Abstract
Programmed ribosomal frameshifting (PRF) is one of the multiple translational recoding processes that fundamentally alters triplet decoding of the messenger RNA by the elongating ribosome. The ability of the ribosome to change translational reading frames in the −1 direction (−1 PRF) is employed by many positive strand RNA viruses, including economically important plant viruses and many human pathogens, such as retroviruses, e.g., HIV-1, and coronaviruses, e.g., the causative agent of severe acute respiratory syndrome (SARS), in order to properly express their genomes. −1 PRF is programmed by a bipartite signal embedded in the mRNA and includes a heptanucleotide “slip site” over which the paused ribosome “backs up” by one nucleotide, and a downstream stimulatory element, either an RNA pseudoknot or a very stable RNA stem–loop. These two elements are separated by six to eight nucleotides, a distance that places the 5′ edge of the downstream stimulatory element in direct contact with the mRNA entry channel of the 30S ribosomal subunit. The precise mechanism by which the downstream RNA stimulates −1 PRF by the translocating ribosome remains unclear. This review summarizes the recent structural and biophysical studies of RNA pseudoknots and places this work in the context of our evolving mechanistic understanding of translation elongation. Support for the hypothesis that the downstream stimulatory element provides a kinetic barrier to the ribosome-mediated unfolding is discussed.
Keywords: Pseudoknot, Ribosomal recoding, Frameshifting, Translational regulation, HIV-1, Luteovirus, NMR solution structure, Cryo-electron microscopy
1. Introduction
Ribonucleic acid (RNA) molecules adopt tertiary structures of rich complexity and functional diversity (Holbrook, 2005). In messenger RNAs, 5′ and 3′ untranslated regions (UTRs) post-transcriptionally control gene expression by regulating RNA processing and translation initiation (Jenner et al., 2005). 5′ UTRs also fold into metabolite-sensing RNA elements termed riboswitches that recognize specific metabolites and thereby regulate transcription termination/anti-termination or translation initiation in bacteria (Dann et al., 2007, Wakeman et al., 2007, Winkler et al., 2002).
Since its discovery in the 3′ ends of certain plant viral genomic RNAs by the Dutch groups of Pleij, Rietveld and Bosch in the early to mid-1980s (Pleij et al., 1985, Rietveld et al., 1984, Rietveld et al., 1983) and subsequent NMR structural characterization (Kolk et al., 1998), the RNA pseudoknot is now recognized as a ubiquitous folding topology that performs a wide range of functions in biology. RNA pseudoknotting is found in both naturally occurring and in vitro evolved RNA catalysts or ribozymes (Cate et al., 1996, Ferre-D’Amare et al., 1998, Serganov et al., 2005), as well as in many complex folded RNAs and ribonucleoprotein particles (RNPs) including the ribosome and telomerase. Here, pseudoknots play a critical structural or scaffolding role in bringing distant regions of single-stranded RNA together in order to form core helices composed of Watson–Crick base pairs (Adams et al., 2004, Brodersen et al., 2002, Golden et al., 2005, Theimer et al., 2005, Torres-Larios et al., 2006). In addition, pseudoknots are known to play important regulatory roles in translation initiation at internal ribosome entry sites (IRESs) (Otto and Puglisi, 2004, Pfingsten et al., 2006, Pfingsten et al., 2007), in autoregulation of translation initiation (ten Dam et al., 1992, Deckman et al., 1987) and in metabolite-sensing RNAs (Wakeman et al., 2007), where the ribosome binding sequence may be sequestered in a pseudoknotted structure (Gilbert et al., 2008). During translation elongation, the pseudoknot-containing transfer-messenger RNA (tmRNA) mediates rescue of stalled ribosomes that reach the 3′ end of an mRNA lacking a termination codon (Haebel et al., 2004, Moore and Sauer, 2007, Nonin-Lecomte et al., 2006). When found within the coding region of messenger RNA itself, pseudoknots are known to stimulate the efficiency of a number of programmed translational recoding events (Baranov et al., 2002), including stop codon redefinition (Howard et al., 2005) and ribosomal frameshifting (Farabaugh, 1996, Giedroc et al., 2000). The subject of this review is RNA pseudoknots and related RNA motifs that simulate −1 programmed ribosomal frameshifting (−1 PRF).
An important goal of these studies is to elucidate inter-relationships between pseudoknot structure, stability, dynamics and folding kinetics, and how they govern the ability of this deceptively simple motif to stimulate −1 frameshifting during ribosomal translocation. Recent cryo-electron microscopy studies of a putative translocation intermediate stalled over a frameshift site (Namy et al., 2006), mechanical unfolding studies of RNA pseudoknots (Green et al., 2008, Hansen et al., 2007), and NMR structural and thermodynamic studies of a family of evolutionarily closely related frameshifting pseudoknots from plants (Cornish and Giedroc, 2006, Cornish et al., 2005, Cornish et al., 2006b) support the hypothesis that the ability of a frameshift-stimulating pseudoknot to resist the force of mechanical unwinding by the ribosomal mRNA helicase (Takyar et al., 2005) may be more strongly correlated with frameshift stimulation, rather than a specific or unique structural feature(s). This review will summarize these data and place them in the context of our maturing understanding of translation elongation (Wen et al., 2008). The reader is also referred to reviews on RNA pseudoknot structure and function in ribosomal frameshifting and RNA virus replication that have recently appeared (Brierley and Dos Ramos, 2006, Brierley et al., 2007, Staple and Butcher, 2005a).
2. The H-type RNA pseudoknot folding topology
An RNA pseudoknot is a simple folding topology that is formed when nucleotides within a single-stranded loop base pair with complementary nucleotides outside of that loop (Fig. 1 ) (ten Dam et al., 1992, ten Dam et al., 1995). If this new base pairing originates with nucleotides within an RNA hairpin loop, the topology is referred to as an H (hairpin)-type pseudoknot, forming a structure composed of two helical stems, S1 and S2, and two non-equivalent loops, L1 and L2 (Fig. 1) (Giedroc et al., 2000). Loop L1 crosses the deep major groove of the lower stem S2, while loop L2 crosses the minor groove side of stem S1. The pseudoknot-forming stem is S2, with S1 considered the hairpin stem. Some H-type pseudoknots contain a third loop (denoted here as L3), often a single unpaired nucleotide, that interrupts the continuous strand at the helical junction; this base can either be extruded from the helix (Egli et al., 2002, Nixon et al., 2002b) or intercalated between the two helical elements (Shen and Tinoco, 1995). Note that two conventions currently exist for the loop–stem nomenclature of hairpin-type pseudoknots that conform to this basic architecture, in which the designations of L2 and L3 are reversed, and are instead identified in order of appearance in the nucleotide sequence (Brierley et al., 2007). In this case, L2 is the loop found at the helical junction between S1 and S2, and L3 is the long loop that straddles the minor groove of the upper stem S1. In this review, we conform to the first convention.
Fig. 1.
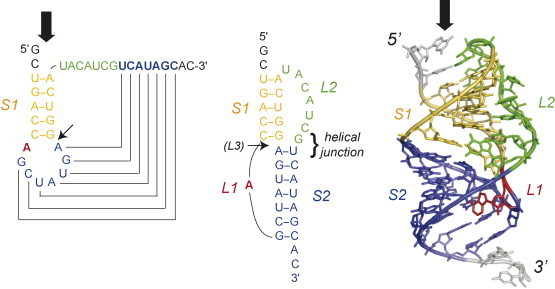
Folding topology and solution structural model of the phage T2 gene 32 autoregulatory pseudoknot (PDB 2TPK) in which the two pseudoknot stems S1 and S2 are coaxially stacked on one another (Du et al., 1996, Holland et al., 1999). The small arrow indicates the position of pseudoknot loop L3, when present, while the large vertical arrow indicates the direction of approach of the translocating ribosome.
3. Translational recoding and −1 programmed ribosomal frameshifting (−1 PRF)
It has long been known that specific regulatory signals in the mRNA can influence the speed and fidelity of ribosomal decoding. For example, elongating ribosomes pause or slow down upon encountering classical secondary structures and this ribosomal pausing is often associated with ribosomal recoding events (Baranov et al., 2002). One such recoding event is frameshifting, when the ribosome is directed to move into the −1, +1 or +2 reading frames from the reference “0” frame (Harger et al., 2002, Stahl et al., 2002). Programmed ribosomal frameshifting (PRF) allows for translation of two proteins encoded in overlapping reading frames from a single translation initiation site upstream of the 5′ open reading frame (Fig. 2A).
Fig. 2.
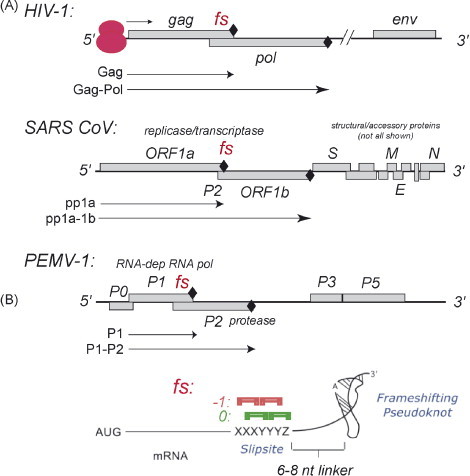
Organization of HIV-1, SARS-CoV and PEMV-1 genomic RNAs highlighting the frameshift sites (fs) in each case. (A) Production of viral fusion proteins, e.g., Gag-Pol in HIV-1, from a single translation initiation event (indicated by the pink ribosome) via a −1 PRF (fs) event. (B) Schematic rendering of a bipartite frameshift signal (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.).
−1 PRF has been documented to occur in bacteria (Tsuchihashi and Kornberg, 1990), yeast (Dinman et al., 1991) and mammals (Clark et al., 2007, Manktelow et al., 2005, Wills et al., 2006); however, the process is particularly exploited by RNA viruses from plants and animals, including HIV-1 and related retroviruses and coronaviruses, including SARS-CoV (Brierley and Dos Ramos, 2006). The efficiency of −1 PRF is not 100%, but instead dictates the molar ratio of downstream and upstream proteins ultimately present in the assembling virus. In HIV-1, this ratio is tightly regulated by the virus; molecules that alter the frameshifting levels have significant effects on virus propagation and infectivity (Biswas et al., 2004); in addition, it is known that changing the frameshifting efficiency downward induces results in significantly lower infectivities in both HIV-1 (Dulude et al., 2006) and in murine Moloney leukemia virus, where the translational readthrough signal was replaced with the frameshifting signal from HIV-1 (Gendron et al., 2005). This, in turn has motivated efforts to develop tightly binding molecules as a potential anti-viral strategy (Biswas et al., 2004, Dulude et al., 2008, Park et al., 2008).
Two mRNA-encoded signals are required for stimulation of efficient frameshifting by an elongating ribosome. One is a heptanucleotide “slippery site” of the general sequence X XXY YYZ over which the ribosome pauses, e.g., G GGA AAC in pea enation mosaic virus-1 (PEMV-1) (Nixon et al., 2002b), and a downstream RNA pseudoknot, positioned six to eight nucleotides from the 3′ edge of the slip-site (Fig. 2B) (Brierley, 1995). The slip-site alone dramatically increases the intrinsic level of frameshift errors from 0.00005 to ≈0.005 per codon depending strongly on the sequence (Stahl et al., 2002), with the pseudoknot further stimulating this process 10–30-fold more. Thus, the pseudoknot induces a subtle, yet critical, perturbation in the kinetic partitioning of the translocating ribosome into the −1 frame from the reference frame (see below). In nearly every case that has been examined in detail, a downstream H-type pseudoknot is the stimulatory element. A prominent exception to the pseudoknot as the downstream stimulator is in lentiviruses HIV-1 and SIV, where a bipartite stem–loop structure appears necessary and sufficient to stimulate −1 PRF at the gag-pol junction (Gaudin et al., 2005, Marcheschi et al., 2007, Staple and Butcher, 2005b). This may be facilitated by the slippery sequence itself, which is UUUUUUA, a particularly shifty sequence (Brierley et al., 1992).
4. The atomic structure of the bacterial ribosome
Although essentially all of the atomic resolution structural information that is currently available for the translating ribosome comes from studies of the bacterial ribosome, it is widely believed that the fundamental features of protein synthesis are evolutionarily conserved from prokaryotes to eukaryotes since the core ribosome structure shows a high degree of conservation (Spahn et al., 2001). For the HIV-1 frameshift signal at least, the bacterial ribosome appears to be fully functional as the eukaryotic ribosome in stimulating frameshifting (Leger et al., 2004). Thus, it is of interest to consider the structure and dynamics (see Section 5 below) of the bacterial 70S ribosome in some detail. Several recent X-ray crystallographic structures of the intact 70S bacterial ribosome either unliganded (Schuwirth et al., 2005) or with various ligands bound (Jenner et al., 2005, Korostelev et al., 2006, Selmer et al., 2006) provide an opportunity to consider −1 PRF at the atomic level. These structures along with the previous Thermus thermophilus 70S structure with bound mRNA, and all three tRNAs to modest (3.9–5.5 Å) resolution in initiation and post-initiation complexes (Jenner et al., 2007, Yusupov et al., 2001, Yusupova et al., 2006, Yusupova et al., 2001) clearly define the path of the mRNA as it threads through the head and platform of the 30S subunit (shaded purple in the initiation complex, Fig. 3 ). They reveal that the relatively short spacer between the slip-site and the pseudoknot places the pseudoknot in direct contact with the mRNA entry channel of the elongating ribosome, as previously modeled (Giedroc et al., 2000, Plant et al., 2003). The mRNA channel is clearly defined and is contained totally within the 30S subunit, between the head and body, lined with ribosomal proteins S3, S4 and S5 (Fig. 3A) (Brodersen et al., 2002). Recent biochemical experiments reveal that the 70S ribosome has helicase activity (Takyar et al., 2005), which may function by passively trapping transiently unfolded secondary structure by dsRNA binding protein S5; alternatively, the S3/S4/S5 proteins might function as a processivity clamp (Jeruzalmi et al., 2002) positioned at the entrance to the mRNA tunnel (Fig. 3B).
Fig. 3.
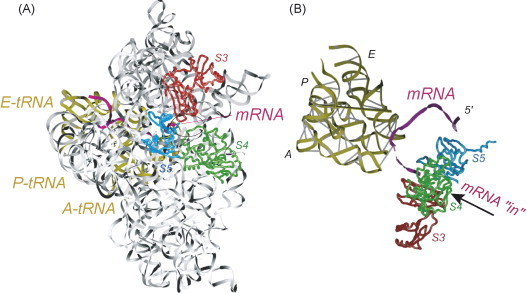
Ribbon representation of the path of the messenger RNA bound to the 30S ribosomal subunit and the three transfer RNAs (PDB 1JGO) (Yusupova et al., 2001). (A) The A-site, P-site and E-site tRNAs are indicated (gold), the mRNA is purple, and S3, S4 and S5 proteins are indicated. (B) Ribbon representation of the structure of the mRNA entry channel indicating a candidate docking site for RNA elements that stimulate −1 PRF (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.).
The atomic (2.8 Å) resolution model of the pre-translocation complex-containing bound mRNA, deacylated initiator tRNAfMet in the P-site, aminoacyl tRNAPhe in the A-site and non-cognate tRNA in the E-site (Selmer et al., 2006) reveals two additional details that likely enable the elongating ribosome to maintain the reading frame. First, the kink in the mRNA between the A- and P-site codons (Yusupov et al., 2001) is stabilized by a bound Mg2+ (shaded green, Fig. 4 A). This Mg2+ ion forms outer sphere coordination bonds with the phosphate oxygens of adjacent nucleotides that define the P- and A-site codon boundaries, as well as nucleotides 1401 and 1402 of helix 44 of the 16S RNA (Fig. 4A). These interactions effectively auger the mRNA frame relative to the 30S subunit and may well prevent translation slippage during normal decoding. Secondly, the junction between the D- and anticodon stems of the deacylated P-site tRNA is distorted or kinked toward the A-site (see also Korostelev et al., 2006); relaxation of this deformation may help drive this tRNA into the E-site upon translocation. To prevent this from happening prematurely, there seems to be a molecular “gate” (Fig. 4B) formed by A-minor hydrogen bonding interactions to the P-site tRNA from the 30S head and platform wedged between the P- and E-site tRNAs (Fig. 4B).
Fig. 4.
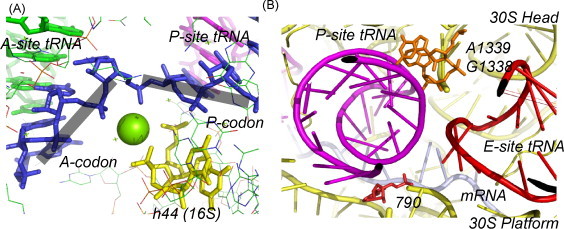
Close-up view of the decoding center of the small ribosomal subunit taken from an atomic resolution structure (2.8 Å) of the a pre-translocation complex of the 70S bacterial ribosome with bound mRNA and aminoacyl tRNAPhe in the A-site, deacylated initiator tRNAfMet in the P-site and non-cognate tRNA in the E-site (Selmer et al., 2006) (PDB 2J02). (A) Aminoacyl A-site and deacylated P-site tRNA anticodon–mRNA codon interactions are shown. A Mg2+ ion is also shown which may help induce a kink if the mRNA and therefore maintain reading frame just prior to translocation. (B) Close-up of the P- and E-site tRNA codon regions within the decoding center, highlighting the displacement of the P-site tRNA codon toward the A-site. See text for details. Adapted from Selmer et al. (2006).
It is noteworthy that despite the fact that the E-site tRNA was stably bound to the ribosome in this pre-translocation model, there are no E-site codon–anticodon base pairing interactions in the structure (Selmer et al., 2006), a finding that differs from what was observed in a post-initiation model at 3.7 Å (Jenner et al., 2007). In this structure, one of the E-site codon nucleotides appears to base pair with the E-site tRNA anticodon. This finding supports the idea that during translation, the tRNAs move with the mRNA while maintaining codon–anticodon hydrogen bonding for all three tRNAs, although this has not yet been fully established (Feinberg and Joseph, 2001). In any case, this structure might reflect an intermediate state just prior to base pair dissociation and finally release of the deacylated E-site tRNA. These findings would appear to be relevant for understanding −1 PRF since it has recently been shown that mutations in the E-site codon alter the efficiency of −1 PRF at least in the context of the HIV-1 frameshift signal; furthermore, mutations in 16S ribosomal RNA that influence HIV-1 frameshifting efficiency map to helices 21 and 22 in the Escherichia coli ribosome, a region previously implicated in translocation and E-site tRNA anticodon binding (Leger et al., 2007). Mutations in this region also influence translocation and in many cases give rise to spontaneous frameshifts (Sergiev et al., 2005). Nonetheless, it remains unclear at this point what precise role the E-site tRNA might play in frameshifting since there is still controversy as to when the E-site tRNA exits the ribosome (Dinos et al., 2005, Spiegel et al., 2007).
5. The ribosome as a dynamic machine: implications for ribosomal frameshifting
Various frameshifting models have been presented that attempt to pinpoint the exact step in a translocation cycle where frameshifting occurs (Harger et al., 2002, Jacks et al., 1988, Leger et al., 2007, Plant et al., 2003). To consider these models, we outline the basic steps of a translational elongation cycle in the context of a ribosome encountering an RNA pseudoknot during decoding (Fig. 5 ) (Joseph, 2003, Moazed and Noller, 1989). During active translation, a ribosome with a vacant A-site accepts a cognate aminoacylated tRNA delivered as a complex with EF-Tu·GTP (eEF1A·GTP in eukaryotes). Almost immediately, the peptide on the P-site tRNA is transferred to the fully accommodated A-site aminoacylated tRNA in a reaction catalyzed by the peptidyl transfer center on the large subunit (50S in bacteria; 60S in eukaryotes). Subsequently, the tRNA acceptor ends translocate on the 50S (60S) subunit resulting in a P/E-A/P hybrid state of the ribosome (Dorner et al., 2006, Moazed and Noller, 1989). EF-G·GTP (eEF2·GTP in eukaryotes) then binds to the ribosome and drives 30S (40S) translocation returning the ribosome to the classical P/P-E/E state with a vacant A-site. Regardless of which mRNA codons are present in the P- and A-sites of the ribosome, there are only three places in an elongation cycle that a ribosome can frameshift: during accommodation (step 2, Fig. 5), during or immediately following 50S translocation to form the hybrid state (step 3), or during 30S translocation to return to classical state (step 4). It is also possible in fact that −1 PRF is capable of occurring at all of these steps, with the precise mechanism perhaps dictated by the frameshifting signal itself.
Fig. 5.
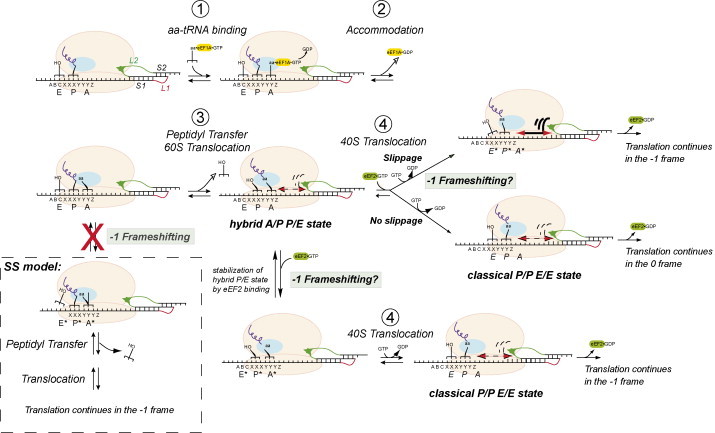
A schematized view of a single translocation cycle of translation that shows three possible points in the cycle that −1 frameshifting could potentially be stimulated by a downstream pseudoknot. In one scenario, −1 PRF is proposed to occur from the hybrid A/P-P/E state which is stabilized by EF-G·GTP (eEF2·GTP) binding; this binding in turn, weakens the interaction of the deacylated P-site tRNA with the mRNA inducing frameshifting before 30S (40S in eukaryotes) translocation to the classical P/P and E/E states accompanied by hydrolysis and EF-G·GDP (eEF2·GDP) release (Ermolenko et al., 2007, Spiegel et al., 2007). Alternatively, −1 PRF occurs during the 30S (40S) translocation step itself (Namy et al., 2006). Both models are distinguished from a previous model that invoked simultaneous slippage (SS) of two tRNAs in the 5′ direction after accommodation and before peptidyl transfer (Jacks et al., 1988, Plant et al., 2003). See text for additional details.
Closer inspection of each step of the elongation process from recent biochemical, bulk and single-molecule experiments clearly reveals that the elongating ribosome is a dynamic machine; this in turn, has clear implications for any proposed mechanisms of −1 PRF. Recent structures of the bacterial ribosome obtained by cryo-electron microscopy prepared in different stages of the translation elongation cycle reveal that the ribosome can adopt at least two different conformations, in which the small subunit is oriented differently relative to the large subunit (Frank and Agrawal, 2000, Gao et al., 2003, Valle et al., 2003). Subsequent single-molecule experiments have shown that the bound tRNAs in the pretranslocation state (pre-30S translocation, see Fig. 5) are conformationally dynamic (Blanchard et al., 2004, Lee et al., 2007, Munro et al., 2007). More recently, bacterial ribosomes assembled from subunits specifically labeled with fluorescent probes (Ermolenko et al., 2007) have been shown to spontaneously fluctuate between two states when prepared in either a pretranslocation state or with a single deacyl-tRNA bound in the P-site and prefer the rotated or hybrid state (Cornish et al., 2008). Upon addition of EF-G and the non-hydrolyzable analogue GDPNP, the ribosomes were further stabilized in a rotated or hybrid state (Cornish et al., 2008, Spiegel et al., 2007). In contrast, when these ribosomes lacked tRNA or contained a peptidyl-tRNA in the P-site of the ribosome, i.e., in a pre-50S translocation state (see Fig. 5), the non-rotated or classical state was favored and the dynamic rotational movement of the ribosome was strongly hindered (Cornish et al., 2008).
What does a dynamic ribosome have to do with frameshifting? Since error-free translation likely requires that the A- and P-site tRNAs maintain hydrogen bonding contact with the mRNA during both 50S and 30S translocation, at some point in an elongation cycle with the paused ribosome positioned over the slip-site, these interactions must be broken for frameshifting to occur (Fig. 5). The above studies suggest that the ribosome is most dynamic in the pre-30S (40S) translocation state, i.e., during 50S (60S) translocation or when the ribosome adopts the hybrid state (see Fig. 5). Indeed, a recent study suggests that the tRNA and mRNA interactions with the ribosome are weakened to the point where a change in reading frame, albeit in the 3′ direction, could readily occur provided the translocating tRNA lacks a peptidyl group and a degenerate codon for repairing by the deacylated P-site is available (McGarry et al., 2005). Another study showed that addition of EF-G·GDPNP further stabilizes the hybrid P/E state allowing for destabilization of the P-site codon–anticodon hydrogen bonding interactions (Spiegel et al., 2007). This is interesting since some tRNAs are known to favor the hybrid state of the ribosome more than others (Cornish et al., 2008, Dorner et al., 2006, Spiegel et al., 2007, Studer et al., 2003); this suggests that different tRNAs may have different propensities to frameshift based on their propensity to form the hybrid conformational state.
The sequence of the heptanucleotide slip-site in a −1 PRF signal is such that during repairing of the P- and A-site tRNAs in the new −1 reading frame, only the wobble codon–anticodon interaction is changed (“0” frame: X XXY YYZ to “−1” frame: XXX YYY Z) (see Fig. 2B) (Brierley et al., 1992). As a result, the new −1 reading frame provides near cognate repairing partners for the bound tRNAs. This suggests that the total free energy of codon–anticodon formation of the −1 frame will be comparable to that of the reference frame. Since frameshifting does in fact occur over such a sequence, but happens infrequently, a sizable transition state energy barrier to shifting reading frames must be present (see Fig. 4A). A downstream secondary structural element could therefore function by lowering this energy barrier, either by playing an active (mechanical) or passive role in this process.
A mechanical model of frameshift stimulation hypothesizes that the downstream pseudoknot functions as a kinetic barrier to normal translocation, and in so doing, lowers the energy barrier to tRNA–mRNA repairing. It can be imagined that this kinetic barrier would come into play either during spontaneous hybrid states formation (Cornish et al., 2008) (50S translocation) and/or during EF-G·GTP driven translocation (30S translocation) (see Fig. 5) (Namy et al., 2006). In both cases, movement of the ribosomal subunits may cause tension to build up in the mRNA strand due to the downstream structural element positioned in the mRNA entry channel. This tension could then be released via −1 PRF (Plant et al., 2003). Consistent with these models, prior studies have demonstrated that tRNAs can translocate on the ribosome in the absence of an mRNA (Belitsina et al., 1981, Belitsina et al., 1982). Alternatively, the downstream RNA element may function simply as a passive barrier, by pausing the ribosome over the slippery sequence for a time sufficient for repairing to occur, as the ribosomal helicase attempts to unwind whatever secondary structure is present in the mRNA entry channel (Takyar et al., 2005). However, such a model would appear to be inconsistent with biochemical experiments that suggested that pausing is necessary (Lopinski et al., 2000) but not sufficient to induce −1 PRF (Kontos et al., 2001, Tu et al., 1992). It is also plausible that the paused ribosome may actually be in equilibrium between the XXY YYZ and XXX YYY reading frame states while in the hybrid state (see Fig. 5). The binding of EF-G·GTP to the ribosome would then trap the ribosome in one of the two reading frames prior to 30S (40S) translocation and a return to the classical state. In this context, it would be of interest to investigate the influence of a downstream structural element(s) on hybrid states formation by the translating ribosome. In any event, these studies taken collectively make it most likely that tRNA–mRNA codon–anticodon interactions are broken when the ribosome is in the hybrid conformational state.
6. A structural model for a mechanical basis of frameshift stimulation by a pseudoknot
Brierley and co-workers have recently used cryo-electron microscopy to image mammalian 80S ribosomes (to ≈16 Å) paused over the infectious bronchitis virus (IBV) pp1a/pp1b pseudoknot frameshift signal (Fig. 6A) (Namy et al., 2006). Although solved at low resolution, these reconstructions reveal several significant features. Firstly, as expected (Yusupova et al., 2001), the pseudoknot lies at the entrance to the mRNA channel apparently making direct contact with elements of the putative ribosomal helicase, including rpos3 (bacterial S3), 16S helix 16, rpS9 (S4) and rpS2 (S5), as well as the ribosome regulatory protein RACK1 (Fig. 6B). More importantly, this structure contains eEF2 trapped in the A-site, with the D-helix of the P-site tRNA strongly bent, relative to control reconstructions with a non-frameshift-stimulating stem–loop RNA. Thus, the movement of the tRNA through the ribosome during translocation appears to be prevented by the inability of the ribosome to unwind the pseudoknot, and blockage of the A-site mRNA codon by eEF2. The P-site tRNA strongly bends toward the 3′ direction (or A-site codon) (Fig. 6B) more so than in the structures above (Namy et al., 2006, Selmer et al., 2006) thought to be due to the opposing forces of translocation and the pseudoknot plug. At some frequency, the P-site tRNA unpairs and repairs in the new −1 frame (in the 5′ direction), a proposal consistent with a major role of specifically the P-site tRNA in this process (Baranov et al., 2004). After this time, the tension built up in the mRNA is relaxed, and the incoming aminoacylated tRNA is delivered to the A-site in the new −1 reading frame (Fig. 6C).
Fig. 6.
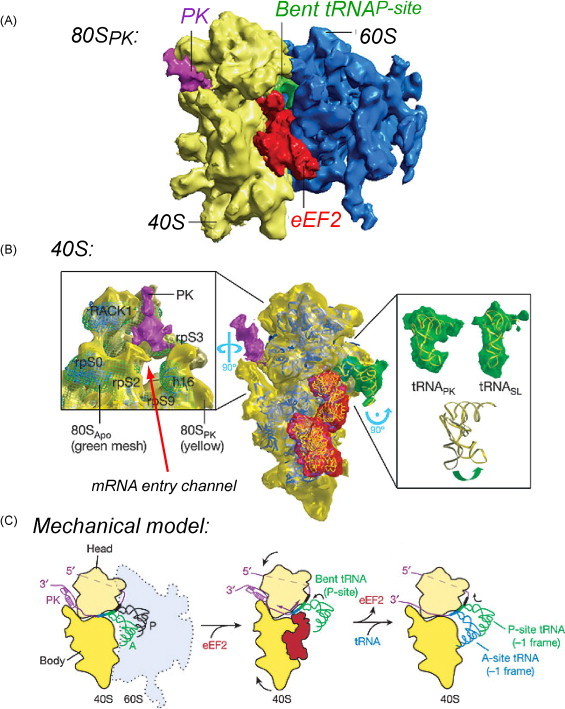
Cryo-electron microscopy (16 Å resolution) of mammalian 80S ribosomes paused at the IBV frameshift signal (Namy et al., 2006). (A) Overall structure of the complex with the large (60S) and small (40S) ribosomal subunits indicated as are the P-site tRNA (green), eukaryotic elongation factor-2 (eEF2; red) and the pseudoknot (PK, purple). (B) Close-up of the 40S subunit focusing on the mRNA entry channel and the electron density for the P-site tRNA in the pseudoknot complex vs. that bound to a non-frameshift-stimulating stem–loop RNA. Note the clear differences in the position of the P-site tRNA (see text for details). (C) A cartoon model of a mechanical model for −1 PRF, in which slippage of the P-site tRNA occurs during translocation mediated by eEF2-GTP hydrolysis. Reproduced with permission from Namy et al. (2006) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.).
This structure would seem to pinpoint the 30S (40S) translocation step in the elongation cycle (see Fig. 5) where ribosomes shift into the −1 reading frame; however, the low resolution of the structure makes this difficult to claim with certainty. Since both eEF2 and a P-site tRNA are bound, this suggests that the ribosome is in the hybrid conformation state, a state that would be favored with a deacylated tRNA in the P-site (Spiegel et al., 2007). At 16 Å resolution, however, it is not clear if the P-site tRNA observed in the structure is deacylated, nor can it be determined what reading frame the mRNA is actually in. On the other hand, the clearly defined electron density observed for the pseudoknot strongly suggests that the pseudoknot is largely folded as the ribosome shifts reading frames. The hypothesis that emerges from this work is that the ability of the downstream pseudoknot to actively lower the energy barrier for unpairing of the P-site codon–anticodon interaction will be more strongly correlated with frameshift stimulation. By extension, RNA motifs more capable of resisting the force of ribosomal helicase-mediated unwinding and eEF2 (EF-G)-catalyzed translocation will thus be more efficient frameshift stimulators. The structural data summarized here further suggests that the helical junction in at least one class of frameshifting pseudoknots is a “hot-spot” or positive determinant for frameshift stimulation (Cornish et al., 2005).
7. Structural studies of RNA motifs that stimulate −1 PRF
A strikingly diverse array of RNA motifs are capable of stimulating −1 PRF when placed ≈6–8 nucleotides downstream of the heptanucleotide slippery sequence or “slip-site” (see Fig. 7 ). The known structures, schematically illustrated as secondary structure diagrams in Fig. 7, can be divided roughly into two groups. The first group is comprised of standard hairpin-type pseudoknots that conform to the overall architecture illustrated in Fig. 1. Pseudoknotted motifs that stimulate −1 PRF can thus far be further subdivided into three distinct structural classes and include the coronaviral (IBV-type), retroviral gag-pro and luteoviral P1–P2 frameshift stimulators (Giedroc et al., 2000). These differ primarily in the distinct functional requirements of the stem and loop lengths within these signals, to be discussed in more detail below.
Fig. 7.
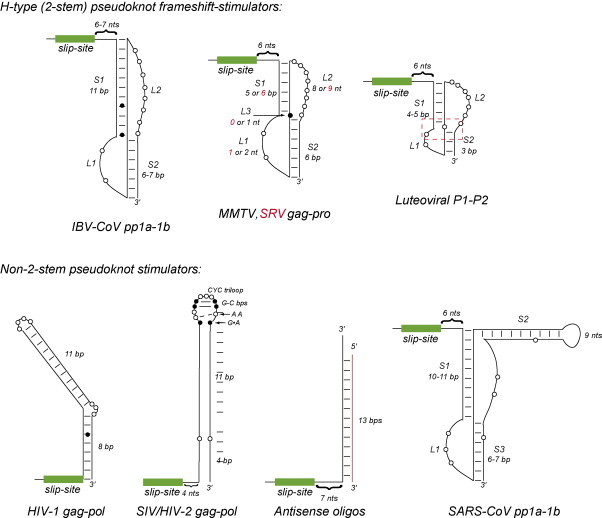
Secondary structural representations of structurally or functionally characterized frameshifting elements. Upper panel: Three distinct structural classes of canonical two-stem containing hairpin (H)-type pseudoknots, with the luteoviral P1–P2 helical junction region boxed. Lower panel: Frameshifting elements that do not conform to the standard H-type pseudoknot paradigm.
The second group contains all other motifs, including the very stable stem–loop structures known to stimulate −1 frameshifting at the gag-pol junctions in HIV-1 and related lentiviruses (Marcheschi et al., 2007, Staple and Butcher, 2005b), as well as the three-stemmed pseudoknot of the frameshift signal positioned between ORFs pp1a and pp1b of the replicase polyprotein (pp) precursor in the SARS coronavirus genome (Baranov et al., 2005, Plant et al., 2005). Note that this latter motif can be thought of an H-type pseudoknot that contains an additional stem–loop embedded in the standard loop L2. Although, a structure of the SARS-CoV signal is not yet available, many of the proposed base pairs predicted to form in each of the three helical stems have been confirmed by NMR methods (Plant et al., 2005). Disruption or deletion of stem S3 has only a small influence on frameshift stimulation in vitro (Plant et al., 2005); this finding is consistent with the fact that this helix can also be deleted from the well-studied pp1a–pp1b pseudoknot from the distantly related group 3 coronavirus, avian infectious bronchitis virus (IBV) (Brierley et al., 1989, Napthine et al., 1999). Interestingly, the P1–P2 pseudoknot from the luteovirus barley yellow dwarf virus (BYDV) is a variation on the SARS-CoV pseudoknot structure, but contains a loop L2 of nearly 4 kb, with S2 formed by long-range base pairing with nucleotides near the 3′ end of the genome (Paul et al., 2001); this provides an elegant mechanism to coordinate translation of the replicase gene and negative strand RNA synthesis (Barry and Miller, 2002). Finally, a number of antisense nucleic acids have also been shown to stimulate ribosomal frameshifting in vitro (Howard et al., 2004, Olsthoorn et al., 2004). Although these duplexes bear some resemblance to the simple stem–loops known to stimulate −1 PRF in lentiviruses (see Fig. 7), their mechanism of action is not known.
7.1. Coronaviral and retroviral gag-pro frameshifting signals
Although early investigations of the frameshifting signal from the retrovirus Rous sarcoma virus (RSV) clearly established −1 PRF as an event programmed by the mRNA sequence and required to express the downstream pol or pro ORF gene products in retroviruses (Jacks et al., 1988, Jacks and Varmus, 1985), the mouse mammary tumor virus (MMTV) gag-pro frameshift-stimulating pseudoknot was the first such −1 PRF stimulatory RNA element investigated in considerable detail as to its solution structure and functional requirements (Chamorro et al., 1992, Chen et al., 1995, Chen et al., 1996, Jacks et al., 1987, Shen and Tinoco, 1995). As such, the MMTV pseudoknot has long provided a basis of comparison with all −1 PRF-stimulating RNA motifs subsequently discovered and characterized (for a review, see Giedroc et al., 2000). These include the coronaviral pp1a–pp1b signals characterized first in avian infectious bronchitis virus (IBV) nearly coincidently with the RSV and MMTV gag-pro signals (Brierley et al., 1987, Brierley et al., 1989, Brierley et al., 1991), followed by other structurally characterized retroviral gag-pol and gag-pro signals, most notably that from HIV-1 (Parkin et al., 1992) and the simian retrovirus-1, SRV-1 (Du et al., 1997, Michiels et al., 2001, ten Dam et al., 1994, ten Dam et al., 1995). The pseudoknot derived from the P1–P2 junction from beet western yellows virus (BWYV) (Egli et al., 2002, Kim et al., 1999, Su et al., 1999) from a family of viruses collectively called Luteoviridae, followed shortly thereafter, with structures of three additional luteoviral pseudoknots solved using either NMR (Cornish et al., 2005, Nixon et al., 2002b) or crystallographic (Pallan et al., 2005) methods.
Although an atomic structure of the minimal IBV pp1a/pp1b pseudoknot remains unavailable 20 years after its discovery (Brierley et al., 1987), the major functional requirements of the IBV frameshifting signal relative to the retroviral-type gag-pro pseudoknot from MMTV could be deduced from mutational studies (Chen et al., 1995, Liphardt et al., 1999, Napthine et al., 1999). The major difference was in the length of the stem S1 (Fig. 7), with 11 bp, or one turn of A-form helix, clearly optimal for the IBV pseudoknot vs. 5–6 bp in the MMTV gag-pro pseudoknot. Remarkably, a 10-bp stem S1 induces a dramatic ≈7-fold decrease in frameshift stimulation (46–7%) in the IBV pseudoknot, while shorter S1 stems are effectively inactive in promoting frameshifting in this context (Napthine et al., 1999). Structural features specific to the MMTV pseudoknot include an unpaired adenosine (A14) that is wedged between the two stems (pseudoknot loop L3; see Fig. 1, Fig. 8 ) thereby creating a bent structure that prevents coaxial stacking of the helical stems, as well as a number of loop L2–stem S1 interactions (Shen and Tinoco, 1995). The presence of this “wedge” adenosine is strongly positively correlated with frameshift stimulation since deletion of the nucleotide reduces frameshifting efficiencies to low levels (Shen and Tinoco, 1995). Atkins and co-workers have made a similar finding in a retroviral-type frameshifter found in the human gene Ma3 (Wills et al., 2006). Thermodynamic studies reveal that the unpaired adenosine is globally stabilizing in the gag-pro pseudoknots from MMTV and mouse intercisternal A-type particle (mIAP), but by just ≈0.5 kcal mol−1 (37 °C) (Theimer and Giedroc, 1999, Theimer and Giedroc, 2000).
Fig. 8.
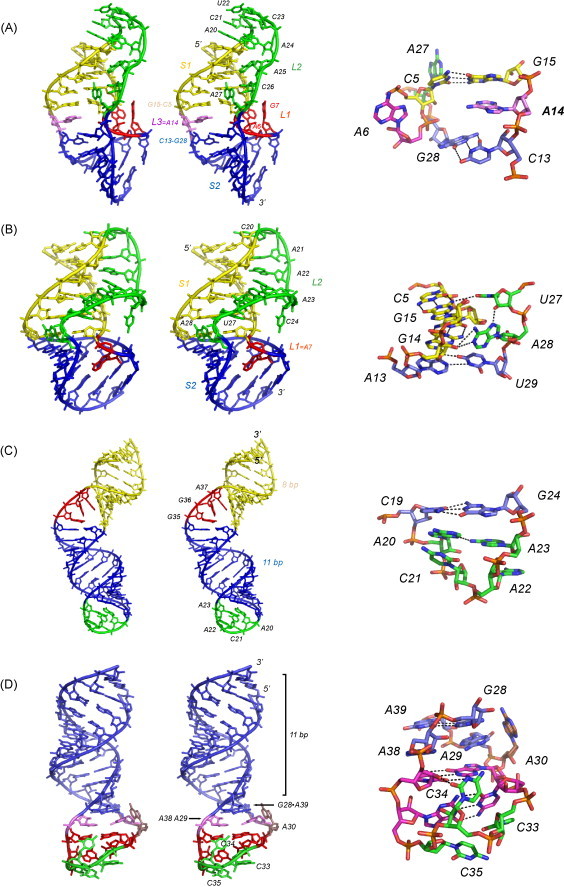
Stereo views of non-luteoviral P1–P2 frameshifting signals of known three-dimensional structure. (A) MMTV gag-pro pseudoknot vpk (1RNK) (Shen and Tinoco, 1995); (B) SRV–1 gag-pro pseudoknot (1E95) (Michiels et al., 2001); (C) HIV-1 gag-pol stem–loop (1Z2J) (Staple and Butcher, 2005b); (D) SIV gag-pol stem–loop (2JTP) (Marcheschi et al., 2007). Close-up views of the helical junction regions for the MMTV and SRV-1 gag-pro pseudoknots and the well-ordered hairpin loops for HIV-1 and SIV gag-pol signals are also shown (right). For the pseudoknots, S1 is shaded yellow, S2 is blue, L1 is red, L2 is green and L3 is purple (see also Fig. 9, Fig. 10). Note that these structures are not drawn to scale (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.).
In a series of mutational experiments, Liphardt et al. (1999) was able to convert a functionally inactive IBV pseudoknot with 6 bps in stem S1 and 8 nucleotides in loop L2, by simply adding an unpaired and presumably intercalated adenosine at the helical junction, and an adenosine in the 3′ terminal position of loop L2, i.e., features that mimic those found in the MMTV gag-pro pseudoknot. These findings argue strongly for two distinct structural classes of frameshifting pseudoknots, one with a full helical turn of stem S1 and few other obvious structural requirements, and one with a much shorter stem S1 that seemed to require favorable interactions at the helical junction region to mediate high levels of frameshift stimulation. Subsequent structural studies with the gag-pro signal from SRV-1 further suggested that it was not the bent conformation of the retroviral gag-pro pseudoknot that was positively correlated with frameshifting since this structure is characterized by coaxially stacked S1 and S2 helices, but rather favorable loop L2–stem S1 interactions, a subset of which are found in the helical junction region (see Fig. 8B) (Michiels et al., 2001).
7.2. Lentiviral gag-pol frameshift signals
In contrast to the pseudoknotted motifs discussed above, it is now well-established that the frameshift stimulators found at the gag-pol junctions in lentiviruses HIV-1 and SIV are simple RNA stem–loops (see Fig. 8C and D) (Gaudin et al., 2005, Marcheschi et al., 2007, Staple and Butcher, 2005b). Each is characterized by an 11-bp helical stem and a highly ordered hairpin loop. The HIV-1 stem–loop contains an ACAA tetraloop characterized by an A20–A23 hydrogen bond, while the 12-nucleotide SIV/HIV-2 loop incorporates a sheared G28-A39 base pair, a cross-strand adenosine stack formed by A29 and A38, two G-C base pairs, and a novel CYC triloop turn sequence (Fig. 8D). Each loop is characterized by multiple hydrogen bonding interactions that stabilize the overall structure (see Fig. 8C and D) (Marcheschi et al., 2007, Staple and Butcher, 2005b) and as such, are predicted to preclude base pairing with the adjacent 3′ region that would be required to form a pseudoknot, as proposed previously for HIV-1 (Dinman et al., 2002). Both the HIV-1 and SIV signals also contain a lower, more dynamic stem (shaded yellow in the HIV-1 RNA, Fig. 8C) that when base paired comes closer to the 3′ edge of the slippery sequence than do the downstream pseudoknots in other systems (see Fig. 7) (Marcheschi et al., 2007); in HIV-1, these two helical stems are interrupted by an asymmetric three-nucleotide G35-G36-A37 bulge (Fig. 8C). Since the lower stem in both HIV-1 and SIV RNAs will be unfolded when the ribosome is positioned over the slippery signal, its function is unclear. In HIV-1, it has been hypothesized that the lower stem functions as a “positioning element” that allows the stem–loop to induce ribosomal pausing which in turn mediates a translocation perturbation; how this occurs at the molecular level is unknown (Staple and Butcher, 2005b). It is also intriguing that both HIV-1 and SIV/HIV-2 stem–loop structures, like stem S1 in the IBV pp1a–pp1b pseudoknot, are 11 bp in length, perhaps suggesting a similar mechanism of frameshift stimulation, but potentially enhanced by a very slippery UUUUUUA slip-site (Brierley et al., 1992).
7.3. Plant luteoviral P1–P2 frameshifting signals
The atomic resolution (1.6 Å) crystallographic structure of the 28-nucleotide P1–P2 pseudoknot from beet western yellows virus (BWYV) clearly established how RNA pseudoknots with a very short 3-bp pseudoknot-forming stem S2 could fold and stimulate −1 PRF to a level of 5–15% (Su et al., 1999) (Fig. 9A). This RNA is compact and adopts a largely triple-stranded conformation, with many of the loop nucleotides making non-canonical base–base and base–sugar edge hydrogen bonds with base pairs in each of the two stems. For example, a protonated C8+ from loop L1 forms a standard Hoogsteen-type base pair with an accepting G12 as part of a C8+·(G12-C26) base triple positioned at the “top” of the S2 helical junction (Fig. 9A). Thermodynamic and structural studies of three other luteoviral RNA pseudoknots confirm that this C+·(C-G) major groove triple base pair is a common feature of all luteoviral RNA pseudoknots (see Fig. 9B–D) (Nixon et al., 2002a, Nixon and Giedroc, 2000), protonation of which is strongly stabilizing. The degree to which protonation of this cytidine affects frameshifting efficiency or mechanical stability (Tinoco et al., 2006) remains unknown.
Fig. 9.
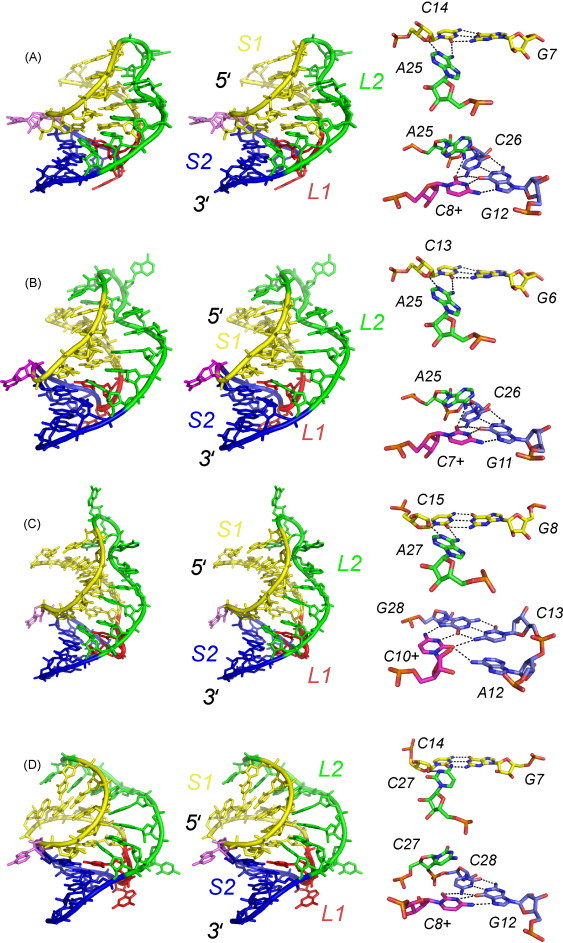
Stereo views of the structures of four related luteoviral P1–P2 pseudoknots. (A) BWYV (PDB 1L2X solved to 1.25 Å resolution) (Egli et al., 2002); (B) PLRV (PDB 2A43 solved to 1.34 Å resolution) (Pallan et al., 2005); (C) PEMV-1 (PDB 2RP1) (this work); (D) ScYLV (PDB 1YG4) (Cornish et al., 2005). Close-ups of the helical junction regions of all four RNAs are shown to the right, with the L2–S1 minor groove base triple highlighted at the top, and the L1–S2 major groove base quadruple shown at the bottom. Nucleotide shading is the same as above.
The other striking feature of the BWYV pseudoknot is a minor groove triplex, in which a run of three consecutive adenosines at the 3′ end of the loop L2 form a series of Watson–Crick-sugar edge hydrogen bonding interactions with lower base pairs of the upper stem S1 (see Fig. 9A) (Su et al., 1999). While reminiscent of classical A-minor interactions found in other complex RNAs and the large ribosomal subunit (Doherty et al., 2001, Nissen et al., 2001), these interactions are characterized by direct adenosine N1-2′OH hydrogen bonding interactions, many of which could be directly detected in solution using NMR methods on related luteoviral pseudoknots (Cornish et al., 2006a, Cornish et al., 2005, Giedroc et al., 2003, Nixon et al., 2002b) (see below). Other distinct L2–S1 hydrogen bonding interactions are found further from the helical junction in all luteoviral RNAs but these tend to be somewhat unique to individual RNAs.
Fig. 9B–D compares the structures of three other luteoviral P1–P2 frameshifting pseudoknots to that from BWYV (Fig. 9A) (Egli et al., 2002, Su et al., 1999). These include a solution structure of the frameshifting signal from RNA-1 of the enamovirus pea enation mosaic virus (PEMV-1) originally reported in 2002 (Nixon et al., 2002b) with the refined structure presented here (see Table 1 for refinement statistics), a crystallographic structure of the “minimal” 22-nucleotide pseudoknot from potato leaf roll virus (PLRV) (Pallan et al., 2005) and a solution structure of the frameshifting motif from sugar cane yellow leaf virus (ScYLV) (Cornish et al., 2005). Fig. 10A shows full and backbone ribbon representations of a global superposition of the 28 lowest energy structures of the PEMV-1 RNA, while Fig. 10B and C highlight the unique helical junction region of this pseudoknot. Note that both Watson–Crick and Hoogsteen faces of the 3′ nucleotide of loop L2, A27, are tied up in hydrogen bonding (Giedroc et al., 2003, Nixon et al., 2002b), much like previously observed for A155 in the HCV IRES four-way junction (Kieft et al., 2002).
Table 1.
Experimental restraints and structure statistics for the refined PEMV-1 RNA pseudoknot structurea
| Number of structures | 28 |
| NOE distance restraints (22/residue) | 704 |
| Intranucleotide | 464 |
| Internucleotide | 240 |
| Dihedral angle restraints | 151 |
| Residual dipolar couplings (1DCH) | 118 |
| Sugar | 78 |
| Base | 40 |
| RMS from experimental restraints | |
| Distance restraints (Å)b, c | 0.029 ± 0.002 |
| Dihedral restraints (°)b, c | n/a |
| Dipolar couplings (Hz) | 2.50 ± 0.03 |
| Deviations from idealized geometry | |
| Bonds (Å) | 0.007 ± 0.001 |
| Angle (°) | 1.57 ± 0.02 |
| Impropers (°) | 0.93 ± 0.02 |
| Heavy-atom RMSD (Å) | |
| Overall (residues 4–30, excluding 14 and 20) | 1.18 ± 0.14 |
| S1 | 1.10 ± 0.18 |
| S2 | 0.86 ± 0.16 |
Refinement of the previous lower resolution structure [PDB codes 1KPZ (average solution structure) and 1KPY (bundle)] through analysis of a 3D 13C-separated NOESY spectrum acquired at 900 Hz 1H frequency (τm = 100 ms) and 2D 1H–1H NOESY spectra (600 MHz; τm = 60 and 280 ms) with the resulting bundle of 28 lowest energy structures refined with 118 sugar and base 1DCH residual dipolar coupling restraints obtained from 2D J-modulated CT-HSQC (Ottiger et al., 1998) and CT-TROSY (Boisbouvier et al., 2000) spectra using a three-step simulated annealing protocol essentially identical to that previously described (Cornish et al., 2005, McCallum and Pardi, 2003). A global overlay of a bundle of 28 lowest energy structures and an average structure have been deposited in the RCSB under PDB accession codes 2RP0 and 2RP1, respectively.
No violations >0.5 Å.
No violations >5°.
Fig. 10.
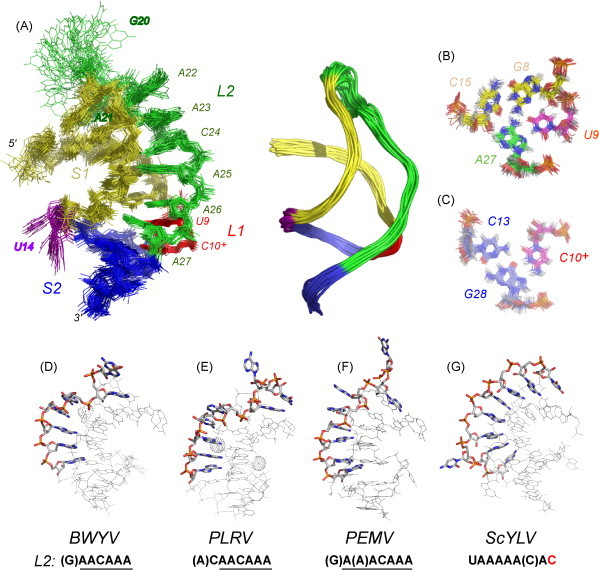
Solution structural characteristics of the refined PEMV-1 P1–P2 pseudoknot (PDB 2RP0). (A) Left: Global all atom superposition (residues 4–30) of 28 lowest energy models of the PEMV-1 pseudoknot (see Table 1 for structure statistics); right, ribbon representation of the same structure bundle color-coded as in Fig. 9. All-atom superposition above (B) and below (C) the helical junction of the PEMV-1 pseudoknot. Models of the BWYV (D), PLRV (E), PEMV (F) and ScYLV (G) pseudoknots emphasizing the structure of the minor-groove spanning L2 loop. The sequence (5′–3′) of L2 is also indicated below the figure.
Since the conformation of loop L2 in the PEMV-1 pseudoknot is now far better defined by the data (Fig. 10A), it is possible to conclude that the structure of the five most 3′ residues of loop L2 (5′-A23-C24-A25-A26-A27) is essentially identical to that found in the BWYV (Egli et al., 2002) and PLRV (Pallan et al., 2005) pseudoknots, each of which contains an identical 5′-ACAAA 3′ L2 sequence (Fig. 10D–F). Briefly, residues C24 through A27 all point their Watson–Crick edges into the S1 minor groove where they are engaged in numerous hydrogen-bonding interactions. In addition to A27 and A25 discussed above, the N1/N6 face of A26 forms two hydrogen bonds with the N3–N2 edge of G7. C24 also points into the S1 minor groove, with the N4 amino group close to the 2′-OH of G17. A23 is stacked on C24, with the Watson–Crick edge rotated out of the stack, leaving the N7 and N6 nitrogens within hydrogen bonding distance of the 2′-OH of G17. At this point, the PEMV-1 structure diverges from the BWYV/PLRV structures, with A22 stacked on A23, and essentially extruded from of the triple helix, with A21 inserted back into the S1 minor groove near the top two S1 base pairs. G20 is extrahelical. In fact, if one considers A22 an extrahelical insertion in the PEMV-1 loop L2, the entire 5′-A(A)ACAAA loop sequence adopts essentially identical conformations in all three pseudoknots (Fig. 10D–F).
The solution structure of the ScYLV P1-P2 pseudoknot is characterized by several unique features relative to the PEMV-1 and BWYV/PLRV RNAs (see Fig. 9) (Cornish et al., 2005). Notably, all of loop L2 is very well-ordered and exhibits continuous stacking of A20 through C27 into the minor groove of S1, with C25 flipped out of the triple-stranded stack. Five consecutive triple base pairs flank the helical junction where the 3′ nucleotide of L2, C27, adopts a cytidine 27 N3-cytidine 14 2′-OH hydrogen bonding interaction with the C14-G7 base pair (Cornish et al., 2006a). This interaction is isosteric with the adenosine N1-2′-OH interaction found in the BWYV (Su et al., 1999), PLRV (Pallan et al., 2005) and PEMV-1 RNAs (cf. Fig. 9); however, the ScYLV and BWYV mRNA structures differ in their detailed L2–S1 hydrogen bonding and L2 stacking interactions (Fig. 10D–G).
Given the isosteric nature of the C27 N3⋯C14 2′ OH (ScYLV) and A25 N1⋯C14 2′ OH (BWYV) hydrogen bonds in these two RNAs by NMR (Cornish et al., 2006a, Giedroc et al., 2003), the extent to which a BWYV-like adenosine in the ScYLV context could functionally substitute for the 3′ L2 cytidine (C27) was investigated. The unexpected finding from these experiments was that the C27A ScYLV RNA is a very poor frameshift stimulator in vitro (Cornish et al., 2005). This surprising finding, in turn, made the prediction that substitution of A25 in the BWYV RNA with cytidine (A25C) would increase frameshifting in this context; this is exactly what was found. Thus, a 3′ L2 cytidine is a positive determinant for frameshift stimulation by luteoviral RNAs (Cornish et al., 2006b). Strikingly, the global structure of the C27A ScYLV RNA is nearly indistinguishable from the wild-type counterpart, despite the fact that the helical junction region is altered and incorporates the anticipated isostructural A27·(G7-C14) minor groove base triple (Cornish et al., 2006b) in adopting a helical junction region that is superimposable with that of the beet western yellows virus (BWYV) pseudoknot (Su et al., 1999), as expected. These results suggest that the global “ground-state” structure is not strongly correlated with frameshift stimulation and point to a reduced stability that derives from an altered helical junction architecture in the C27A ScYLV RNA as of significant functional importance, for which there is some evidence (Cornish and Giedroc, 2006, Cornish et al., 2005).
Studies in other frameshifting systems have also uncovered some correlation between thermodynamic stability of the downstream element and frameshifting efficiency (Bidou et al., 1997, ten Dam et al., 1995), although it is not obvious why this has to be the case (Cao and Chen, 2008, Theimer and Giedroc, 2000). In fact, in luteoviral pseudoknots, insertion or deletion of an unpaired and extrahelical residue(s) near the “top” of loop L2 often strongly increases frameshifting efficiency, a finding often attributed to a direct interaction with the ribosome (Kim et al., 2000, Kim et al., 1999). We have argued that distinct unfolding thermodynamics measured for closely related RNAs, e.g., WT vs. C27A ScYLV RNAs, may in fact be reporting on different kinetics of pseudoknot unfolding, with increased rates of unfolding, i.e., unfolding at lower applied forces, negatively correlated with frameshift stimulation (Giedroc et al., 2000, Theimer and Giedroc, 2000). In other words, these studies suggest the hypothesis that the helical junction in luteoviral RNAs is mechanically stable and functions as a classic kinetic barrier (Onoa et al., 2003) to force-induced unfolding, essentially placing the unfolding of the entire pseudoknot under kinetic control. This barrier is predicted to be altered in functionally compromised RNAs. Mechanical force-induced unfolding/refolding experiments will be required to obtain direct evidence for this proposal (see below).
8. Mechanical unfolding studies with RNA pseudoknots
Given that the ground-state or lowest energy structure of a frameshifting pseudoknot may be a poor predictor of frameshifting efficiency (Cornish et al., 2006b), this has motivated efforts to identify other physical features of the RNA that might be more strongly positively correlated with setting frameshifting efficiency. Since the translocating ribosome places force on the downstream mRNA (Wen et al., 2008) while engaging in either active or passive unwinding of secondary structure, understanding the mechanical stability of pseudoknots toward force-induced unfolding over a force regime thought to be achievable by the ribosome is of considerable interest. Two groups have now investigated the extent to which various IBV pp1a–pp1b derived frameshifting pseudoknots that differ in their frameshifting efficiencies are unfolded as a function of force in an optical trapping or tweezers experiment (Green et al., 2008, Hansen et al., 2007). Here, the pseudoknot is attached to two microbeads via annealing to long ≈500 nucleotide ssDNA handles immobilized on the bead, with one bead held in an optical trap and the other held on the tip of a micropipette by suction (Liphardt et al., 2001). Both groups compared 11- and 10-bp S1-containing IBV pseudoknots which stimulate frameshifting to very different degrees (Napthine et al., 1999); loop L2 lengths were also different in the two studies. Note that these experiments investigated the intrinsic mechanical properties of the pseudoknot itself in isolation and in the absence of bound or stalled ribosomes.
In the more limited of the two studies, Hansen et al. (2007) measured simple repetitive stretching-relaxation unfolding–folding trajectories and showed that the RNA with the 11-bp stem S1 appear to require approximately two-fold greater unfolding force than the 10-bp construct, with the 10-bp construct more often than not unfolding in multiple steps relative to the 11-bp RNA. In contrast, Green et al. (2008) saw no clear correlation between frameshifting efficiency and the mechanical force required to unfold the RNAs; however, the work made several striking observations. Mg2+ strongly stabilizes the kinetic and thermodynamic stability of the “wild-type” IBV pseudoknot, with an unfolding transition state very close to the folded structure in the presence of Mg2+ (Green et al., 2008). This means that the pseudoknot is a “brittle” structure (Onoa et al., 2003) with a correspondingly very shallow dependence of the unfolding rate on applied force (measured at three different pulling rates) (Li et al., 2006a, Li et al., 2006b); the rate of unfolding is also much slower than component hairpin structures. Both features are in striking contrast to the component hairpins and the unfolding of the partially folded intermediates that lack pseudoknot tertiary pairing (Green et al., 2008). Thus, the pseudoknot tertiary structure is clearly characterized by unique mechanical properties relative to component RNA hairpins (Green et al., 2008). It will be interesting to determine the degree to which the tertiary structure of frameshifting pseudoknots from other structural classes (Cornish et al., 2005, Michiels et al., 2001) and the stem–loop structures from HIV-1 and SIV/HIV-2 gag-pol shift sites (Marcheschi et al., 2007, Staple and Butcher, 2005b) (see Fig. 7, Fig. 8, Fig. 9) form “brittle” structures distinct from compliant RNA hairpins (Li et al., 2006b).
Although these studies focus on the unfolding rate as critical for frameshift stimulation by individual ribosome-pseudoknot encounters, the number of translocating ribosomes on a single mRNA molecule might also have a detectable influence on ensemble average frameshifting efficiency (Lopinski et al., 2000). In this model, the rate of refolding the downstream stimulatory element after unfolding and decoding by the first ribosome may have a significant impact on overall frameshift efficiency because a lagging ribosome(s) may encounter a stimulatory element that remains unfolded. This model predicts that the rate of translation initiation, or loading of successive ribosomes, on a mRNA containing a frameshift signal might have a detectable influence on overall frameshifting efficiency. Recent support for this idea has recently been published for HIV-1-infected cells in culture (Gendron et al., 2008), in which the presence of a low concentration of the upstream transactivation response element (TAR) RNA, known to be present in the cytoplasm of virus-infected cells as a free RNA, activates protein kinase R (PKR) which in turn phosphorylates the translation initiation factor eIF2α, effectively halting translation initiation (Sadler and Williams, 2007). Although it is not known precisely how HIV-1 manipulates translation initiation to appropriately express the genome, this suggests that frameshifting efficiency may well be determined by the number of ribosomes engaged in translation elongation on a single mRNA.
9. Future prospective
Future experiments capable of investigating the high resolution structure (Beardsley et al., 2006, Namy et al., 2006, Selmer et al., 2006) and/or real-time conformational dynamics (Blanchard et al., 2004, Cornish et al., 2008, Lee et al., 2007, Munro et al., 2007) of single ribosome-mRNA–tRNA complexes paused over a frameshifting signal hold considerable promise for enhancing our mechanistic understanding of how programmed ribosomal recoding events function (for a recent review on single-molecule methods as applied to translation, see Munro et al., 2008). For example, a recent study reports the use of optical trapping methods to observe single ribosomes translocating through an RNA stem–loop structure via continuous measurement of the end-to-end length of the mRNA as it is unwound by the ribosome (Wen et al., 2008). The most striking observation from these studies, among many, is that while the ribosome moves in discrete translocation steps of three nucleotides or one codon over 0.1 s on average, this step-wise movement is punctuated by pauses of a very wide time duration (seconds to minutes) (Wen et al., 2008). Thus, under these conditions, ribosomal pausing is rate-determining for protein synthesis; this in turn, creates a mechanistic scenario that can be directly exploited by recoding signals embedded in the mRNA that are dependent on ribosomal pausing, like −1 PRF (Blanchard, 2008). Indeed, of the two rate-determining steps for pausing, one step that preceded translocation was force-dependent (Wen et al., 2008); it seems possible that this step might be reporting on the action of the ribosomal helicase as it attempts to unwind the stem–loop. In any case, this finding provides support for the hypothesis that mRNA tension implicit in any mechanical model of ribosomal frameshifting might strongly influence ribosomal movement in a functionally relevant way.
Future single-molecule studies that employ more natural sequence mRNAs that are capable of forming frameshift-stimulating RNA hairpins or RNA pseudoknots like those discussed here to directly measure translation, ribosome movement or conformational switching of ribosomal subunits in real time (Cornish et al., 2008) will shed dramatic new light on how downstream RNA elements mediate programmed ribosomal frameshifting. Recent developments of ultrasensitive dual optical trap instrumentation (Greenleaf et al., 2008) as well as other instrumentation that combines single-molecule fluorescence with force manipulation in a single experiment (Hohng et al., 2007) will permit the investigation of more subtle structural changes in essentially every phase of translation, thereby shedding direct insight on the ribosome as a motor protein (Myong et al., 2007), much like the seminal work on bacterial RNA polymerase has provided for our understanding of mechanics of transcription (Abbondanzieri et al., 2005) and transcriptional regulation by riboswitches (Greenleaf et al., 2008).
Acknowledgements
We thank Dr. Mirko Hennig of the Medical University of South Carolina for help in acquiring some of the NMR experiments on the PEMV-1 pseudoknot. P.V.C. acknowledges support by a postdoctoral fellowship (PF-07-123-01-GMC) from the American Cancer Society in Professor Taekjip Ha's laboratory. D.P.G. gratefully acknowledges the US National Institutes of Health (AI040184 and AI067416) for financial support of this and related projects on mechanisms of viral replication in his laboratory.
References
- Abbondanzieri E.A., Greenleaf W.J., Shaevitz J.W., Landick R., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland R.F., Atkins J.F. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J.K., Miller W.A. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardsley R.L., Running W.E., Reilly J.P. Probing the structure of the Caulobacter crescentus ribosome with chemical labeling and mass spectrometry. J. Proteome Res. 2006;5:2935–2946. doi: 10.1021/pr060170w. [DOI] [PubMed] [Google Scholar]
- Belitsina N.V., Tnalina G.Z., Spirin A.S. Template-free ribosomal synthesis of polylysine from lysyl-tRNA. FEBS Letters. 1981;131:289–292. doi: 10.1016/0014-5793(81)80387-0. [DOI] [PubMed] [Google Scholar]
- Belitsina N.V., Tnalina G.Z., Spirin A.S. Template-free ribosomal synthesis of polypeptides from aminoacyl-tRNAs. Biosystems. 1982;15:233–241. doi: 10.1016/0303-2647(82)90008-9. [DOI] [PubMed] [Google Scholar]
- Bidou L., Stahl G., Grima B., Liu H., Cassan M., Rousset J.P. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem–loop stimulatory signal. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- Biswas P., Jiang X., Pacchia A.L., Dougherty J.P., Peltz S.W. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard S.C. Breaking the barriers of translation. Nat. Chem. Biol. 2008;4:275–276. doi: 10.1038/nchembio0508-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard S.C., Kim H.D., Gonzalez R.L., Jr., Puglisi J.D., Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisbouvier J., Brutscher B., Pardi A., Marion D., Simorre J.P. NMR determination of sugar puckers in nucleic acids from CSA-dipolar cross-correlated relaxation. J. Am. Chem. Soc. 2000;122:6779–6780. [Google Scholar]
- Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76(Pt 8):1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Dos Ramos F.J. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Pennell S., Gilbert R.J. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen D.E., Clemons W.M.J., Carter A.P., Wimberly B.T., Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Cao S., Chen S.J. Predicting ribosomal frameshifting efficiency. Phys. Biol. 2008;5:16002. doi: 10.1088/1478-3975/5/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H.E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr., Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Kang H., Shen L.X., Chamorro M., Varmus H.E., Tinoco I., Jr. A characteristic bent conformation of RNA pseudoknots promotes -1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Clark M.B., Janicke M., Gottesbuhren U., Kleffmann T., Legge M., Poole E.S., Tate W.P. Mammalian gene PEG10 expresses two reading frames by high efficiency −1 frameshifting in embryonic-associated tissues. J. Biol. Chem. 2007;282:37359–37369. doi: 10.1074/jbc.M705676200. [DOI] [PubMed] [Google Scholar]
- Cornish P.V., Ermolenko D.N., Noller H.F., Ha T.J. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P.V., Giedroc D.P. Pairwise coupling analysis of helical junction hydrogen bonding interactions in luteoviral RNA pseudoknots. Biochemistry. 2006;45:11162–11171. doi: 10.1021/bi060430n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P.V., Giedroc D.P., Hennig M. Dissecting non-canonical interactions in frameshift-stimulating mRNA pseudoknots. J. Biomol. NMR. 2006;35:209–223. doi: 10.1007/s10858-006-9033-x. [DOI] [PubMed] [Google Scholar]
- Cornish P.V., Hennig M., Giedroc D.P. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P.V., Stammler S.N., Giedroc D.P. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann C.E., 3rd, Wakeman C.A., Sieling C.L., Baker S.C., Irnov I., Winkler W.C. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Deckman I.C., Draper D.E., Thomas M.S. S4-alpha mRNA translation repression complex. I. Thermodynamics of formation. J. Mol. Biol. 1987;196:313–322. doi: 10.1016/0022-2836(87)90692-9. [DOI] [PubMed] [Google Scholar]
- Dinman J.D., Icho T., Wickner R.B. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U.S.A. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D., Richter S., Plant E.P., Taylor R.C., Hammell A.B., Rana T.M. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5331–5336. doi: 10.1073/pnas.082102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinos G., Kalpaxis D.L., Wilson D.N., Nierhaus K.H. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty E.A., Batey R.T., Masquida B., Doudna J.A. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–433. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Dorner S., Brunelle J.L., Sharma D., Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Giedroc D.P., Hoffman D.W. Structure of the autoregulatory pseudoknot within the gene 32 messenger RNA of bacteriophages T2 and T6: a model for a possible family of structurally related RNA pseudoknots. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- Du Z., Holland J.A., Hansen M.R., Giedroc D.P., Hoffman D.W. Base-pairings within the RNA pseudoknot associated with the simian retrovirus-1 gag-pro frameshift site. J. Mol. Biol. 1997;270:464–470. doi: 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- Dulude D., Berchiche Y.A., Gendron K., Brakier-Gingras L., Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345:127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Dulude D., Theberge-Julien G., Brakier-Gingras L., Heveker N. Selection of peptides interfering with a ribosomal frameshift in the human immunodeficiency virus type 1. RNA. 2008;14:981–991. doi: 10.1261/rna.887008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M., Minasov G., Su L., Rich A. Metal ions and flexibility in a viral RNA pseudoknot at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4302–4307. doi: 10.1073/pnas.062055599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko D.N., Majumdar Z.K., Hickerson R.P., Spiegel P.C., Clegg R.M., Noller H.F. Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Annu. Rev. Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- Feinberg J.S., Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Gao H., Sengupta J., Valle M., Korostelev A., Eswar N., Stagg S.M., Van Roey P., Agrawal R.K., Harvey S.C., Sali A., Chapman M.S., Frank J. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- Gaudin C., Mazauric M.H., Traikia M., Guittet E., Yoshizawa S., Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- Gendron K., Charbonneau J., Dulude D., Heveker N., Ferbeyre G., Brakier-Gingras L. The presence of the TAR RNA structure alters the programmed −1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36:30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron K., Dulude D., Lemay G., Ferbeyre G., Brakier-Gingras L. The virion-associated Gag-Pol is decreased in chimeric Moloney murine leukemia viruses in which the readthrough region is replaced by the frameshift region of the human immunodeficiency virus type 1. Virology. 2005;334:342–352. doi: 10.1016/j.virol.2005.01.044. [DOI] [PubMed] [Google Scholar]
- Giedroc D.P., Cornish P.V., Hennig M. Detection of scalar couplings involving 2′-hydroxyl protons across hydrogen bonds in a frameshifting mRNA pseudoknot. J. Am. Chem. Soc. 2003;125:4676–4677. doi: 10.1021/ja029286t. [DOI] [PubMed] [Google Scholar]
- Giedroc D.P., Theimer C.A., Nixon P.L. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.D., Rambo R.P., Van Tyne D., Batey R.T. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Golden B.L., Kim H., Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- Green L., Kim C.H., Bustamante C., Tinoco I., Jr. Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf W.J., Frieda K.L., Foster D.A., Woodside M.T., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haebel P.W., Gutmann S., Ban N. Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr. Opin. Struct. Biol. 2004;14:58–65. doi: 10.1016/j.sbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Hansen T.M., Reihani S.N., Oddershede L.B., Sorensen M.A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger J.W., Meskauskas A., Dinman J.D. An ‘integrated model’ of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- Hohng S., Zhou R., Nahas M.K., Yu J., Schulten K., Lilley D.M., Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S.R. RNA structure: the long and the short of it. Curr. Opin. Struct. Biol. 2005;15:302–308. doi: 10.1016/j.sbi.2005.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J.A., Hansen M.R., Du Z., Hoffman D.W. An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. RNA. 1999;5:257–271. doi: 10.1017/s1355838299981360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.T., Aggarwal G., Anderson C.B., Khatri S., Flanigan K.M., Atkins J.F. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.T., Gesteland R.F., Atkins J.F. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H.E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl. Acad. Sci. U.S.A. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H.E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Jenner L., Rees B., Yusupov M., Yusupova G. Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep. 2007;8:846–850. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L., Romby P., Rees B., Schulze-Briese C., Springer M., Ehresmann C., Ehresmann B., Moras D., Yusupova G., Yusupov M. Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science. 2005;308:120–123. doi: 10.1126/science.1105639. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., O’Donnell M., Kuriyan J. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- Joseph S. After the ribosome structure: how does translocation work? RNA. 2003;9:160–164. doi: 10.1261/rna.2163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J.S., Zhou K., Grech A., Jubin R., Doudna J.A. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Maas S., Wang S.C., Rich A. Mutational study reveals that tertiary interactions are conserved in ribosomal frameshifting pseudoknots of two luteoviruses. RNA. 2000;6:1157–1165. doi: 10.1017/s1355838200000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G., Su L., Maas S., O’Neill A., Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk M.H., van der Graaf M., Wijmenga S.S., Pleij C.W., Heus H.A., Hilbers C.W. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- Kontos H., Napthine S., Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A., Trakhanov S., Laurberg M., Noller H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Lee T.H., Blanchard S.C., Kim H.D., Puglisi J.D., Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger M., Dulude D., Steinberg S.V., Brakier-Gingras L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed −1 ribosomal frameshift. Nucleic Acids Res. 2007;35:5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger M., Sidani S., Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.T., Bustamante C., Tinoco I., Jr. Unusual mechanical stability of a minimal RNA kissing complex. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15847–15852. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.T., Collin D., Smith S.B., Bustamante C., Tinoco I., Jr. Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump, and force-ramp methods. Biophys. J. 2006;90:250–260. doi: 10.1529/biophysj.105.068049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J., Napthine S., Kontos H., Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J., Onoa B., Smith S.B., Tinoco I.J., Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- Lopinski J.D., Dinman J.D., Bruenn J.A. Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol. Cell Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manktelow E., Shigemoto K., Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheschi R.J., Staple D.W., Butcher S.E. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;373:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum S.A., Pardi A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- McGarry K.G., Walker S.E., Wang H., Fredrick K. Destabilization of the P site codon–anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels P.J., Versleijen A.A., Verlaan P.W., Pleij C.W., Hilbers C.W., Heus H.A. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Moore S.D., Sauer R.T. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Munro J.B., Altman R.B., O’Connor N., Blanchard S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J.B., Vaiana A., Sanbonmatsu K.Y., Blanchard S.C. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S., Bruno M.M., Pyle A.M., Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S., Liphardt J., Bloys A., Routledge S., Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.L., Cornish P.V., Suram S.V., Giedroc D.P. Thermodynamic analysis of conserved loop–stem interactions in P1-P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry. 2002;41:10665–10674. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- Nixon P.L., Giedroc D.P. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- Nixon P.L., Rangan A., Kim Y.G., Rich A., Hoffman D.W., Hennig M., Giedroc D.P. Solution structure of a luteoviral P1–P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- Nonin-Lecomte S., Felden B., Dardel F. NMR structure of the Aquifex aeolicus tmRNA pseudoknot PK1: new insights into the recoding event of the ribosomal trans-translation. Nucleic Acids Res. 2006;34:1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R.C., Laurs M., Sohet F., Hilbers C.W., Heus H.A., Pleij C.W. Novel application of sRNA: stimulation of ribosomal frameshifting. RNA. 2004;10:1702–1703. doi: 10.1261/rna.7139704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoa B., Dumont S., Liphardt J., Smith S.B., Tinoco I., Jr., Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Otto G.A., Puglisi J.D. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Pallan P.S., Marshall W.S., Harp J., Jewett F.C., 3rd, Wawrzak Z., Brown B.A., 2nd, Rich A., Egli M. Crystal structure of a luteoviral RNA pseudoknot and model for a minimal ribosomal frameshifting motif. Biochemistry. 2005;44:11315–11322. doi: 10.1021/bi051061i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jung Y.H., Kim Y.G., Park H.J. Identification of novel ligands for the RNA pseudoknot that regulate −1 ribosomal frameshifting. Bioorg. Med. Chem. 2008 doi: 10.1016/j.bmc.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N.T., Chamorro M., Varmus H.E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C.P., Barry J.K., Dinesh-Kumar S.P., Brault V., Miller W.A. A sequence required for −1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 2001;310:987–999. doi: 10.1006/jmbi.2001.4801. [DOI] [PubMed] [Google Scholar]
- Pfingsten J.S., Costantino D.A., Kieft J.S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten J.S., Costantino D.A., Kieft J.S. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Jacobs K.L., Harger J.W., Meskauskas A., Jacobs J.L., Baxter J.L., Petrov A.N., Dinman J.D. The 9-A solution: how mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Perez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C.W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Linschooten K., Pleij C.W., Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984;3:2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Pleij C.W., Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M., Dunham C.M., Murphy F.V.T., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Serganov A., Keiper S., Malinina L., Tereshko V., Skripkin E., Hobartner C., Polonskaia A., Phan A.T., Wombacher R., Micura R., Dauter Z., Jaschke A., Patel D.J. Structural basis for Diels-Alder ribozyme-catalyzed carbon–carbon bond formation. Nat. Struct. Mol. Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev P.V., Lesnyak D.V., Kiparisov S.V., Burakovsky D.E., Leonov A.A., Bogdanov A.A., Brimacombe R., Dontsova O.A. Function of the ribosomal E-site: a mutagenesis study. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.X., Tinoco I., Jr. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spiegel P.C., Ermolenko D.N., Noller H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl G., McCarty G.P., Farabaugh P.J. Ribosome structure: revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 2002;27:178–183. doi: 10.1016/S0968-0004(02)02064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staple D.W., Butcher S.E. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staple D.W., Butcher S.E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Studer S.M., Feinberg J.S., Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- Su L., Chen L., Egli M., Berger J.M., Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- ten Dam E., Brierley I., Inglis S., Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E., Pleij K., Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- ten Dam E.B., Verlaan P.W., Pleij C.W. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Blois C.A., Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Theimer C.A., Giedroc D.P. Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed −1 ribosomal frameshifting. J. Mol. Biol. 1999;289:1283–1299. doi: 10.1006/jmbi.1999.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Giedroc D.P. Contribution of the intercalated adenosine at the helical junction to the stability of the gag-pro frameshifting pseudoknot from mouse mammary tumor virus. RNA. 2000;6:409–421. doi: 10.1017/s1355838200992057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr., Li P.T., Bustamante C. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev. Biophys. 2006;39:325–360. doi: 10.1017/S0033583506004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A., Swinger K.K., Pan T., Mondragon A. Structure of ribonuclease P—a universal ribozyme. Curr. Opin. Struct. Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Tzeng T.H., Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wakeman C.A., Winkler W.C., Dann C.E., 3rd. Structural features of metabolite-sensing riboswitches. Trends Biochem. Sci. 2007;32:415–424. doi: 10.1016/j.tibs.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J.D., Lancaster L., Hodges C., Zeri A.C., Yoshimura S.H., Noller H.F., Bustamante C., Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N.M., Moore B., Hammer A., Gesteland R.F., Atkins J.F. A functional −1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]
- Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova G., Jenner L., Rees B., Moras D., Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]


