Abstract
Phospholemman (PLM) belongs to the FXYD family of small ion transport regulators. When phosphorylated at Ser68, PLM inhibits cardiac Na+/Ca2+ exchanger (NCX1). We previously demonstrated that the cytoplasmic tail of PLM interacts with the proximal intracellular loop (residues 218–358), but not the transmembrane (residues 1–217 and 765–938) or Ca2+-binding (residues 371–508) domains, of NCX1. In this study, we used intact Na+/Ca2+ exchanger with various deletions in the intracellular loop to map the interaction sites with PLM. We first demonstrated by Western blotting and confocal immunofluorescence microscopy that wild-type (WT) NCX1 and its deletion mutants were expressed in transfected HEK-293 cells. Cotransfection with PLM and NCX1 (or its deletion mutants) in HEK-293 cells did not decrease expression of NCX1 (or its deletion mutants). Coexpression of PLM with WT NCX1 inhibited NCX1 current (INaCa). Deletion of residues 240–679, 265–373, 250–300, or 300–373 from WT NCX1 resulted in loss of inhibition of INaCa by PLM. Inhibition of INaCa by PLM was preserved when residues 229–237, 270–300, 328–330, or 330–373 were deleted from the intracellular loop of NCX1. These results suggest that PLM mediated inhibition of INaCa by interacting with two distinct regions (residues 238–270 and 300–328) of NCX1. Indeed, INaCa measured in mutants lacking residues 238–270, 300–328, or 238–270 + 300–328 was not affected by PLM. Glutathione S-transferase pull-down assays confirmed that PLM bound to fragments corresponding to residues 218–371, 218–320, 218–270, 238–371, and 300–373, but not to fragments encompassing residues 250–300 and 371–508 of NCX1, indicating that residues 218–270 and 300–373 physically associated with PLM. Finally, acute regulation of INaCa by PLM phosphorylation observed with WT NCX1 was absent in 250–300 deletion mutant but preserved in 229–237 deletion mutant. We conclude that PLM mediates its inhibition of NCX1 by interacting with residues 238–270 and 300–328.
Keywords: FXYD1, ion transport, sodium-calcium exchange
phospholemman (PLM), the first cloned member of the FXYD family of regulators of ion transport (35), is a 72-amino acid sarcolemmal protein with a single transmembrane (TM) domain (27). The signature PFXYD motif is located in the extracellular NH2 terminus. The cytoplasmic tail of human, rat, and dog PLM contains three serines (at residues 62, 63, and 68) and one threonine (at residue 69), but Thr69 is replaced by serine in mouse PLM. NMR (10) and infrared spectroscopy (2) showed that the TM domain of PLM reconstituted in liposomes is an α-helix with a maximum tilt of 15–17°. Specifically, NMR spectroscopic studies of highly purified PLM in model micelles indicate that the molecule consists of four α-helices: H1 (residues 12–17) is in the extracellular NH2 terminus, H2 (residues 22–38) is the main TM helix followed by the short H3 (residues 39–45), and H4 (residues 60–68) in the COOH terminus is connected to H3 by a flexible linker (36). PKA phosphorylates Ser68, whereas PKC phosphorylates Ser63 and Ser68, of PLM (37). In addition, PLM is also a substrate for myotonic dystrophy protein kinase (23) and never in mitosis A kinase (19). In transfected Madin-Darby canine kidney cells expressing mouse PLM, phosphorylation of Ser69 is important in shifting the distribution of PLM from the endoplasmic reticulum to the plasma membrane (16).
PLM regulates the activities of Na+-K+-ATPase (4, 6, 7, 11, 32) and Na+/Ca2+ exchanger (NCX1) (1, 40) in the heart. Phosphorylation of Ser68 of PLM by PKA or PKC relieves its inhibition on Na+-K+-ATPase (7, 11, 12, 28, 32). By contrast, PLM phosphorylated at Ser68 is the active species that inhibits NCX1 (34, 39). PLM and its unphosphorylatable Ser68-to-alanine (S68A) mutant coimmunoprecipitate with NCX1 in transfected HEK-293 cells (34). Similarly, phosphorylated and unphosphorylated PLM coimmunoprecipitate with Na+-K+-ATPase in rabbit ventricular myocytes (4). These observations suggest that although changes in PLM phosphorylation are critical in regulating Na+-K+-ATPase and NCX1 activities, phosphorylation at Ser68 does not appear to affect association of PLM with these two ion transporters as detected by coimmunoprecipitation techniques.
Among the many transporters and ion channels involved in cardiac Ca2+ fluxes, NCX1 is unique, in that it mediates Ca2+ efflux (3 Na+ in:1 Ca2+ out; forward mode) and Ca2+ influx (3 Na+ out:1 Ca2+ in; reverse mode) during an excitation-contraction cycle (3). NCX1 is a 938-amino acid (939 amino acids in the rat) consisting of an NH2-terminal domain comprising the first five TM segments, a large intracellular loop (residues 218–764), and a COOH-terminal domain consisting of the last four TM segments (25, 29). The NH2 terminus of NCX1 is extracellular; the COOH terminus is intracellular. The α-repeats in TM segments 2, 3, and 7 of NCX1 are important in ion transport activity (15, 24), while the large intracellular loop contains the regulatory domains of the exchanger (17, 18, 20). Specifically, the exchange inhibitory peptide (XIP) region (residues 219–238) (18) associated with regulation of NCX1 activity by Na+, Ca2+, and phosphatidylinositol 4,5-bisphosphate (13, 21), the proximal linker domain (residues 259–370), the two consecutive Ca2+-binding domains 1 (residues 371–508) (17) and 2 (residues 501–650) (14), and the interaction site for endogenous XIP (residues 562–679) (20) reside within the intracellular loop. Using glutathione S-transferase (GST) pull-down assays and split NCX1 exchangers consisting of NH2- or COOH-terminal domains with varying lengths of intracellular loop (26), we previously demonstrated that PLM interacts and associates with a region encompassing residues 218–358 of the intracellular loop of NCX1 (38).
Here, we coexpressed intact Na+/Ca2+ exchanger modified by deletions of varying lengths of the intracellular loop and PLM in HEK-293 cells. Wild-type (WT) and NCX1 loop deletion mutants were correctly targeted to the plasma membrane and resulted in functional NCX1 current (INaCa). PLM inhibited INaCa only when residues 238–270 and 300–328 of the intracellular loop were present in NCX1 deletion mutants. Protein constructs of GST fused to various segments of the intracellular loop of NCX1 confirmed that PLM physically associated with fragments encompassing residues 218–270 and 300–373, but not residues 250–300. PLM phosphorylation (Ser68) by forskolin resulted in enhanced inhibition of INaCa in cells cotransfected with WT NCX1 or a deletion mutant (residues 229–237) in which regulation by PLM was preserved, but not in cells cotransfected with another deletion mutant (residues 250–300) in which regulation by and association with PLM was disrupted. We conclude that PLM inhibited INaCa by interacting with regions encompassing residues 238–270 and 300–328 of NCX1.
METHODS
NCX1 deletion mutants.
The full-length rat NCX1 (41) was used to produce discrete regions of the NCX1 open reading frame by PCR using Pfu Ultra II polymerase. Number designations for each fragment refer to the amino acid position along the mature protein sequence. Forward and reverse primers containing restriction endonuclease sites used for ligation (underlined) were designed, and their identities are indicated after each sequence: 5′-dAGATCTGGTACCATGCTTCGACTAAGTCTCCCA-3′ (forward) KpnI and 5′-dTCTAGAGTCGACCTGCTTAAGTTCCTTCAGAAT-3′ (reverse) SalI for NCX1/1–300; 5′-dAGATCTGGTACCATGCTTCGACTAAGTCTCCCA-3′ (forward) KpnI and 5′-dTCTAGAGTCGACAACGTGGGAGTTGACTACTTT-3′ (reverse) SalI for NCX1/1–265; 5′-dAGATCTGGTACCATGCTTCGACTAAGTCTCCCA-3′ (forward) KpnI and 5′-dTCTAGAGTCGACGGAAGCTGGTCTGTCTCCTCC-3′ (reverse) SalI for NCX1/1–250; 5′-dAGATCTGGTACCATGCTTCGACTAAGTCTCCCA-3′ (forward) KpnI and 5′-dTCTAGAGTCGACATCTAAGAAATTGTCAACGTG-3′ (reverse) SalI for NCX1/1–270; 5′-dTCTAGAGTCGACAAGGTCTTCTTTGAGCAAGGG-3′ (forward) SalI and 5′-dGGTACCAAGCTTTTAGAAGCCTTTTATGTGGCA-3′ (reverse) HindIII for NCX1/373–939; 5′-dTCTAGAGTCGACCAGAAGCATCCCGACAAAGAG-3′ (forward) SalI and 5′-dGGTACCAAGCTTTTAGAAGCCTTTTATGTGGCA-3′ (reverse) HindIII for NCX1/300–939.
The PCR fragments were ligated at their SalI sites and cloned into pAdTrack-CMV using the KpnI and HindIII sites to produce the following rat NCX1 deletion mutants: Δ265–373, Δ300–373, Δ250–300, and Δ270–300. After transformation into XL2-Blue Ultracompetent cells (Stratagene), clones were confirmed by restriction endonuclease mapping and by DNA sequencing.
In some experiments, dog NCX1 deletion mutants in BlueScript were repackaged into pAdTrack-CMV vector. Dog NCX1 Δ240–679 (22) and Δ229–237 (21) deletion mutants were constructed as previously described. For the Δ330–373 deletion mutant, BamHI restriction sites were introduced at residues 330 and 373, and the resultant fragment was excised. Dog NCX1 Δ328–330 deletion mutant was constructed by the QuickChange site-directed mutagenesis kit (Stratagene). Dog NCX1 inserts (Δ229–237, Δ328–330, Δ330–373, and Δ240–679 deletion mutants) in BlueScript were released by restriction digest using BamHI and HindIII, while the BlueScript was subjected to further digestion with FspI to reduce it to smaller fragments to facilitate identification of the NCX1 insert. The fragments were then ligated into pAdTrack-CMV at the BglII and HindIII sites. After transformation into XL2-Blue Ultracompetent cells, the clones were confirmed by restriction endonuclease mapping using HindIII and PacI and by DNA sequencing.
For production of the Δ238–270 + Δ300–328 double-deletion NCX1 mutant, three fragments containing restriction endonuclease sites used for ligation were constructed by PCR, with the full-length rat NCX1 used as template. The NCX1 fragments were as follows: KpnI 1–238 SalI, SalI 270–300 MluI, and MluI 328–939 HindIII. The three fragments were ligated and cloned into pAdTrack-CMV using KpnI and HindIII restriction sites. After transformation into XL2-Blue Ultracompetent cells, clones were confirmed by restriction endonuclease mapping and by DNA sequencing. Forward and reverse primers containing the desired restriction endonuclease sites (underlined) were designed, and their identities are indicated after each sequence: 5′-dAGATCTGGTACCATGCTTCGACTAAGTCTCCCA-3′ (forward) KpnI and 5′-dAGATCTGTCGACCCCCCTCTGCTTGCCAGCCCT-3′ (reverse) SalI for NCX1/1–238; 5′-dAGATCTGTCGACGATGGCGCTCTGGTTTTGGAA-3′ (forward) SalI and 5′-dAGATCTACGCGTCTGCTTAAGTTCCTTCAGAAT-3′ (reverse) MluI for NCX1/270–300; 5′-dAGATCTACGCGTTTTTACCGAATTCAAGCTACT-3′ (forward) MluI and 5′-dAGATCTAAGCTTTTAGAAGCCTTTTATGTGGCA-3′ (reverse) HindIII for NCX1/328–939.
For construction of Δ238–270 and Δ300–328 NCX1 deletion mutants, full-length rat NCX1 in pAdTrack-CMV vector was used as the template. Two complementary nucleotides containing the desired deletion flanked by unmodified nucleotide sequences were synthesized. Purified oliognucleotide primers were used to construct the NCX1 deletion mutants using the QuickChange II XL site-directed mutagenesis kit. Amplified products were treated with DpnI restriction enzyme to digest the nonmutated supercoiled double-stranded DNA. The resultant product was used to transform XL10-Gold Ultracompetent cells. Clones were confirmed by restriction endonuclease mapping and by DNA sequencing. Complementary oligonucleotide primers were as follows: 5′-dGGCAAGCAGAGGGGAGATGGCGCTCTGG-3′ and 5′-dCCAGAGCGCCATCTCCCCTCTGCTTGCC-3′ for the Δ238–270 deletion mutant and 5′-dGATTCTGAAGGAACTTAAGCAGTTTTACCGAATTCAAGCTACTC-3′ and 5′-dGAGTAGCTTGAATTCGGTAAAACTGCTTAAGTTCTTTCAGAATC-3′ for the Δ300–328 deletion mutant.
The species origin of the NCX1 deletion mutants is indicated by inclusion of r (rat) or d (dog) in the nomenclature. For example, dΔ328–330 indicates an NCX1 mutant derived from dog NCX1 with residues 328–330 deleted.
Transfection of HEK-293 cells.
HEK-293 cells were transfected with control pAdTrack-CMV vector alone (3 μg), vector (2 μg) + pAdTrack-CMV-NCX1 (WT or deletion mutant, 1 μg), or vector + pAdTrack-CMV-PLM + pAdTrack-CMV-NCX1 (WT or deletion mutant, 1 μg each) as described in detail previously (1, 38, 39). Empty vector was used to control for the amount of DNA (3 μg for each plate) used in transfection. For patch-clamp experiments, cells were trypsinized 24 h after transfection using trypsin-EDTA, transferred to 35-mm dishes containing sterile glass coverslips, and incubated for an additional 24 h before experiments. Cells for Western blotting were left in 100-mm dishes until 48 h after transfection.
Western blotting.
After 48 h, cells transfected with WT NCX1 or its deletion mutants, with or without PLM cotransfection, were washed three times with ice-cold PBS and lysed with 300 μl of ice-cold lysis buffer containing (in mM) 50 Tris (pH 8.0), 150 NaCl, 1 Na+ orthovanadate, 1 PMSF, 100 NaF, and 1 EGTA, with 0.5% NP-40, a Complete Mini protease inhibitor mixture tablet (Roche Applied Science), and phosphatase inhibitor I and II (1:100 dilution; Sigma). The cell lysate was sonicated three times, snap frozen in dry ice-ethanol, and stored at −80°C.
For detection of NCX1 and its deletion mutants, cell lysates (60 μg/lane) in SDS sample buffer containing 10 mM N-ethylmaleimide were subjected to 7.5% PAGE. Fractionated proteins were transferred onto polyvinylidene difluoride membranes. NCX1 and its deletion mutants were detected with R3F1 (1:1,000 dilution; Swant, Bellinzona, Switzerland), a monoclonal antibody that recognizes the epitope formed by residues 560–629 and 649–705 of the intracellular loop (30); π11-13 antibody (1:500 dilution; Swant), a polyclonal antibody the epitope of which is unknown; or monoclonal 6H2 antibody (1:1,000; R & D Systems, Minneapolis, MN), which recognizes the extracellular NH2-terminal motif. Secondary antibody was sheep anti-mouse IgG-conjugated horseradish peroxidase (1:2,000 dilution; Amersham Biosciences, Uppsala, Sweden) or donkey anti-rabbit IgG (1:5,000 dilution; Amersham Biosciences). For detection of PLM, cell lysates (15 μg/lane) in SDS sample buffer containing 5% β-mercaptoethanol were subjected to electrophoresis on a 12% polyacrylamide gel. Primarily unphosphorylated PLM was detected with polyclonal C2 antibody (1:10,000 dilution) (33). For detection of PLM phosphorylated at Ser68, a specific anti-phospho-PLM antibody (CP68, 1:10,000 dilution) (31) was used. For detection of the catalytic subunit of PKA in transfected HEK-293 cells, a monoclonal antibody (1:1,000 dilution; BD Transduction Laboratories) was used. Immunoreactivity was detected using enhanced chemiluminescence as described previously (1, 38, 39).
Confocal microscopy.
HEK-293 cells transiently transfected for 24 h with pAdTrack-CMV vector, WT rat NCX1, rΔ265–373, rΔ300–373, dΔ328–330, or dΔ330–373 NCX1 deletion mutants were plated on laminin-coated glass slide chambers (Nunc, Lab-Tek Division, Naperville, IL) and cultured for an additional 24 h. Adherent cells were washed three times with PBS, fixed and permeabilized, and incubated with R3F1 (1:500 dilution) as described previously (1). After further incubation with Alexa Fluor 546-labeled goat anti-mouse IgG (1:100 dilution; Invitrogen), the slide was removed from the chamber and a coverslip was applied. Images of HEK-293 cells were acquired with a Zeiss LSM confocal microscope using a ×60 oil objective [excitation at 488 nm and emission at 515 nm for green fluorescent protein (GFP); excitation at 546 nm and emission at 570 nm for R3F1].
INaCa measurements.
Whole cell patch-clamp recordings were performed at 30°C as described previously (1, 38, 39). Briefly, fire-polished pipettes (2–3 μm tip diameter; 2.5–3 MΩ resistance) were filled with a buffered Ca2+ solution containing (in mM) 100 Cs+ glutamate, 7.25 Na+-HEPES, 1 MgCl2, 12.75 HEPES, 2.5 Na2ATP, 10 EGTA, and 6 CaCl2 (pH 7.2); free Ca2+ concentration in the pipette solution was 205 nM. Cells were bathed in an external solution containing (in mM) 130 NaCl, 5 CsCl, 1.2 MgSO4, 1.2 NaH2PO4, 5 CaCl2, 10 HEPES, 10 Na+-HEPES, and 10 glucose (pH 7.4). K+ (Cs+ substitution), Ca2+ (1 μM verapamil), Na+-K+-ATPase (1 mM ouabain), and Cl− (30 μM niflumic acid) currents were minimized. In addition, cells were held at the calculated equilibrium potential for Na+/Ca2+ exchanger (−73 mV under our ionic conditions) for 5 min before stimulation. Only cells that fluoresced green (excitation at 380 nm and emission at 510 nm), indicating successful transfection, were selected for current measurements. Cells were stimulated with a descending-ascending voltage ramp (from +100 to −120 and +100 mV), in the absence and presence of 1 mM CdCl2. The difference current measured during the descending voltage ramp was taken to be INaCa. INaCa of each cell was divided by its whole cell capacitance to account for variation in cell sizes.
In a separate series of experiments, HEK-293 cells were cotransfected with pAdTrack-CMV vector (1.5 μg), PLM (0.5 μg), and WT rat NCX1, dΔ229–237, or rΔ250–300 deletion mutant (1 μg each). INaCa was measured in these cells before and 5 min after addition of forskolin (1 μM) to activate PKA and phosphorylate PLM at Ser68 (39).
GST pull-down assays.
GST-NCX1 fusion proteins were synthesized, purified, and used in GST pull-down assays as described in detail previously (38). Briefly, PCR fragments of the intracellular loop of rat NCX1 (218–371, 218–320, 218–270, 238–371, 250–300, 300–373, and 371–508) were cloned in pCR2.1-TOPO (Invitrogen). The verified cDNA sequence was released from pCR2.1-TOPO and subcloned into pGEX-6P-1 (Amersham). After transformation into BL21 Escherichia coli, constructs were screened under the control of an isopropyl 1-thio-β-d-galactopyranoside-inducible promoter for the expression of GST-NCX1 fusion proteins at the predicted molecular weights. GST fusion proteins were purified as follows (9, 38). After centrifugation, bacterial pellets were resuspended in 25 ml of STE buffer [in mM: 10 Tris (pH 8.0), 150 NaCl, 2 EDTA, and 1 PMSF] containing protease inhibitor cocktail (1 tablet per 10 ml). Bacteria were lysed by incubation with 100 μg/ml lysozyme at 4°C for 15 min, and 5 mM DTT and 1.5% N-laurylsarcosine were added. After brief sonication on ice, 2% Triton X-100 was added, and the lysate was centrifuged at 10,000 rpm for 10 min at 4°C. Then ∼25 ml of supernatant were incubated at 4°C for 4 h with 400 μl of GSH-Sepharose (50% slurry; Pharmacia LKB Biotechnology). The bead matrices were recovered by centrifugation, washed five times with wash buffer [50 mM Tris (pH 8), 150 mM NaCl, 0.05% Tween, and complete protease and phosphatase inhibitor cocktails], and washed twice with 500 mM KCl. The recovered beads were resuspended in 800 μl of Tween-free wash buffer.
Resuspended beads (5 μl) were subjected to SDS-PAGE followed by Coomassie blue staining to check for GST fusion protein integrity as well as different levels of expression of the GST fusion proteins. Protein concentrations of GST fusion proteins were determined by Lowry assay (Bio-Rad). To account for varying levels of expression of the different GST-protein constructs, different volumes of GST fusion protein-attached beads (2–4 μl) were used for GST pull-down assays.
For pull-down assays, His-tagged PLM obtained from transformed BL21(DE3)pLysS bacteria (33) was used. Briefly, varying volumes of GST fusion protein-attached beads (to match the protein concentrations of GST fusion protein constructs) were incubated with His-tagged PLM (1 μg) for 30 min at 4°C. Then the beads were centrifuged and washed five times with buffer (15 mM Tris, 150 mM NaCl, 0.05% Tween, and 0.1% NP-40). Washed beads were heated at 95°C for 5 min with 40 μl of SDS sample buffer. Half of the content was loaded on 7.5% SDS-polyacrylamide gel, and proteins were separated. Biosafe Coomassie blue staining was used to detect GST fusion proteins. The other half was loaded on 12% SDS-polyacrylamide gel, and proteins were separated. C2 antibody (1:10,000 dilution) was used to detect His-tagged PLM in these experiments. We previously demonstrated that WT PLM (expressed in HEK-293 cells) and His-tagged PLM gave similar results in GST pull-down experiments (38).
Statistical analysis.
Values are means ± SE. For the analysis of INaCa as a function of group (e.g., NCX1 vs. NCX1 + PLM) and voltage, two-way ANOVA was used to determine statistical significance. For analysis of protein contents of NCX1 or its deletion mutants, unpaired Student's t-test was used. A commercial software package (JMP version 7, SAS Institute, Cary, NC) was used. P ≤ 0.05 was taken to be statistically significant.
RESULTS
Expression of NCX1 and intracellular loop deletion mutants in HEK-293 cells.
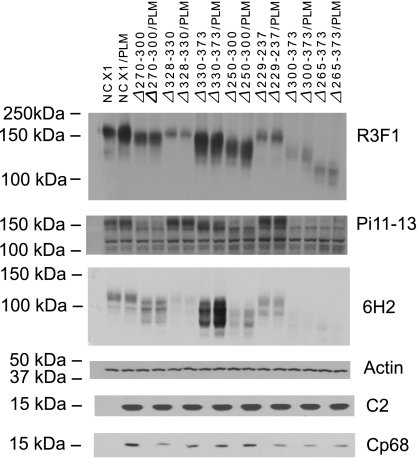
In agreement with our previous observations (1, 38, 39), WT NCX1 was expressed in the plasma membrane of transfected HEK-293 cells (Figs. 1 and 2B). Cells transfected with the control vector pAdTrack-CMV only exhibited intracellular GFP fluorescence (Fig. 2A). Compared with the signal intensity of WT NCX1, the signal intensities of various NCX1 deletion mutants depended on the anti-NCX1 antibody used for detection (Fig. 1). For example, although the monoclonal R3F1 signal for the dΔ328–330 mutant was weaker than that for WT NCX1, the polyclonal π11-13 signal for the dΔ328–330 mutant was more intense. Another example is the rΔ270–300 mutant: although the R3F1 signal was similar when compared with WT NCX1, the π11-13 signal was quite weak. INaCa amplitudes, however, were similar among WT NCX1, dΔ328–330, and rΔ270–300 deletion mutants (see Fig. 4). As in the case with R3F1, it was difficult to detect signals from rΔ300–373 and rΔ265–373 deletion mutants using π11-13 antibody.
Fig. 1.
Heterologous expression of phospholemman (PLM) and Na+/Ca2+ exchanger (NCX1) loop deletion mutants in HEK-293 cells. Equal amounts of total plasmid DNA were transiently transfected in HEK-293 cells with wild-type (WT) NCX1 or its loop deletion mutants, with or without PLM. Cell lysates from indicated cell cultures were subjected to immunoblot analysis 48 h after transfection using monoclonal anti-NCX1 (R3F1), polyclonal anti-NCX1 [π11-13 (Pi11-13)], monoclonal anti-NCX1 (6H2), polyclonal anti-PLM (C2), and polyclonal anti-phospho-PLM (CP68) antibodies. Under nonreducing conditions (10 mM N-ethylmaleimide) used for R3F1 and π11-13, WT NCX1 is detected as a 160-kDa band. Under reducing conditions (5% β-mercaptoethanol) used for 6H2, WT NCX1 is detected as a 120-kDa band. R3F1 signal intensities of NCX1 and its deletion mutants, with or without PLM coexpression, are summarized in Table 1.CP68 signal intensities of PLM phosphorylated at Ser68 in cells cotransfected with PLM and NCX1 or NCX1 deletion mutants are summarized in Table 2.
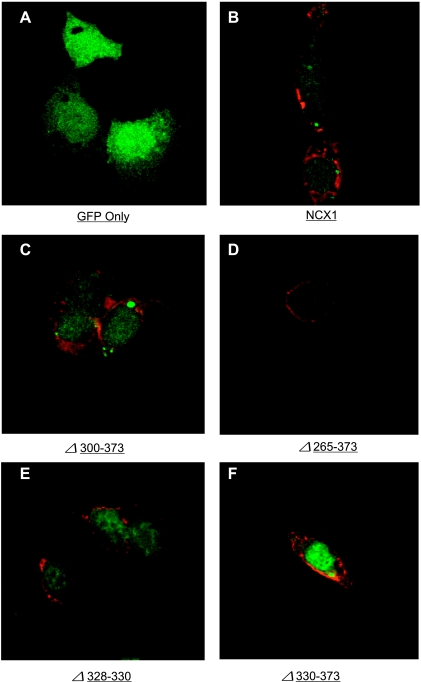
Fig. 2.
Localization of NCX1 deletion mutants by confocal microscopy in transfected HEK-293 cells. Cells were transiently transfected with control plasmid vector pAdTrack-CMV expressing green fluorescent protein (GFP; A), WT rat NCX1 (B), rΔ300–373 (C), rΔ265–373 (D), dΔ328–330 (E), or dΔ330–373 (F) loop deletion mutants. After 48 h, cells were fixed, permeabilized, labeled with anti-NCX1 monoclonal antibody R3F1, and visualized with Alexa Fluor 546-labeled goat anti-mouse IgG using a Zeiss LSM confocal microscope and ×60 oil objective. GFP expressed under control of a second CMV promoter is localized to the cytosol (A). NCX1 and its loop deletion mutants (red) are largely confined to the plasma membrane. R3F1 signals in images of cells expressing rΔ300–373, rΔ265–373, or dΔ328–330 mutants are weaker than raw signals of R3F1 in cells expressing WT NCX1 or dΔ330–373 mutant, and adjustment of image contrast and intensity was necessary so that the red rim is more clearly visible.
Because of the different Western blot signal patterns of NCX1 deletion mutants obtained with R3F1 and π11-13 antibodies, we used a third anti-NCX1 antibody, 6H2, which was raised against an immunogen consisting of a fusion protein from the extracellular NH2 terminus of rabbit renal NCX1. We reasoned that the sensitivity of 6H2 to detect various NCX1 deletion mutants should not be influenced by cytoplasmic loop deletions. Under nonreducing conditions (10 mM N-ethylmaleimide), 6H2 detected WT NCX1 as a 160-kDa band, but the signals for the various NCX1 deletion mutants were smeared and were not suitable for quantitative analysis (blot not shown). Under reducing conditions (5% β-mercaptoethanol), 6H2 detected WT NCX1 at the expected 120 kDa, but the signal intensities for the NCX1 deletion mutants were variable (Fig. 1). In agreement with the R3F1 and π11-13 blots, we were unable to unequivocally detect rΔ300–373 and rΔ265–373 deletion mutants using 6H2 antibody.
Using confocal immunofluorescence microscopy, we confirmed the presence of WT rat NCX1 and rΔ300–373, rΔ265–373, dΔ328–330, and dΔ330–373 deletion mutants in the plasma membrane of successfully transfected HEK-293 cells (Fig. 2). Similar to the results obtained with Western blotting, R3F1 signals in confocal images were much easier to detect and stronger in cells expressing WT rat NCX1 and dΔ330–373 NCX1 deletion mutant, but the R3F1 signals in cells expressing rΔ300–373, rΔ265–373, or dΔ328–330 NCX1 deletion mutants were much weaker, such that adjustment of the intensity and contrast of the images was necessary to make the R3F1 signal (red rim) more easily visible. In addition, despite very “weak” R3F1 signals in cells expressing the dΔ328–330 mutant (Fig. 1, Table 1), INaCa was the highest among all the NCX1 deletion mutants (see Fig. 4). The results of Western blotting using three different anti-NCX1 antibodies, confocal imaging, and INaCa measurements, when interpreted together, suggest that the three NCX1 antibodies likely have varying sensitivities to detect different NCX1 deletion mutants. Paired comparisons of R3F1 signal intensities indicate that PLM coexpression did not significantly decrease the level of expression of NCX1 or its deletion mutants in transfected HEK-293 cells (Fig. 1, Table 1).
Table 1.
Effects of PLM coexpression on protein levels of NCX1 or its deletion mutants
| NCX1 or Mutant | Without PLM | With PLM |
|---|---|---|
| WT | 95.0±10.7 | 70.9±7.0 |
| dΔ229–237 | 46.2±13.6 | 72.6±6.0 |
| rΔ250–300 | 61.4±12.4 | 62.5±7.6 |
| rΔ270–300 | 419.8±19.6 | 356.3±6.2 |
| dΔ328–330 | 90.1±7.0 | 134.0±12.3* |
| dΔ330–373 | 334.4±8.8 | 338.7±1.2 |
| rΔ238–270 | 299.8±9.7 | 256.7±15.7 |
| rΔ238–270 + Δ300–328 | 251.3±26.9 | 210.4±8.1 |
| rΔ300–328 | 97.1±4.2 | 74.4±19.5 |
Values (means ± SE) are arbitrary units; n = 3 separate transfections for each group, except rΔ300-328, where n = 6. Protein lysates from HEK-293 cells transfected with wild-type (WT) rat Na+/Ca2+ exchanger (NCX1), dΔ229-237, and rΔ250-300 mutants were separated on one gel, those from rΔ270-300, dΔ328-330, and dΔ330-373 mutants on a 2nd gel, those from rΔ238-270 and rΔ238-270 + Δ300-328 mutants on a 3rd gel, and those from rΔ300-328 mutants on a 4th gel. Analysis on separate gels accounts for differences in R3F1 signals (arbitrary units) among the 4 groups. PLM, phospholemman.
P < 0.04.
Fraction of expressed PLM in HEK-293 cells is phosphorylated at Ser68.
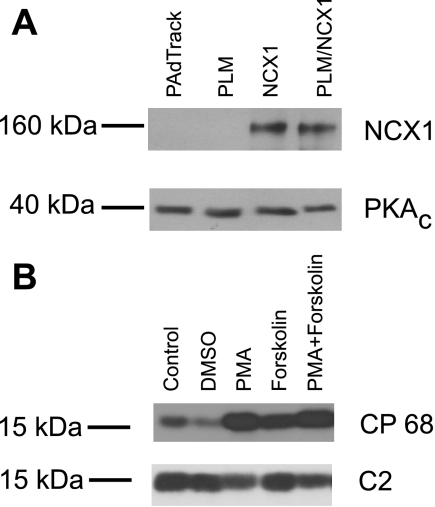
PLM phosphorylated at Ser68 is the active species that inhibits NCX1 (34, 39). Therefore, it is important to demonstrate that a fraction of the expressed PLM is phosphorylated. We first showed the presence of the catalytic subunit of PKA in transfected HEK-293 cells (Fig. 3A). When CP68, a polyclonal antibody specific for PLM phosphorylated at Ser68 (31), was used, stimulation of PKA by forskolin or PKC by PMA resulted in expected increases in phosphorylation of Ser68 (Fig. 3B). Levels of unphosphorylated PLM as detected by C2 antibody showed the corresponding decrease. If it is assumed that the CP68 signal obtained after PMA treatment was 100% phosphorylation of Ser68, ∼29% of expressed PLM was phosphorylated in resting transfected HEK-293 cells. In addition, the signal intensities for PLM phosphorylated at Ser68 were similar whether cells were cotransfected with WT NCX1 or its deletion mutants (Table 2).
Fig. 3.
Phosphorylation of exogenous PLM in transfected HEK-293 cells. A: cells were transfected with control plasmid vector pAdTrack-CMV, WT PLM, WT rat NCX1, or PLM + NCX1. At 48 h after transfection, cell lysates were subjected to immunoblot analysis using monoclonal antibodies against the catalytic subunit of PKA (PKAC) and R3F1 against NCX1. Note similar expression of NCX1 whether cells were transfected with NCX1 alone or NCX1 + PLM. B: HEK-293 cells were transfected with WT PLM. After 48 h, they were treated with PBS (control), DMSO, forskolin (10 μM), PMA (0.5 μM), or PMA + forskolin for 10 min at 37°C. Cell lysates were subjected to immunoblot analysis using CP68 antibody to specifically detect PLM phosphorylated at Ser68 and C2 antibody to detect predominantly unphosphorylated PLM.
Table 2.
PLM Ser68 phosphorylation among NCX1 deletion mutants
| NCX1 or Mutant | CP68 signal |
|---|---|
| WT | 1.0 |
| rΔ270–300 | 0.53±0.07 |
| dΔ328–330 | 1.56±0.67 |
| dΔ330–373 | 1.46±0.32 |
| rΔ250–300 | 1.51±0.61 |
| dΔ229–237 | 0.90±0.46 |
| rΔ300–373 | 1.34±0.44 |
| rΔ265–373 | 1.04±0.43 |
Values (means ± SE) are arbitrary units; n =3 separate transfections. Protein lysates from HEK-293 cells transfected with WT NCX1 or its deletion mutants, with and without PLM, were separated on SDS-PAGE, transferred, and blotted with CP68 anti-phospho-PLM antibody (see Fig. 1). Since there are 3 separate Western blots, for statistical analysis CP68 signals of various NCX1 deletion mutants on a blot were normalized to WT NCX1 on the same blot. One-way ANOVA indicates no significant differences in CP68 signals among the various NCX1 deletion mutants.
Expression of NCX1 and intracellular loop deletion mutants results in functional INaCa.
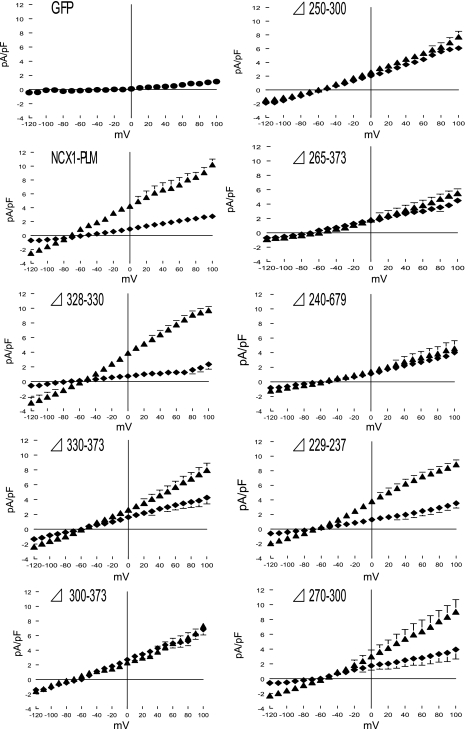
We previously demonstrated that, under the conditions used in our patch-clamp experiments, the Cd2+-sensitive current is INaCa (1, 38, 39). HEK-293 cells transfected with the control pAdTrack-CMV vector expressing GFP exhibited little to no background current (Fig. 4), in agreement with previous findings that these cells do not express NCX1 (1) and are electrically silent (8). By contrast, cells transfected with NCX1 or its intracellular loop deletion mutants displayed the characteristic INaCa (Fig. 4). In general, the magnitudes of INaCa measured in HEK-293 cells transfected with NCX1 deletion mutants appeared to be inversely related to the lengths of the deleted proximal intracellular loop segments. For example, although the magnitude of INaCa exhibited by the dΔ328–330 NCX1 deletion mutant was similar to that measured for WT rat NCX1, mutants with large intracellular loop deletions (dΔ240–679 and rΔ265–373) had the smallest INaCa magnitudes.
Fig. 4.
Effects of PLM on NCX1 current (INaCa) in HEK-293 cells expressing NCX1 or its loop deletion mutants. INaCa was measured in HEK-293 cells transfected with pAdTrack-CMV plasmid expressing WT rat NCX1 or various NCX1 loop deletion mutants, with PLM (⧫; n = 8, 5, 5, 8, 11, 6, 6, 8, and 7 for WT rat NCX1, dΔ328–330, dΔ330–373, rΔ300–373, rΔ250–300, rΔ265–373, dΔ240–679, dΔ229–237, and rΔ270–300 deletion mutants, respectively) or without PLM (▴; n = 8, 12, 10, 7, 9, 9, 8, 12, and 4 for WT rat NCX1, dΔ328–330, dΔ330–373, rΔ300–373, rΔ250–300, rΔ265–373, dΔ240–679, dΔ229–237, and rΔ270–300 deletion mutants, respectively). Top left: background currents in HEK-293 cells transfected with control plasmid vector pAdTrack-CMV expressing GFP (•; n = 5). Symbols represent means ± SE; error bars that fall within boundaries of symbols are not shown.
PLM inhibits INaCa by interacting with residues 238–270 and 300–328 of the intracellular loop.
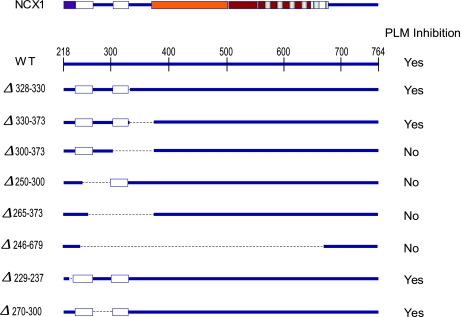
Coexpression of PLM with WT rat NCX1 resulted in significant (P < 0.0001) suppression of INaCa in HEK-293 cells (Fig. 4), in agreement with our previous observations (1, 38, 39). Deletion of large segments of the intracellular loop of NCX1 (dΔ240–679) abolished the ability of PLM to inhibit INaCa (Fig. 4; P > 0.05), in agreement with our previous observations using split Na+/Ca2+ exchangers in which almost the entire intracellular loop was absent (38). Deleting residues 229–237, 270–300, 328–330, or 330–373 did not interfere with the ability of PLM to inhibit INaCa (Fig. 4; P < 0.0001 in all 4 mutant cases). By contrast, INaCa measured in the presence or absence of PLM were not different in the case of rΔ250–300, rΔ265–373, and rΔ300–373 NCX1 deletion mutants (Fig. 4; P > 0.05 in all 3 mutant cases). Figure 5 shows the schematic representation of the intracellular loops of WT NCX1 and its deletion mutants, together with the experimental results with and without inhibition of INaCa by PLM. It is readily apparent that the two regions encompassing residues 238–270 and 300–328 (Fig. 5) in the intracellular loop must be present together in order for NCX1 to be regulated by PLM.
Fig. 5.
Deduction of functional interaction sites between PLM and NCX1. Top: schematic representation of intracellular loop of NCX1 (residues 218–764) containing exchange inhibitory peptide (XIP) region (purple box; residues 219–238), Ca2+-binding domain (CBD) 1 (orange box; residues 371–508), CBD 2 (brown box; residues 501–650), and putative interaction site for endogenous XIP (gray striped box; residues 562–679). There is significant overlap between CBD2 and interaction site for endogenous XIP. Two putative interaction sites with PLM are shown as white boxes (residues 238–270 and 300–328). Bottom: various NCX1 loop deletion mutants shown as preserved regions (thick blue line) and deleted segments (dotted black line). Ability of PLM to inhibit INaCa when coexpressed with WT NCX1 or its deletion mutants is shown at right. Alignment of deletion mutants indicates that regions encompassing residues 238–270 and 300–328 (white boxes) must be present in the intracellular loop for PLM to exert its modulatory effects on NCX1.
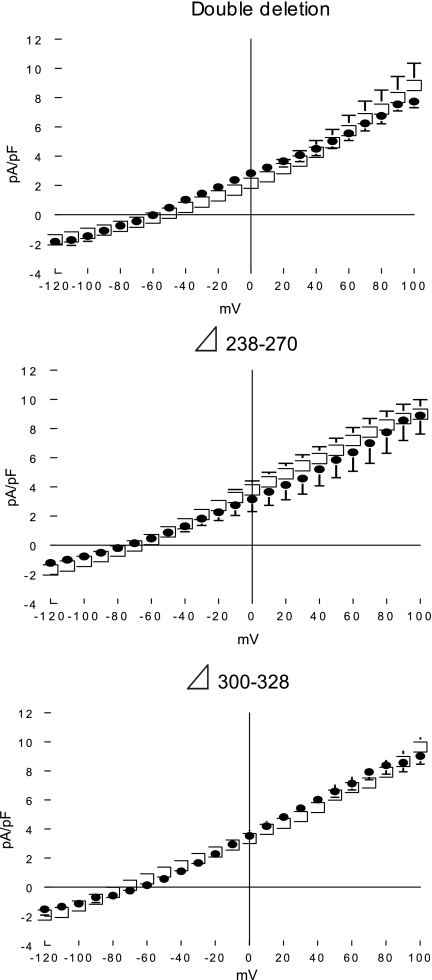
To confirm that regions encompassing residues 238–270 and 300–328 in the intracellular loop of NCX1 are required for modulation of its activity by PLM, we subsequently constructed three additional NCX1 deletion mutants: rΔ238–270, rΔ300–328, and a double-deletion mutant rΔ238–270 + Δ300–328. As shown in Fig. 6, all three new rat NCX1 deletion mutants exhibited the characteristic baseline INaCa. PLM coexpression did not alter protein levels of the mutant exchangers (Table 1) or INaCa in cells expressing rΔ238–270, rΔ300–328, or rΔ238–270 + Δ300–328 deletion mutants (Fig. 6).
Fig. 6.
Inhibition of INaCa by PLM requires regions encompassing residues 238–270 and 300–328 in the intracellular loop of NCX1. INaCa was measured in HEK-293 cells expressing NCX1 loop deletion mutants, with PLM (•; n = 9, 4, and 4 for double-deletion rΔ238–270 + Δ300–328, rΔ238–270, and rΔ300–328 mutants, respectively) and without PLM (□; n = 12, 7, and 5 for double-deletion rΔ238–270 + Δ300–328, rΔ238–270, and rΔ300–328 mutants, respectively). Symbols represent means ± SE; error bars that fall within boundaries of symbols are not shown.
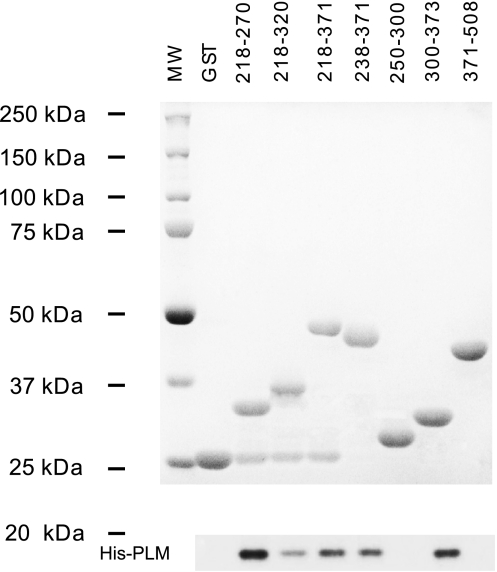
GST pull-down assays confirm that fragments corresponding to residues 218–270 and 300–373 of the intracellular loop associate with PLM.
We previously showed by GST pull-down assays that PLM associates with a fragment corresponding to the intracellular loop (residues 218–764) of NCX1, specifically regions encompassing residues 218–371 and 508–674, but not Ca2+-binding domain 1 (residues 371–508) (38). Further dissection of the proximal linker domain of NCX1 demonstrated that PLM associated with GST-NCX1/218–270, GST-NCX1/218–320, GST-NCX1/218–371, GST-NCX1/238–371, and GST-NCX1/300–373, but not GST-NCX1/250–300, GST-NCX1/371–508, or GST alone (Fig. 7). These observations indicate that fragments encompassing residues 218–270 and 300–373 in the proximal linker domain of NCX1 physically associated with PLM. The GST pull-down results are consistent with findings from our electrophysiological studies on NCX1 deletion mutants that PLM interacted with residues 238–270 and 300–328 of NCX1.
Fig. 7.
Glutathione S-transferase (GST) pull-down demonstrates physical association of PLM with residues 218–270 and 300–373, but not residues 250–300, of NCX1. Purified GST or GST-NCX1 fusion proteins linked to GSH-Sepharose beads were incubated with His-tagged PLM (1 μg), and GST pull-down assay was performed. Top: GST and GST-NCX1 fusion proteins detected by Coomassie blue staining. MW, molecular weight marker. Bottom: His-tagged PLM detected by C2 antibody. Experiment was performed 4 times using new sets of GST fusion proteins prepared from 4 independent bacterial cultures, and similar results were obtained.
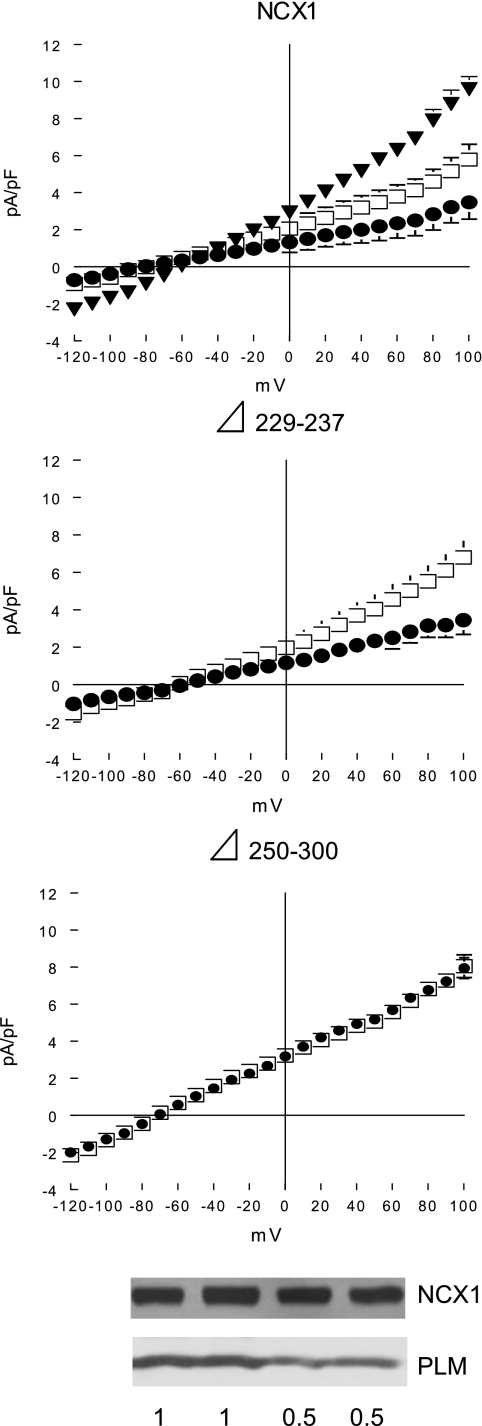
Acute regulation of INaCa by PLM phosphorylation is absent in rΔ250–300 but preserved in dΔ229–237 NCX1 deletion mutants.
We previously demonstrated in transfected HEK-293 cells expressing PLM and WT rat NCX1 that stimulation of PKA by forskolin resulted in further suppression of INaCa (39). To demonstrate that regulation of INaCa by PLM phosphorylation requires interaction between PLM and NCX1, we examined the effects of forskolin on INaCa in cells coexpressing PLM and WT rat NCX1 or dΔ229–237 (positive association with PLM) or rΔ250–300 (negative association with PLM) NCX1 deletion mutants. In order for additive inhibition of INaCa by phosphorylated PLM to be clearly observable, it was necessary to decrease the magnitude of inhibition of INaCa in HEK-293 cells coexpressing PLM before forskolin exposure (Fig. 4). Therefore, in this series of experiments, the amount of PLM DNA used in transfection was decreased from 1 to 0.5 μg. Western blots demonstrated ∼58% of PLM in cells transfected with 0.5 μg of PLM DNA compared with cells transfected with 1 μg of PLM DNA, with no differences in NCX1 expression (Fig. 8, bottom). In agreement with our previous results (39), phosphorylation of PLM by forskolin (Fig. 3) resulted in additional inhibition of INaCa in cells expressing WT rat NCX1 (Fig. 8). Forskolin treatment also enhanced inhibition of INaCa by PLM in cells expressing dΔ229–237 but not rΔ250–300 NCX1 deletion mutants (Fig. 8). The observation that acute regulation of NCX1 by PLM phosphorylation was absent in a NCX1 deletion mutant in which association/interaction was disrupted provides additional evidence that regulation of NCX1 by PLM is not simply due to altered protein trafficking or degradation.
Fig. 8.
Effects of forskolin on INaCa in HEK-293 cells coexpressing PLM and NCX1 or its deletion mutants. HEK-293 cells were transfected with PLM + WT rat NCX1 (n = 5), PLM + dΔ229–237 (n = 5), or PLM + rΔ250–300 (n = 4) loop deletion mutants. INaCa was measured before (□) and 5 min after (•) addition of forskolin (1 μM). For comparison, INaCa in cells expressing WT rat NCX1 alone (top, ▾; n = 4) is also shown. Symbols represent means ± SE; error bars that fall within boundaries of symbols are not shown. To decrease magnitude of INaCa inhibition by PLM (Fig. 4) before forskolin exposure, pAdTrack-CMV vector expressing PLM used in transfection was decreased from 1 to 0.5 μg. Bottom: Western blots demonstrate ∼58% of PLM (detected with C2 antibody) in cells transfected with 0.5 μg of PLM, 1 μg of NCX1, and 1.5 μg of empty vector compared with cells transfected with 1 μg of PLM, 1 μg of NCX1, and 1 μg of empty vector. There were no differences in NCX1 expression between the 2 groups of transfected cells.
DISCUSSION
Our previous studies to map the functional interaction sites between PLM and NCX1 utilize split Na+/Ca2+ exchangers (38). Split exchangers consist of NH2- or COOH-terminal TM domains of NCX1 with varying lengths of intracellular loop (26). Coexpression of paired NH2- and COOH-terminal split exchangers results in correct membrane targeting and functional NCX1 activity (26, 38). Using split exchangers, we found that the cytoplasmic tail of PLM interacts with and coimmunoprecipitates the proximal end of the intracellular loop (region encompassing residues 218–358) of NCX1 (38). Association of PLM with part of the intracellular loop of NCX1 is confirmed by GST pull-down assays. GST-NCX1/218–764 (entire intracellular loop), GST-NCX1/218–371 (XIP region + proximal linker domain), and GST-NCX1/508–674 (encompassing Ca2+-binding domain 2), but not GST-NCX1/1–220 (1st 5 NH2-terminal TM domains), GST-NCX1/371–508 (Ca2+-binding domain 1), and GST-NCX1/764–939 (last 4 COOH-terminal TM domains), bind PLM (38). Because of the bulky nature of the fluorescent indicator proteins attached to split exchangers, it is technically difficult to use this approach to further refine the interaction sites between PLM and NCX1. This consideration led us to use NCX1 mutants with varying lengths of the intracellular loop deleted. We focused on the proximal intracellular loop encompassing residues 218–358 and constructed overlapping, relatively small (<100 residues) deletion mutants for the present mapping study.
Expression of NCX1 loop deletion mutants in HEK-293 cells resulted in measurable INaCa, even when almost all the intracellular loop (dΔ240–679 mutant) had been deleted. This result is consistent with the experimental findings of paired NH2- and COOH-terminal split exchangers in which little to no intracellular loop was present (26, 38). Coexpression of PLM with WT rat NCX1 or dΔ229–237, rΔ270–300, dΔ328–330, and dΔ330–373 deletion mutants resulted in inhibition of INaCa. An unlikely but formal explanation for the inhibition of INaCa by PLM is that cotransfection of PLM and NCX1 in HEK-293 cells decreases NCX1 expression compared with cells transfected with NCX1 alone. We previously showed that NCX1 protein levels are similar whether cells are expressing PLM + NCX1 or NCX1 alone (1). Figure 1 and Table 1 demonstrate that, with the exception of the dΔ328–330 mutant, cells coexpressing PLM and NCX1 deletion mutants had similar NCX1 protein levels compared with cells expressing NCX1 deletion mutants alone. In the case of the dΔ328–330 mutant, the NCX1 mutant protein level was higher, rather than lower, when PLM was coexpressed. Therefore, inhibition of INaCa in HEK-293 cells coexpressing NCX1 (or its deletion mutants) and PLM can be unambiguously assigned to the effects of PLM.
The major finding of the present study is mapping of the functional interaction sites between PLM and NCX1 to regions encompassing residues 238–270 and 300–328 in the intracellular loop by electrophysiological techniques. Using GST pull-down assays, we demonstrated a physical association between PLM and fragments corresponding to residues 218–270 and residues 300–373, but not residues 250–300, of NCX1. With respect to PLM, it is its cytoplasmic tail that interacts with the XIP region and proximal linker domain of NCX1 (38). The cytoplasmic helix H4 (residues 60–68) of PLM is highly basic and favors association with the negatively charged plasma membrane surface. Indeed, NMR spectroscopy indicates that H4 of PLM is at the cytoplasmic lipid-water interface (36). Therefore, in order for PLM to interact and associate with NCX1, the proximal intracellular loop of NCX1 must also lie very close to the cytoplasmic side of the plasma membrane. In support of this hypothesis, recent NMR data coupled with modeling of the intracellular loop of NCX1 indicate that regions corresponding to residues 238–270 and 300–328 in the proximal linker domain are close to the cytoplasmic membrane surface compared with Ca2+-binding domains 1 and 2 (14).
The sites of interaction between PLM and NCX1 also provide a logical explanation for the negative fluorescence resonance energy transfer (FRET) between PLM and NCX1 (5). In the study of Bossuyt et al. (5), the donor fluorophore (CFP) was linked to the COOH terminus of PLM (and, therefore, very close to Ser68), while the acceptor fluorophore (YFP) was inserted into the large intracellular loop at residue 358 in the full-length NCX1. Since detectable FRET requires the molecules to be physically very close (<9 nm with the CFP-YFP pair), insertion of bulky fluorophores so close to the interaction sites may result in conditions not conducive to FRET.
Although the unphosphorylatable S68A PLM mutant can coimmunoprecipitate NCX1 (34), PLM phosphorylated at Ser68 is the active moiety that inhibits INaCa (39). In the present study, using CP68 antibody, we estimated that ∼29% of the expressed PLM was phosphorylated at Ser68 under resting conditions. In our previous experiments using the phosphomimetic Ser68-to-glutamic acid (S68E) PLM mutant, INaCa (+100 mV, 5 mM extracellular Ca2+) was 9.1 ± 1.2, 7.8 ± 0.8, and 6.2 ± 0.6 pA/pF in cells expressing NCX1 alone, NCX1 + PLM, and NCX1 + S68E mutant, respectively (39). If it is assumed that the S68E mutant faithfully mimics PLM phosphorylated at Ser68, ∼46% of PLM is estimated to be phosphorylated at Ser68 in transfected HEK-293 cells under resting conditions. These two fundamentally different approaches suggest that, under resting conditions, ∼30–45% of PLM is phosphorylated at Ser68 in transfected HEK-293 cells. In adult rat cardiac myocytes, on the basis of percent inhibition of INaCa in cells overexpressing PLM or its S68A or S68E mutant, the estimated fraction of PLM phosphorylated at Ser68 under resting conditions is also ∼46% (34).
There are caveats to the present study. 1) Deletion of varying segments of the intracellular loop of NCX1 may alter its secondary and tertiary structures, such that it may no longer interact with PLM. Although we cannot formally exclude this possibility, the fact that congruent results are obtained using fundamentally different experimental techniques (electrophysiology vs. GST pull-down) and different molecular approaches [split NCX1 exchangers (38) vs. deletion mutants] should largely allay this concern. In addition, the observation that closely spaced deletion mutant pairs (e.g., rΔ300–373 and dΔ330–373, rΔ250–300, and rΔ270–300) exhibited preservation of PLM inhibitory effects in one but not the other suggests that potential distortion of NCX1 intracellular loop structure by deletion of some residues is not too severe to totally abrogate the ability of PLM to inhibit INaCa. 2) The lack of correlation between INaCa amplitude and apparent protein expression levels of NCX1 mutants as detected by R3F1 is a concern. The different Western blot patterns of various NCX1 deletion mutants detected with R3F1, π11-13, and 6H2 anti-NCX1 antibodies, when interpreted together with results from confocal imaging and INaCa measurements, suggest that, compared with WT NCX1, the low R3F1 signals on Western blot and confocal images of some, but not all, NCX1 deletion mutants may be an artifact due to different sensitivities of the R3F1 antibody to detect various NCX1 deletion mutants. 3) Although both regions encompassing residues 238–270 and 300–328 are required for regulation of NCX1 activity by PLM, physical association between PLM and NCX1 appears to require only a single region encompassing residues 218–270 or 300–373. One hypothetical model of the proximal linker domain encompassing residues 259–370 suggests that Val261 is in close proximity with Ala314 (14). We speculate that the cytoplasmic tail of PLM interacts within the spatial domain in which Val261 and Ala314 reside. This hypothesis accommodates our INaCa data (both regions spanning residues 238–270 and 300–328 are required for inhibition of NCX1 by PLM) and GST pull-down results (either region is sufficient to bring down PLM). Definitive proof of our speculation will require detailed knowledge of the three-dimensional structure of the proximal linker domain of NCX1 as well as the critical residues involved in regulation of NCX1 activity by PLM.
In summary, by expressing NCX1 and its deletion mutants in HEK-293 cells, we found that PLM inhibited INaCa by interacting with residues 238–270 and 300–328 of NCX1. PLM physically associated with fragments encompassing residues 218–270 and 300–373 but not residues 250–300 of NCX1. On the basis of the results of this study, the three-dimensional structure of PLM, and the modeled structure of intracellular loop of NCX1, we suggest that the cytoplasmic tail of PLM associates with the proximal linker domain of NCX1 at the cytoplasmic surface of the plasma membrane.
GRANTS
This work was supported in part by National Heart, Lung, and Blood Institute Grants HL-58672 and HL-74854.
Acknowledgments
We thank Drs. Kenneth D. Philipson and Debra A. Nicoll of the David Geffen School of Medicine at UCLA for generously providing the dog NCX1 deletion mutants Δ240–679, Δ229–237, Δ330–373, and Δ328–330.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Ahlers BA, Zhang XQ, Moorman JR, Rothblum LI, Carl LL, Song J, Wang J, Geddis LM, Tucker AL, Mounsey JP, Cheung JY. Identification of an endogenous inhibitor of the cardiac Na+/Ca2+ exchanger, phospholemman. J Biol Chem 280: 19875–19882, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Beevers AJ, Kukol A. Secondary structure, orientation, and oligomerization of phospholemman, a cardiac transmembrane protein. Protein Sci 15: 1127–1132, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bers DM Cardiac excitation-contraction coupling. Nature 415: 198–205, 2002. [DOI] [PubMed] [Google Scholar]
- 4.Bossuyt J, Ai X, Moorman JR, Pogwizd SM, Bers DM. Expression and phosphorylation of the Na-pump regulatory subunit phospholemman in heart failure. Circ Res 97: 558–565, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Bossuyt J, Despa S, Martin JL, Bers DM. Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J Biol Chem 281: 32765–32773, 2006. [DOI] [PubMed] [Google Scholar]
- 6.Crambert G, Fuzesi M, Garty H, Karlish S, Geering K. Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc Natl Acad Sci USA 99: 11476–11481, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Despa S, Bossuyt J, Han F, Ginsburg KS, Jia LG, Kutchai H, Tucker AL, Bers DM. Phospholemman-phosphorylation mediates the β-adrenergic effects on Na/K pump function in cardiac myocytes. Circ Res 97: 252–259, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Dong H, Dunn J, Lytton J. Stoichiometry of the cardiac Na+/Ca2+ exchanger NCX1.1 measured in transfected HEK cells. Biophys J 82: 1943–1952, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem 210: 179–187, 1993. [DOI] [PubMed] [Google Scholar]
- 10.Franzin CM, Gong XM, Thai K, Yu J, Marassi FM. NMR of membrane proteins in micelles and bilayers: the FXYD family proteins. Methods 41: 398–408, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller W, Eaton P, Bell JR, Shattock MJ. Ischemia-induced phosphorylation of phospholemman directly activates rat cardiac Na/K-ATPase. FASEB J 18: 197–199, 2004. [DOI] [PubMed] [Google Scholar]
- 12.Han F, Bossuyt J, Despa S, Tucker AL, Bers DM. Phospholemman phosphorylation mediates the protein kinase C-dependent effects on Na+/K+ pump function in cardiac myocytes. Circ Res 99: 1376–1383, 2006. [DOI] [PubMed] [Google Scholar]
- 13.He Z, Feng S, Tong Q, Hilgemann DW, Philipson KD. Interaction of PIP2 with the XIP region of the cardiac Na/Ca exchanger. Am J Physiol Cell Physiol 278: C661–C666, 2000. [DOI] [PubMed] [Google Scholar]
- 14.Hilge M, Aelen J, Vuister GW. Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors. Mol Cell 22: 15–25, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Iwamoto T, Uehara A, Imanaga I, Shigekawa M. The Na+/Ca2+ exchanger NCX1 has oppositely oriented reentrant loop domains that contain conserved aspartic acids whose mutation alters its apparent Ca2+ affinity. J Biol Chem 275: 38571–38580, 2000. [DOI] [PubMed] [Google Scholar]
- 16.Lansbery KL, Burcea LC, Mendenhall ML, Mercer RW. Cytoplasmic targeting signals mediate delivery of phospholemman to the plasma membrane. Am J Physiol Cell Physiol 290: C1275–C1286, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Levitsky DO, Nicoll DA, Philipson KD. Identification of the high affinity Ca2+-binding domain of the cardiac Na+-Ca2+ exchanger. J Biol Chem 269: 22847–22852, 1994. [PubMed] [Google Scholar]
- 18.Li Z, Nicoll DA, Collins A, Hilgemann D, Filoteo A, Penniston J, Weiss J, Tomich J, Philipson KD. Identification of a peptide inhibitor of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem 266: 1014–1020, 1991. [PubMed] [Google Scholar]
- 19.Lu KP, Kemp BE, Means AR. Identification of substrate specificity determinants for the cell cycle-regulated NIMA protein kinase. J Biol Chem 269: 6603–6607, 1994. [PubMed] [Google Scholar]
- 20.Maack C, Ganesan A, Sidor A, O'Rourke B. Cardiac sodium-calcium exchanger is regulated by allosteric calcium and exchanger inhibitory peptide at distinct sites. Circ Res 96: 91–99, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka S, Nicoll DA, He Z, Philipson KD. Regulation of cardiac Na+-Ca2+ exchanger by the endogenous XIP region. J Gen Physiol 109: 273–286, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka S, Nicoll DA, Reilly RF, Hilgemann DW, Philipson KD. Initial localization of regulatory regions of the cardiac sarcolemmal Na+-Ca2+ exchanger. Proc Natl Acad Sci USA 90: 3870–3874, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mounsey JP, John J 3rd, Helmke S, Bush E, Gilbert J, Roses A, Perryman M, Jones LR, Moorman JR. Phospholemman is a substrate for myotonic dystrophy protein kinase. J Biol Chem 275: 23362–23367, 2000. [DOI] [PubMed] [Google Scholar]
- 24.Nicoll DA, Hryshko LV, Matsuoka S, Frank JS, Philipson KD. Mutation of amino acid residues in the putative transmembrane segments of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem 271: 13385–13391, 1996. [DOI] [PubMed] [Google Scholar]
- 25.Nicoll DA, Ottolia M, Lu L, Lu Y, Philipson KD. A new topological model of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem 274: 910–917, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Ottolia M, John S, Qiu Z, Philipson KD. Split Na+-Ca2+ exchangers. Implications for function and expression. J Biol Chem 276: 19603–19609, 2001. [DOI] [PubMed] [Google Scholar]
- 27.Palmer CJ, Scott BT, Jones LR. Purification and complete sequence determination of the major plasma membrane substrate for cAMP-dependent protein kinase and protein kinase C in myocardium. J Biol Chem 266: 11126–11130, 1991. [PubMed] [Google Scholar]
- 28.Pavlovic D, Fuller W, Shattock MJ. The intracellular region of FXYD1 is sufficient to regulate cardiac Na/K ATPase. FASEB J 21: 1539–1546, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Philipson KD, Nicoll DA. Sodium-calcium exchange: a molecular perspective. Annu Rev Physiol 62: 111–133, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Porzig H, Li Z, Nicoll DA, Philipson KD. Mapping of the cardiac sodium-calcium exchanger with monoclonal antibodies. Am J Physiol Cell Physiol 265: C748–C756, 1993. [DOI] [PubMed] [Google Scholar]
- 31.Rembold CM, Ripley ML, Meeks MK, Geddis LM, Kutchai HC, Marassi FM, Cheung JY, Moorman JR. Serine 68 phospholemman phosphorylation during forskolin-induced swine carotid artery relaxation. J Vasc Res 42: 483–491, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman BZ, Fuller W, Eaton P, Deng J, Moorman JR, Cheung JY, James AF, Shattock MJ. Serine 68 phosphorylation of phospholemman: acute isoform-specific activation of cardiac Na/K ATPase. Cardiovasc Res 65: 93–103, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Song J, Zhang X, Carl L, Qureshi A, Rothblum L, Cheung J. Overexpression of phospholemman alter contractility and [Ca2+]i transients in adult rat myocytes. Am J Physiol Heart Circ Physiol 283: H576–H583, 2002. [DOI] [PubMed] [Google Scholar]
- 34.Song J, Zhang XQ, Ahlers BA, Carl LL, Wang J, Rothblum LI, Stahl RC, Mounsey JP, Tucker AL, Moorman JR, Cheung JY. Serine 68 of phospholemman is critical in modulation of contractility, [Ca2+]i transients, and Na+/Ca2+ exchange in adult rat cardiac myocytes. Am J Physiol Heart Circ Physiol 288: H2342–H2354, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics 68: 41–56, 2000. [DOI] [PubMed] [Google Scholar]
- 36.Teriete P, Franzin CM, Choi J, Marassi FM. Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles. Biochemistry 46: 6774–6783, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waalas SI, Czernik AJ, Olstad OK, Sletten K, Walaas O. Protein kinase C and cyclic AMP-dependent protein kinase phosphorylate phospholemman, an insulin and adrenaline-regulated membrane phosphoprotein, at specific sites in the carboxy terminal domain. Biochem J 304: 635–640, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Zhang XQ, Ahlers BA, Carl LL, Song J, Rothblum LI, Stahl RC, Carey DJ, Cheung JY. Cytoplasmic tail of phospholemman interacts with the intracellular loop of the cardiac Na+/Ca2+ exchanger. J Biol Chem 281: 32004–32014, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang XQ, Ahlers BA, Tucker AL, Song J, Wang J, Moorman JR, Mounsey JP, Carl LL, Rothblum LI, Cheung JY. Phospholemman inhibition of the cardiac Na+/Ca2+ exchanger. Role of phosphorylation. J Biol Chem 281: 7784–7792, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang XQ, Qureshi A, Song J, Carl LL, Tian Q, Stahl RC, Carey DJ, Rothblum LI, Cheung JY. Phospholemman modulates Na+/Ca2+ exchange in adult rat cardiac myocytes. Am J Physiol Heart Circ Physiol 284: H225–H233, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Zhang XQ, Song J, Rothblum LI, Lun M, Wang X, Ding F, Dunn J, Lytton J, McDermott PJ, Cheung JY. Overexpression of Na+/Ca2+ exchanger alters contractility and SR Ca2+ content in adult rat myocytes. Am J Physiol Heart Circ Physiol 281: H2079–H2088, 2001. [DOI] [PubMed] [Google Scholar]