Abstract
Although aging is associated with increased proliferation and decreased apoptosis in the colonic mucosa of Fischer 344 rats, the regulatory mechanisms are poorly understood. Gene expression profiling (Illumina platform) was carried out in freshly isolated colonic mucosal cells from young (4–6 mo old) and aged (22–24 mo old) Fischer 344 rats. Sixty-six genes were differentially expressed in the colonic mucosa between young and old animals (P < 0.05). In particular, the expression of schlafen 3, a negative regulator of proliferation, was decreased by 8- to 10-fold in the colonic mucosa of aged rats. Administration of wortmannin, which inhibited colonic mucosal proliferation in the colonic mucosa of aged rats, stimulated the expression of schlafen 3, indicating a growth regulatory role of this gene. To further determine the growth regulatory properties of schlafen 3 gene, schlafen 3 cDNA was transfected in colon cancer HCT-116 cells. This resulted in a 30–40% inhibition of cellular growth, accompanied by decreased expression of PCNA and cyclin D1 and reduced phosphorylation of retinoblastoma protein. In conclusion, our present study demonstrates that several genes involved in proliferation and apoptosis are differentially expressed in the colonic mucosa of young and aged rats. Schlafen 3, a novel negative regulator of growth, which is markedly downregulated in the colonic mucosa of the aged, may play a role in regulating colonic mucosal growth during aging.
Keywords: aging, gastrointestinal mucosa, cancer, proliferation, microarrays
it is estimated that the number of elderly will continue to increase and by 2050 nearly a quarter of Americans will be 65 years of age (14). An improved understanding of age-related changes of the structural and functional properties of different organ systems, including the gastrointestinal tract, is essential to counteract various diseases, particularly cancer, whose incidence increases sharply with aging.
Aging has been shown to be associated with increased incidence of premalignant lesions in the colon and stomach (14). In fact, the occurrence of both nonmalignant and malignant colorectal neoplasm increases with advancing age (14). To better understand the biochemical events associated with the age-related rise in premalignant lesions in the gastrointestinal tract, we and others have examined changes in mucosal proliferation and apoptosis, the events that maintain the homeostasis of the gastrointestinal mucosa in healthy adults. Morphological and biochemical studies from this and other laboratories have demonstrated increased proliferation and decreased apoptosis in the colonic mucosa of Fischer 344 rats, events that are seen during the development and progression of carcinogenesis (1, 10, 14–16). Moreover, in colonic mucosa, the age-related decrease in apoptosis (increase in cell survival) is evident throughout the entire length of the crypt (15). Morphological studies of the colonic mucosa of human volunteers have further revealed that, whereas cell proliferation in young is confined to the lower two-thirds of the crypt, with aging there is a major shift from the base to the middle and upper third of the gland (6, 18), a pattern commonly observed in colorectal cancer. These and other relevant observations led us to postulate that aging may predispose the gastrointestinal tract to cancer (14). In support of this postulation, we have observed that the susceptibility of the colon to carcinogens increases with advancing age. We reported that the formation of aberrant crypt foci (considered to be precursors of adenoma and carcinoma) in response to the colonic carcinogen dimethylhydrazine in the colon of aged Fischer 344 rats is higher than in young rats (20). Furthermore, our recent observation that in humans the number of polyps, the most frequent premalignant colorectal lesion, increases linearly with advancing age provides further support for the contention that aging predisposes the gastrointestinal tract to neoplasia (17). Additional support for this postulation comes from the observation that the age-related rise in colonic polyps is associated with increased expression of CD44, CD166, and epithelial specific antigen, the markers of colon cancer stemlike cells in macroscopically normal mucosa (17).
Although the regulatory mechanisms for the age-related increase in proliferation and decrease in apoptosis in the colonic mucosa remain to be fully elucidated, we have observed that these changes are associated with increased expression and activation of EGF receptor and some of its family members, particularly ErbB-2/HER-2 and ErbB-3/HER-3 (15, 16, 25). Others have reported increased activation of insulin-like growth factor receptor (9). These changes are also associated with decreased expression of proapoptotic protein Bak and increased expression antiapoptotic protein BcLxL in the colonic mucosa of aged rats (15). Although these biochemical changes may explain part of the morphological changes observed in the aging colon, further studies are needed for a better and more comprehensive understanding of various age-associated changes leading to deregulated growth in the colonic mucosa. In the present investigation, we have employed microarray-based gene expression profiling to better understand the molecular changes associated with aging in the colonic mucosa of Fischer 344 rats. Although several genes involved in proliferation and apoptosis were differentially expressed in the colonic mucosa between young and aged rats, the expression of schlafen 3, a negative regulator of proliferation (3, 21), was found to be decreased by 8- to 10-fold in the colonic mucosa of aged than in young Fischer 344 rats.
Schlafen 3 belongs to a multigene family in mice that has 10 members. They are classified into three subgroups (short, intermediate, and long) on the basis of overall sequence homology and size of the encoded proteins. Schlafen 3 belongs to intermediate subgroup and possess the following characteristics: a unique domain, common to all members of the family, referred to as “Slfn box,” which lies adjacent to a GTP/ATP binding AAA domain and the highly conserved “SWADL” domain, defined by a five-amino-acid sequence (Ser-Trp-Ala-Asp-Leu) (8). Schlafen 1 to 3 have been shown to regulate cell growth in vitro, thought to be through the inhibition of cyclin D1 (3, 8, 21). Transgenic mice expressing schlafen 1 or schlafen 8 within the T cell lineages showed an overall decrease in thymocyte number (21). In addition, inflammatory mediator proteins such as AP-1 and NF-κB regulate schlafen 2 expression, suggesting an important role of this gene in immune response (22). However, role of Schlafen family of proteins, particularly schlafen 3, in regulating gastrointestinal mucosal function is unknown. The present investigation was undertaken to examine the role that schlafen 3 might play in regulating colonic mucosal growth during aging.
MATERIALS AND METHODS
Animals and collection of tissues.
Male Fischer 344 rats, aged 4–6 (young) and 21–24 (old) mo, were used. The animals were purchased from the National Institute on Aging (Bethesda, MD) at least 2 mo before the experiment. They were housed two per cage and had access to Purina chow and water ad libitum. The reasons for using Fischer 344 rats for aging studies are because of purity of breeding, low susceptibility to spontaneous colorectal cancer, and their ability to maintain body weight. All animals were fasted overnight before being killed. The overnight-fasted animals were either used without any intervention or injected intraperitoneally with wortmannin [0.1 mg/kg body wt in 15% DMSO (15)] or vehicle 6 h before being killed. The entire colon (∼18 cm) was removed, cut along the longitudinal median, and rinsed thoroughly in cold normal saline. The mucosa was obtained by scraping with glass slides. Mucosal aliquots were either processed immediately or stored at −80°C. In some experiments, the colon was used immediately to isolate cells from the mucosa, as described below.
Isolation of colonic mucosal epithelial cells.
Cells were isolated from the entire colon by as described previously (15). Briefly, the contents of the colon were washed with PBS. The colon was everted and ligated at both ends after being filled with a 3- to 5-ml protease solution [5 mg/ml in buffer A composed of (in mM) 0.5 NaH2PO4, 1 Na2HPO4, 70 NaCl, 5 KCl, 11 glucose, 50 HEPES, 20 NaHCO3, and 2 EDTA with 2% BSA]. The colon was placed in pronase-free buffer A and incubated for 30 min at 37°C and gassed with O2. The colonic bags were then transferred into 50 ml buffer B containing 1.0 mM CaCl2 and 1.5 mM MgCl2 instead of EDTA in buffer A and gently agitated at room temperature for 30 min. The dispersed cells were discarded by centrifugation. The colonic bag was transferred into 50 ml buffer B and incubated initially for 60 min at room temperature to obtain the cells from the upper part of the colonic crypt. The colonic tissue was then incubated at room temperature for another 45 min to obtain the cells from the middle region of the crypt, which were discarded. The colon was then incubated further for 45 min at room temperature to obtain the cells from the lower part of the crypt. The dispersed mucosal cells were collected by centrifugation at 500 g for 10 min, washed with PBS, and used immediately for isolation of RNA. Although the mucosal cells isolated from the upper and lower part of the colonic crypts were heterogeneous, we have reported that the levels of alkaline phosphatase, an indicator of differentiation and maturation, were substantially higher in cells isolated from the upper one-third of the colonic crypt than those from lower region of the crypts (15).
Isolation of RNA.
Total RNA was extracted from mucosal cells isolated from the upper and lower regions of the colonic crypts by using RNA-STAT solution (Tel Test, Friendswood, TX) according to the manufacturer's instruction. The total RNA was treated with DNase I to remove contaminating genomic DNA, subsequently purified with RNAeasy Mini Kit (Qiagen, Valencia, CA).
Microarray analysis.
Age-related changes in gene expression in mucosal cells isolated from upper and lower regions of the colon of young and aged rats were performed at the Genomic Core Facility, Karmanos Cancer Institute by utilizing Illumina Rat Sentrix-12 BeadChip Arrays essentially according to manufacturer's instruction (Illumina). Briefly, 0.5 μg total RNA was biotinylated, hybridized with BeadChips. The signal was detected with streptovadin-Cy3 according to manufacturer's instruction (Illumina). BeadChips were imaged by use of a Bead Array Reader and raw data were obtained with Bead Studio software (Illumina). Data were normalized by a quantile based-approach that transforms the raw data so that the resulting normalized expression values of each sample have the same distribution (23). An unsupervised cluster analysis was performed to detect similarities among samples on the basis of gene expression profiles. The genes retained to perform the clustering were those varying the most regardless their group membership as described elsewhere (24). Differential expression among the four different groups was tested by using a moderated t-test, which allows computing P values for the significance of gene changes. P values were adjusted by the False Discovery Rate method (24) to allow for multiple hypothesis testing. A corrected P value less than 0.05 was considered significant provided that the fold change in expression was also larger than 1.5-fold. This rule was applied for the two comparisons with two samples per group. However, a more stringent fold change threshold of 2.0 was used for the remaining two comparisons involving the old, lower-crypt group, in which only one sample was available.
RT-PCR.
Two-step RT-PCR was performed by using the GeneAmp Gold RNA PCR kit (Applied Biosystems, Foster City, CA). Briefly, 1 μg of purified RNA was reverse transcribed in the presence of 2.5 mM MgCl2, 1× RT-PCR buffer, 1 mM dNTPs, 10 mM dithiothreitol, 10 units RNase inhibitor, 1.25 μM random hexamers, and 15 units Multiscribe Reverse Transcriptase in a final reaction volume of 20 μl. The mixture was incubated at 25°C for 10 min for hybridization, subsequently at 42°C for 15 min in a Gene Amp PCR system 9600 (Perkin-Elmer), and then by cooling to 4°C. The RT reactions were subjected to PCR amplification. Five microliters of cDNA products were amplified with 2.5 units of Ampli-Taq Gold Polymerase (Applied Biosystems), 1× RT-PCR buffer, 1.75 mM MgCl2, 0.8 mM dNTPs, 0.15 μM of forward (5′-ATTCTGCTGTGCAGTGTTCG-3′) and reverse (5′-TTGCTTGGAGAAACATGCTG-3′) primers for rat schlafen 3 that resulted in a 127-bp product. β-Actin forward (5′-CCCAGCACAATGAAGATCAA-3′) and reverse (5′-ACATCTGCTGGAAGGTGGAC-3′) primers (107 bp product) were used as internal controls. Reactions were carried out in the Gene Amp PCR system 9600, first hold of 10 min at 95°C for activated Ampli-Taq Gold DNA Polymerase, and were followed by 20 s at 94°C, 60 s at 62°C for 40 cycles for amplification target gene.
Cloning of rat schlafen 3 cDNA and recombinant plasmid construct.
The total RNA from rat colonic mucosal cells was reverse transcribed with schlafen 3 sequence-specific oligos, 5′-TGGTAGAGCGCTTGCCTAGT-3′, and the schlafen 3 cDNA was amplified by use of primers designed against the data base sequence of coding region (NM_053687): forward primer, 5′-CTCAAGCTTGGATTTCATCTGGGAAGCAG-3′, reverse primer, 5′-GTGGATCCCTAGGCTCTGGGTTCAGTCCCC-3′. Briefly, 1 μg of purified RNA was reverse transcribed in the presence of 2.5 mM MgCl2, 1× RT-PCR buffer (1 mM dNTPs, 10 mM dithiothreitol, 10 units RNase inhibitor, 1.25 μM schlafen 3) sequence-specific oligo and 15 units Multiscribe Reverse Transcriptase in a final reaction volume of 20 μl; the components were mixed, briefly spun down, and incubated at 25°C for 10 min for hybridization. Reactions were carried out at 42°C for 30 min in a Gene Amp PCR system 9600 (Perkin-Elmer) and then by cooling to 4°C. The RT reactions were subjected to PCR amplification, and 5 μl of cDNA products were amplified with 25 μl of PfuUltra hotstart 2× PCR Master Mix (Stratagene, La Jolla, CA), 0.3 μM upstream primers, and 0.3 μM downstream primers in 50 μl reaction volume. Reactions were carried out in the Gene Amp PCR system 9600, first hold of 2 min at 95°C for activated PfuUltra Hotstart DNA Polymerase, followed by 40 cycles of 95°C for 30 s, 62°C for 60 s, 72°C for 2 min, and final extension at 72°C for 10 min. The PCR products were separated by electrophoresis in a 1% agarose gel and visualized by ethidium bromide staining. The 1,816 bp of target DNA fragments were recovered, digested, and cloned into HindIII and BamHI sites of plasmid, pEGFP-N1 (Clontech, CA) to generate Slfn3-GFP protein. The constructs was sequenced and confirmed to contain Rattus norvegicus schlafen 3.
Cellular growth.
This was assessed by 3-(4,5-dimethyl-thiazol-2yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay according to our standard protocol. Briefly, the cells were dispersed by trypsin-EDTA treatment, and 2.5 × 104 cells/ml, resuspended in DMEM containing 10% of FBS, were seeded into 96-well culture plates with six replicates. At the end of the 24-h incubation period, the reaction was terminated by adding 20 μl of 5 mg/ml stock MTT to each well. The reaction was allowed to proceed for 3 h at 37°C. The culture medium was then removed. The formazan crystals were then dissolved by adding 0.1 ml of DMSO. The intensity of the color developed, reflecting the number of live cells, was measured at a wavelength of 570 nm. All the values were compared with the corresponding controls.
Cell cycle analysis.
Twenty-four hours after vector and schlafen 3 transfected HCT-116 cells were fixed in 70% ethanol and treated with the staining solution (10 μg/ml propidium iodide, 50 μg/ml RNAse, 0.1% Triton X-100 and 0.1 mM EDTA) for at least 30 min at 4°C before being subjected to a BD Biosciences FAC Scan cytometer (BD Biosciences, San Jose, CA) and analyzed by Modfit software.
Western blot analysis.
Western blot analysis was performed essentially according to our standard protocol (15, 16). Briefly, aliquots of cell lysates containing 50 μg of protein were separated by SDS-PAGE. Following electrophoresis, proteins were transferred electrophoretically onto nitrocellulose membranes (Osmonics, Gloucester, MA) and subsequently incubated for 1 h at room temperature with blocking buffer, TBS-T (20 mM Tris, pH 7.6, 100 nM NaCl, 0.1% Tween-20), and 5% nonfat dry milk with gentle agitation. After the membranes were washed with TBS-T, they were incubated overnight at 4°C in TBS-T buffer containing 2.5% milk with goat polyclonal schlafen 3 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies at 1:500 dilution. The membranes were washed three times with TBS-T, subsequently incubated with appropriate secondary antibodies (1:5,000 dilution) in TBS-T-2.5% milk for 2 h at room temperature. The membranes were washed again with TBS-T. The protein bands were visualized by enhanced chemiluminescence detection system (Amersham Bioscience). The membranes were then stripped and reprobed with either β-actin or α-tubulin as a loading control.
Immunohistochemistry.
For immunohistochemical staining, an immunoperoxidase method was used with a streptavidin biotinylated horseradish peroxidase complex (Dako, Carpenteria, CA). The rat colonic tissues were formalin fixed and paraffin embedded, and 5-μm serial sections were generated. The tissue sections were deparaffinized and microwaved for 15 min in citrate buffer (0.1 M citrate acid and 0.1 M sodium citrate, pH 6.0) for antigen retrieval. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide and subsequently incubated with 5% horse serum to block nonspecific binding. The slides were then incubated at room temperature for 2 h with polyclonal antibodies to PCNA at 1:50 dilution. All slides were slightly counterstained with Harris's hematoxylin. At least ten well-oriented crypts on each slide and five slides from each sample were examined under high power. At least 750 cells/slide were counted with a ×40 objective. Labeling index = number of total labeled cells × 100/total cells per high-power focus.
Statistical analysis.
Unless otherwise mentioned, data are expressed as means ± SE. Where applicable, the results were analyzed either by Student's t-test or by ANOVA, taking P < 0.05 as the level of significance.
RESULTS
Unsupervised and supervised analysis of gene expression patterns in colonic mucosa.
Upper and lower thirds of the colonic crypts are involved in different physiological processes, namely apoptosis and proliferation, respectively, that maintain homeostasis of the colonic mucosa. Hence, we studied gene expression in two different sites (upper and lower) of the colonic mucosa with an additional variable of age (young: 4–6 mo; old 21–24 mo). This yielded four groups with two major comparisons: namely, upper third of the crypt in young vs. old and lower third of the crypt in young vs. old.
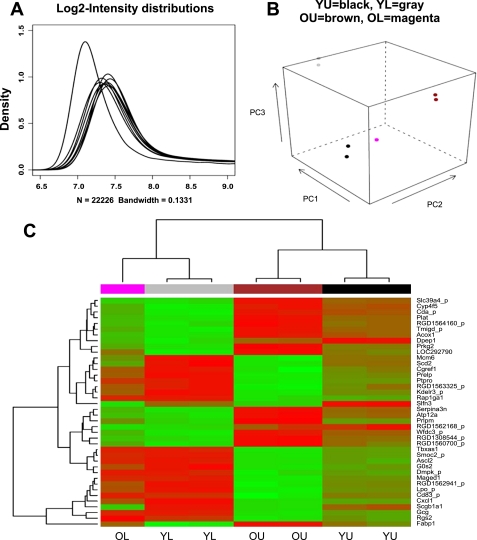
The quality control, based on probe intensity distributions, revealed that an artifact affected one sample of old lower third group in which the intensities of most of the probes were found to be close to the lower level of the detection ranges (Fig. 1A). This sample was therefore removed from further preprocessing and analysis.
Fig. 1.
Distribution of microarray probe log-intensity in each sample profile by use of Illumina Rat Sentrix-12 BeadChip Arrays (A). The leftmost distribution corresponds to a sample whose hybridization reaction was perturbed by an array sealing problem. A principal component (PC) plot (B) shows small within-group variability and large between-group variability. A hierarchical clustering retrieved correctly the known membership of the samples into the 4 groups, on the basis of the expression level of a few tens of most varying genes (C). U, upper third of colonic crypt; L, lower third of colonic crypt; Y, young rats, 4–6 mo old; O, old rats, 22–24 mo old.
The unsupervised analyses depicted in Fig. 1, B and C, show that samples in the same group produce a very similar gene expression pattern and that most of the differences are between the groups. This result is reassuring since high within-group similarity and large between-group dissimilarity is the key for reliable identification of differentially expressed genes. The supervised analysis based on a moderated t-test and fold change calculations revealed hundreds of genes as being differentially expressed in the two major comparisons we have performed.
On the basis of the supervised analysis we also found that 66 genes were differentially expressed in the same direction (up- or downregulation) in both upper and lower thirds of the colonic crypt between young and old animals. Of these genes, 44 were downregulated and 22 were upregulated in old animals compared with their young counterparts. Some of the differentially expressed genes involved in proliferation and apoptosis are shown in Table 1. Schlafen 3, a recently isolated negative regulator of proliferation (3, 21), was found to be most differentially expressed in the colonic mucosa between young and aged rats. We have observed that the expression schlafen 3 was decreased by 10- and 4-fold in mucosal cells isolated from upper and lower region of the crypt of aged rats, respectively, compared with their corresponding younger counterparts (Table 1).
Table 1.
List of selected growth and apoptosis regulatory genes differentially expressed in colonic mucosa of old compared with young rats
| Number | Gene | ID | Fold Change | Putative Growth Regulatory Function | ||||
|---|---|---|---|---|---|---|---|---|
| Genes with similar change in expression in both sites | ||||||||
| 1 | Schlafen 3 | ILMN_50410 | U: −10.0 | Growth inhibition (G1 arrest) | ||||
| L: −4 | (Downregulation of cyclin D1) | |||||||
| 2 | Secretoglobulin family 1A | ILMN_49298 | U: −3.5 | Proliferation and apoptosis | ||||
| L:-6.0 | Inhibition of phospholipase A2 | |||||||
| 3 | Dnase 1/3 | ILMN_62202 | U: −3.02 | Apoptosis | ||||
| L: −1.98 | Common pathway of apoptosis | |||||||
| 4 | Granzyme B | ILMN_49667 | U: −2.94 | Apoptosis | ||||
| L: −1.95 | (?Nonmitochondrial) | |||||||
| 5 | Annexin A1 | ILMN_60211 | U: −1.78 | Inhibition of phospholipase A2 | ||||
| L: −3.0 | ||||||||
| 6 | Granzyme G | ILMN_57532 | U: −3.1 | Apoptosis | ||||
| L: −1.44 | (?Nonmitochondrial) | |||||||
| 7 | Adrenomedullin | ILMN_60494 | U: 2.3 | Growth stimulation | ||||
| L: 2.2 | NF-κB and AKT activation | |||||||
| 8 | Granzyme A | ILMN_69553 | U: −1.95 | Apoptosis | ||||
| L: −1.95 | (?Nonmitochondrial) | |||||||
| 9 | DOCK11 | ILMN_54582 | U: −1.59 | ?Stress-mediated apoptosis | ||||
| L: −1.65 | GEF for cdc42 (activation of p38) | |||||||
| Genes with differential expression only in the upper third of the crypt | ||||||||
| 1 | Melanoma antigen, family D, 1 | ILMN_69307 | −2 | Apototic response to NGF stimulation | ||||
| p38 activation | ||||||||
| 2 | Rho GTPase activating protein 9 | ILMN_60901 | −2 | ?Inhibition of migration/proliferation | ||||
| Inhibition of rac-1 | ||||||||
| 3 | Rho, GDP disassociation inhibitor (GDI) beta | ILMN_66176 | −2 | Inhibition of Rho GTP binding proteins | ||||
| 4 | Annexin A3 | ILMN_57519 | −2 | Proliferation and apoptosis | ||||
| Inhibition of phospholipase A2 | ||||||||
| Genes with differential expression only in the lower third of the crypt | ||||||||
| 1 | PDZ binding kinase (predicted) | ILMN_48491 | −1.9 | Stress-associated apoptosis | ||||
| p38 MAPK activation | ||||||||
U, upper third of the colonic crypt; L, lower third of the colonic crypt.
Age-related changes in expression of schlafen-3.
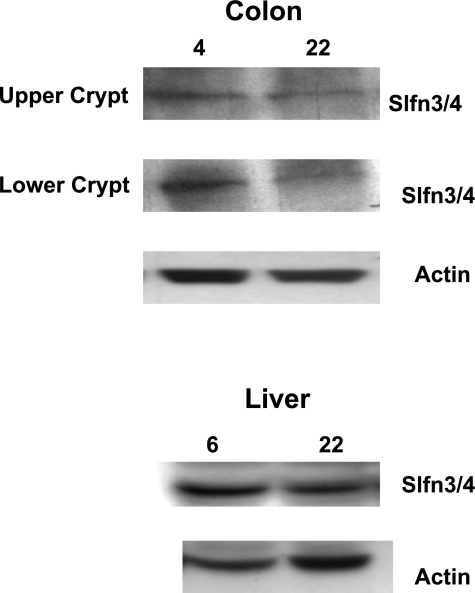
To examine age-related changes in expression of schlafen 3 gene in the colonic mucosa, we carried out real-time RT-PCR analysis. There was a 12- and 3-fold decrease, respectively, in the mucosa from upper and lower third of the colonic crypt of old, compared with young rats (Table 2). To further determine whether the age-related decrease in schlafen 3 mRNA expression would be reflected in the protein levels, Western blot analysis was also carried out. The age-related decline in schlafen 3 mRNA levels in the colonic mucosa was also reflected in protein levels of the gene in that the levels were found to be substantially lower in mucosal cells isolated from both upper and lower third of the colonic crypt of aged than in young rats (Fig. 2). In contrast to what we observed in the colonic mucosa, schlafen 3 expression remained unchanged in the relatively stable organ such as liver of young and aged rats (Fig. 2).
Table 2.
Schlafen 3 mRNA expression by real-time RT-PCR in colonic mucosa of old compared with young rats in both upper and lower thirds of the crypt
|
Schlafen 3 Expression (Real-Time RT-PCR) |
||
|---|---|---|
| Young (4–6 mo) | Old (22–24 mo) | |
| Upper (third) colon crypt | 0.16 (12) | 0.01 (1) |
| Lower (third) colon crypt | 0.03 (3) | 0.01 (1) |
Data are expressed as relative fold increase over the corresponding β-actin.
Fig. 2.
Representative Western blot showing changes in protein expression of schlafen 3 in the upper and lower fraction of colonic mucosa as well as in the liver of young (4-mo-old) and old (22-mo-old) Fisher 344 rats. A small part of the liver and epithelial cells isolated from the colonic mucosa of the young and old rats and were subjected to Western blot analysis. The experiment was repeated at least 3 times. Slfn3/4, schlafen 3. The commercial antibodies for schlafen 3 used in this experiment also cross-react with schlafen 4. 4, 6, and 22 represent age in months.
Wortmannin, a specific PI3K inhibitor, stimulates schlafen-3 expression in the colonic mucosa during aging.
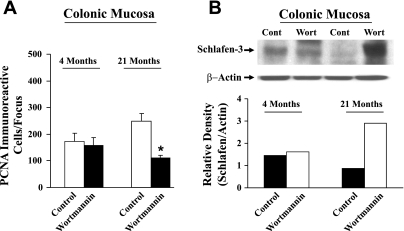
Previously, we have reported that inhibition of PI3K activity in the colonic mucosa of aged rats by wortmannin greatly stimulates apoptosis (15). To determine whether schlafen 3 might be involved in regulating colonic mucosal growth during aging, young and old rats were injected with wortmannin or vehicle (controls) and the levels of schlafen 3 were determined in the colonic mucosa after 6 h. Interestingly, we observed that wortmannin treatment produced more than threefold increase in schlafen 3 levels in the colonic mucosa of aged rats, accompanied by a concomitant reduction in proliferative activity, as evidenced decreased PCNA (proliferating cell nuclear antigen) immunoreactivity, compared with their corresponding younger counterparts (Fig. 3, A and B).
Fig. 3.
Changes in mucosal proliferation as measured by PCNA immunoreactivity (A) and schlafen 3 protein expression (B) in colonic mucosa of young (4-mo-old) and old (21-mo-old) Fisher 344 rats 6 h after a single injection of wortmannin (wort; 0.1 mg/kg ip in 15% DMSO or vehicle). Cont, control. *P < 0.01, compared with controls.
Overexpression of schlafen-3 gene inhibits cellular growth.
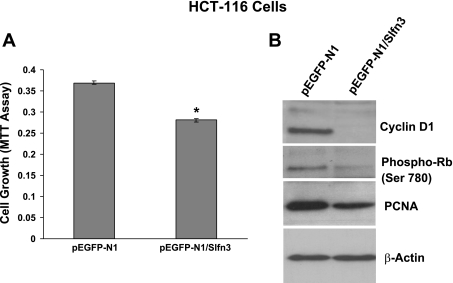
To determine the role of schlafen 3 in regulating cellular growth, human colon cancer HCT-116 cells, that are devoid of rat schlafen 3 gene, were transfected with schlafen 3 cDNA. Twenty-four hours after transfection, cellular growth was determined by MTT assay. Results revealed that transfection of schlafen 3 caused a significant 25–30% reduction in cell growth compared with the vector-transfected controls (Fig. 4A).
Fig. 4.
Cell growth inhibition [3-(4,5-dimethyl-thiazol-2yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay] (A) and expression of selected cell cycle and proliferation regulatory proteins (B) at 24 h in vector and schlafen 3-transfected HCT-116 cells. Phospho-Rb, phosphorylated retinoblastoma protein. *P < 0.01, compared with controls.
To determine regulatory mechanisms of schlafen 3-induced inhibition of cell growth, HCT-116 cells transfected with schlafen 3 cDNA were selected by G418 following 2 wk of transfection. Flow cytometric analysis showed that transfection of schlafen 3 gene in colon cancer HCT-116 cells caused a 57% increase in accumulation of cells at the G0/G1 phase and corresponding reduction in S phase, suggesting a block in G1 to S progression (Table 3). To further determine regulatory mechanism of blockage of G1 to S phase progression, expression of cyclin D1, a critical regulator of this process, was examined. Levels of cyclin D1 in schlafen 3 transfected cells were greatly reduced, compared with the vector transfected controls (Fig. 4B). In fact, no detectable levels of cyclin D1 were observed in schlafen 3 transfected HCT-116 cells (Fig. 4B).
Table 3.
Effect of schlafen 3 overexpression on HCT-116 cell cycle distribution
| Treatment |
Cell Cycle Distribution (%) |
|||
|---|---|---|---|---|
| G0 | G0/1 | S | G2/M | |
| pEGFP-N1 (Control) | 18.27 | 35.12 | 21.43 | 22.31 |
| pEGFP-N1/ Slfn-3 | 9.98 | 54.49 | 16.5 | 18.25 |
Retinoblastoma protein (Rb) is known to be differentially phosphorylated during the cell cycle. In its hypophosphorylated state, Rb binds different cellular proteins, including the transcription factor E2F. The latter is, however, released when Rb is hyperphosphorylated during progression of the cell cycle through the S and G2 (19). As expected, the levels of the phosphorylated form of Rb (pRb) were found to be markedly lower in schlafen 3-transfected HCT-116 cells, compared with the vector transfected controls (Fig. 4B). These changes were also accompanied by a concomitant reduction in the levels of PCNA (Fig. 4B), an auxiliary protein for DNA polymerase δ that is expressed during S phase of the cell cycle (4, 5) and has been shown its levels to correlate positively with proliferation.
DISCUSSION
Aging is associated with changes in the structural and functional properties of the gastrointestinal mucosa as reflected by alterations in growth, differentiation, and immunity (14). However, a consistent pathological observation with aging is the increased incidence of gastrointestinal cancer, particularly in the colon, a leading cause of morbidity and mortality. Although colorectal cancer is a multistage process, increased proliferation and decrease in apoptosis are considered to be critical early events in the progression of cancer (13). Although earlier observations in the mouse and a recent report in rats suggest that the proliferative activity of the intestine either decreases or remains unchanged with aging (7, 9, 12), several morphological and biochemical studies from our and other laboratories have demonstrated that, in barrier-reared Fischer 344 rats, aging is associated with increased proliferative activity and decreased apoptosis in the colonic mucosa (1, 10, 11, 14–16, 26). On the basis of the observations that aging is associated with increased proliferation and decreased apoptosis in the colonic mucosa, it has been postulated that aging may predispose the colon to malignant transformation (1, 14). However, the underlying regulatory mechanisms for the age-related changes gastrointestinal mucosal growth are poorly understood. Gene expression profiling was, therefore, carried out to better understand the regulatory mechanisms.
Many of the genes that are differentially expressed in the colonic mucosa of young and old rats can be grouped into growth suppressors, growth promoters, and proapoptotic and antiapoptotic genes. Of various genes identified in such fashion, schlafen 3 was most differentially expressed in the colonic crypt. Schlafen 3 is a member of expanding family of proteins that includes at least eight members in mice (8, 21). They are classified into three groups on the basis of COOH-terminal length as short (schlafen 1 and 2), intermediate (schlafen 3 and 4), and long (schlafen 5, 8, 9 and 10) forms. The common NH2-terminal domain has a GTP/ATP binding site, whereas COOH-terminal domain in long form of schlafens possesses helicase-like domain, which is thought to play a role in DNA repair or transcriptional regulation (8, 28).
Although the functional properties of murine schlafen family of proteins have not been fully elucidated, they have been suggested to play a critical role in cell proliferation as a negative regulator by downregulating cyclin D1 (3, 8). It has been demonstrated that overexpression of schlafen 3 results in growth inhibition and greatly impairs anchorage-independent growth of various cells (21). Furthermore, schlafen 1, a short-form member of the schlafen family, has been shown to be a negative regulator of cell cycle progression in response to stimulation with a variety of mitogens including EGF, PDGF, and G protein-coupled receptors (3). However, these observations were made in lymphocytes and fibroblasts. Our present data, for the first time, demonstrate a growth regulatory role of schlafen 3 in epithelial cells. The basis for this inference comes from the observation that transfection of schlafen 3 in human colon cancer HCT-116 cells, which are devoid of schlafen 3 (present only in rodents), inhibits cellular growth as determined by MTT assay, accompanied by a concomitant reduction in expression of PCNA, an auxiliary protein for DNA polymerase δ that is expressed during S phase of the cell cycle (4, 5) and has been shown its levels to correlate positively with proliferation.
Although the underlying mechanisms of schlafen 3-induced inhibition of cellular growth are not fully elucidated, downregulation of cyclin D1, an important cyclin involved in G1-S transition, is thought to be a major contributing factor. Our present data also support this postulation in that transfection of schlafen 3 in HCT-116 cells results in decreased expression of cyclin D1. This could be a contributory factor for blockage of G1 to S transition in schlafen 3-transfected cells. Furthermore, our observation that the levels of pRB are decreased in schlafen 3 transfected HCT-116 cells than in controls lends supports to the contention that schlafen 3 could be a negative regulator of proliferation. Further support for this postulation comes from the observation that aging, which has been shown increase proliferative activity in the colonic mucosa of Fischer 344 rats (1, 10, 11, 14–16, 26), is associated with decreased expression of schlafen 3 in the colonic mucosa. In fact, the expression of schlafen 3 was decreased throughout the crypt. On the other hand, administration of wortmannin, which inhibited proliferation in the colonic mucosa, resulted in a marked stimulation in the expression of schlafen 3. Hence the decreased schlafen 3 expression in the colonic mucosa with aging may be partly responsible for the loss of cell cycle control leading to increased G1-S transition and proliferation observed in old rats (27). Interestingly, there was a higher differential expression of schlafen 3 in the upper third of the crypt by quantitative RT-PCR. This might suggest that either schlafen 3 is involved in the prevention of expansion of proliferative zone in the upper third of the crypt during aging or it may have an undetermined role in regulating differentiation and/or apoptosis.
As opposed to the murine family of schlafen, human schlafen consists of seven known members that are located on long arm of chromosome 17. Of these, schlafen 12 shows 47% sequence homology with the murine schlafen 3 gene. However, the functional significance of human family of schlafens including schlafen 12, remains to be elucidated (http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Hs&CID=663548; http://www.ihop-net.org/UniPub/iHOP/?search=SCHLAFEN&field=all&ncbi_tax_id=0&organism_syn=).
In conclusion, our data show that aging is associated with downregulation of genes involved in suppression of proliferation and induction of apoptosis. In particular, the expression of schlafen 3, a negative regulator of proliferation, was decreased by 8- to 10-fold in the colonic mucosa of aged than in young rats. That schlafen 3 could be a negative regulator of growth was supported by the observation that transfection of schlafen 3 in colon cancer cells leads to inhibition of proliferation, which could be attributed to downregulation of cyclin D1 and PCNA and attenuation of phosphorylation of Rb.
GRANTS
This work was supported by grants to A. P. N. Majumdar from the National Institutes of Health/National Institute on Aging (AG014343) and the Department of Veterans Affairs.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Atillasoy E, Holt PR. Gastrointestinal proliferation and aging. J Gerontol 48: B43–B49, 1993. [DOI] [PubMed] [Google Scholar]
- 2.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57: 289–300, 1995. [Google Scholar]
- 3.Brady G, Boggan L, Bowie A, O'Neill LAJ. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J Biol Chem 280: 30723–30734, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA-polymerase-δ. Nature 326: 515–517, 1987. [DOI] [PubMed]
- 5.Chang CD, Ottavio L, Travalli S, Lipson KE, Baserga R. Transcriptional and posttranscriptional regulation of the proliferating cell nuclear antigen gene. Mol Cell Biol 10: 3289–3296, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deschner EE, Lynch HT. The influence of age and genetic risk for colon cancer on colonic epithelial cell proliferation. Proc Annl Mtg Am Assoc Cancer Res 28: 197, 1987. [Google Scholar]
- 7.Fry RJM, Lesher S, Kohn HI. Age effect on cell-transit time in the mouse jejunal epithelium. Am J Physiol 201: 213–216, 1961. [DOI] [PubMed] [Google Scholar]
- 8.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol 16: 1535–1548, 2004. [DOI] [PubMed] [Google Scholar]
- 9.Heung-Man L, Greeley GH Jr, Englander EW. Effects of aging on expression of genes involved in regulation of proliferation and apoptosis in the colonic epithelium. Mech Ageing Dev 155: 139–155, 2000. [DOI] [PubMed] [Google Scholar]
- 10.Holt PR, Yeh KY. Colonic proliferation is increased in senescent rats. Gastroenterology 95: 1556–1563, 1988. [DOI] [PubMed] [Google Scholar]
- 11.Holt PR, Yeh KY, Kolter DP. Altered controls of proliferation in proximal small intestine of the senescent rat. Proc Natl Acad Sci USA 85: 2771–2775, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lesher S, Fry RJM, Kohn HI. Age and generation time of the mouse duodenal epithelium cell. Exp Cell Res 24: 334–343, 1961. [DOI] [PubMed] [Google Scholar]
- 13.Lipkin M Biomarkers of increased susceptibility to gastrointestinal cancer. Their development and application to studies of cancer prevention. Gastroenterology 92: 1083–1086, 1987. [DOI] [PubMed] [Google Scholar]
- 14.Majumdar APN, Basson MD. Effect of aging on the gastrointestinal tract. In: Physiology of the Gastrointestinal Tract, edited by Johnson LR, Barrett K, Ghishan F, Merchant JI, Said HM, Wood JD. New York: Academic, 2006, p. 405–433.
- 15.Majumdar APN, Du J. Phosphatidylinositol 3 kinase/Akt signaling stimulates colonic mucosal cell survival during aging. Am J Physiol Gastrointest Liver Physiol 290: G49–G55, 2006. [DOI] [PubMed] [Google Scholar]
- 16.Majumdar APN, Du J, Yu Y, Xu H, Levi E, Patel BB, Rishi AK. Cell-cycle and apoptosis regulatory protein-1: a novel regulator of apoptosis in the colonic mucosa during aging. Am J Physiol Gastrointest Liver Physiol 293: G1215–G1222, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Patel BB, Yu Y, Du J, Levi E, Phillip PA, Majumdar APN. Age-related increase in colorectal cancer stem cells in macroscopically normal mucosa of patients with adenomas: a risk factor for colon cancer. Biochem Biophys Res Commun 378: 344–347, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roncucci L, Ponz de Leon M, Scalmati A, Malagoli G, Pratissoli S, Perini M, Chahin NJ. The influence of age on colonic epithelial cell proliferation. Cancer 62: 2373–2377, 1988. [DOI] [PubMed] [Google Scholar]
- 19.Rustgi A Oncogene and tumor suppressor genes. In: Gastrointestinal Cancers: Biology, Diagnosis, and Therapy, edited by Rustgi AK. Philadelphia, PA: Lippincott-Raven, 1995, p. 65–76.
- 20.Schmelz EM, Levi E, Du J, Majumdar APN:. Loss of expression of EGF-receptor related peptide (ERRP) in the aging colon: a risk factor for colorectal cancer. Mech Ageing Dev 125: 917–922, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9: 657–668, 1998. [DOI] [PubMed] [Google Scholar]
- 22.Sohn WJ, Kim D, Lee KW, Kim MS, Kwon S, Lee Y, Kim DS, Kwon HJ. Novel transcriptional regulation of the Schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol Immunol 44: 3273–3282, 2007. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK, Limma L. Liner models for microarray data. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor, edited by Gentleman R, Carey V, Duoit S, Irizarry R, Huber W. New York: Springer, 2005, p. 397–420.
- 24.Tarca AL, Carey VJ, Chen XW, Romero R, Drãghici S. Machine learning and its applications to biology. PLoS Comput Biol 3: e116, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tureaud J, Sarkar FH, Fligiel SE, Kulkarni S, Jaszewski R, Reddy K, Yu Y, Majumdar AP. Increased expression of EGFR in gastric mucosa of aged rats. Am J Physiol Gastrointest Liver Physiol 273: G389–G398, 1997. [DOI] [PubMed] [Google Scholar]
- 26.Xiao ZQ, Moragoda L, Jaszewski R, Hatfield JA, Fligiel SE, Majumdar APN. Aging is associated with increased proliferation and decreased apoptosis in the colonic mucosa. Mech Ageing Dev 122: 1849–1864, 2001. [DOI] [PubMed] [Google Scholar]
- 27.Xiao ZQ, Jaszewski R, Majumdar APN. Aging enhances G1 phase in the colonic mucosa of rats. Mech Ageing Dev 116: 1–14, 2000. [DOI] [PubMed] [Google Scholar]
- 28.Yan N, Shi Y. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol 21: 35–56, 2005. [DOI] [PubMed] [Google Scholar]