10.1158/0008-5472.CAN-07-6720. [DOI] [PubMed] [Google Scholar]
- 216.Liu YF, Herschkovitz A, Boura-Halfon S, et al. Serine phosphorylation proximal to its phosphotyrosine binding domain inhibits insulin receptor substrate 1 function and promotes insulin resistance. Mol Cell Biol. 2004;24:9668–81. doi: 10.1128/MCB.24.21.9668-9681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 218.Van Dyke T. p53 and tumor suppression. N Engl J Med. 2007;356:79–81. doi: 10.1056/NEJMcibr066301. [DOI] [PubMed] [Google Scholar]
- 219.Earp HS, Dawson TL, Li X, Yu H. Heterodimerization and functional interaction between EGF receptor family members: a new signaling paradigm with implications for breast cancer research. Breast Cancer Res Treat. 1995;35:115–32. doi: 10.1007/BF00694752. [DOI] [PubMed] [Google Scholar]
- 220.Jones HE, Gee JM, Barrow D, et al. Inhibition of insulin receptor isoform-A signalling restores sensitivity to gefitinib in previously de novo resistant colon cancer cells. Br J Cancer. 2006;95:172–80. doi: 10.1038/sj.bjc.6603237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–27. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 223.Pirker R, Szczesna A, von Pawel J, et al. FLEX: A randomized, multicenter, phase III study of cetuximab in combination with cisplatin/vinorelbine (CV) versus CV alone in the first-line treatment of patients with advanced non-small cell lung cancer (NSCLC) J Clin Oncol (Meeting Abstracts) 2008;26:3. [Google Scholar]
- 224.Van Cutsem E, Lang I, D’Haens G, et al. KRAS status and efficacy in the first-line treatment of patients with metastatic colorectal cancer (mCRC) treated with FOLFIRI with or without cetuximab: The CRYSTAL experience. J Clin Oncol (Meeting Abstracts) 2008;26:2. [Google Scholar]
- 225.Punt CJ, Tol J, Rodenburg CJ, et al. Randomized phase III study of capecitabine, oxaliplatin, and bevacizumab with or without cetuximab in advanced colorectal cancer (ACC), the CAIRO2 study of the Dutch Colorectal Cancer Group (DCCG) J Clin Oncol (Meeting Abstracts) 2008;26:LBA4011. [Google Scholar]
- 226.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a Phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–6. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 227.Jimeno A, Kulesza P, Wheelhouse J, et al. Dual EGFR and mTOR targeting in squamous cell carcinoma models, and development of early markers of efficacy. Br J Cancer. 2007;96:952–9. doi: 10.1038/sj.bjc.6603656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hsieh AC, Moasser MM. Targeting HER proteins in cancer therapy and the role of the non-target HER3. Br J Cancer. 2007;97:453–7. doi: 10.1038/sj.bjc.6603910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bokemeyer C, Bondarenko I, Hartmann JT, et al. KRAS status and efficacy of first-line treatment of patients with metastatic colorectal cancer (mCRC) with FOLFOX with or without cetuximab: The OPUS experience. J Clin Oncol (Meeting Abstracts) 2008;26:4000. [Google Scholar]