Abstract
Mammalian circadian rhythms are orchestrated by the suprachiasmatic nuclei (SCN) of the hypothalamus. The SCN are composed of circadian clock neurons, but the mechanisms by which these populations of neuronal oscillators encode rhythmic behavior are incompletely understood. We have used ex vivo real-time gene expression imaging of the neural correlates of circadian behavior, combined with genetic disruption of vasoactive intestinal polypeptide, a key SCN signaling molecule, to examine the neural basis of circadian organization in the SCN. We show that the coherence and timing of clock neuron rhythms are correlated with the coherence and timing of behavioral rhythms within individual mice and that the degree of disruption of SCN neuronal organization correlates with the degree of behavioral disruption within individuals. Our results suggest that the SCN encode circadian phase as a temporal population vector of its constituent neurons; such that as the neuronal population becomes desynchronized, phase information becomes ambiguous.
Keywords: VIP, Per1, suprachiasmatic nucleus, light, circadian, imaging
Introduction
The master circadian clock in mammals is a gene-driven neural network that temporally regulates behavior and physiology. It is composed of ∼20,000 neurons and is located in the suprachiasmatic nuclei of the hypothalamus (SCN) (Weaver, 1998). At a gross level, the SCN′s control of circadian behavior is clearly established as SCN lesions result in behavioral circadian arrhythmicity (Stephan and Zucker, 1972; Moore and Eichler, 1976; Rusak, 1977) that can be restored by SCN transplants (Lehman et al., 1987). At a more refined level, it is unclear how neurons within the SCN neural network act to control or regulate circadian behavior.
Vasoactive intestinal polypeptide (VIP) is a key signaling molecule expressed in neurons of the retino-recipient region of the SCN, where it is rhythmically released in a light cycle and can reset the phase of the clock in response to nighttime light pulses (Takahashi et al., 1989; Albers et al., 1990; Shinohara et al., 1993; Yang et al., 1993; Piggins et al., 1995; Ban et al., 1997). Mice lacking VIP-ergic signaling show marked circadian abnormalities such as an inability to phase shift in response to light pulses and blunted endogenous behavioral circadian rhythms (Harmar et al., 2002; Colwell et al., 2003). In addition, neonatal SCN from VIP−/− mice exhibit reduced synchrony of neuronal spike frequency rhythms (Aton et al., 2005) and of neuronal clock gene expression of rhythms (Maywood et al., 2006). Manipulation of this SCN signaling molecule offers the opportunity to perturb SCN organization and to test the relationship between clock network organization and the control of behavior.
Using ex vivo real-time clock gene expression imaging of SCN networks and neurons in which SCN are acutely explanted and characterized from behaviorally characterized VIP-deficient mice, we sought to ascertain the aspects of SCN network organization that define behavioral circadian organization. Our findings indicate that within individual mice the degree of behavioral circadian disruption resulting from loss of VIP is correlated with the degree of desynchrony in the SCN neural network and that the onset of behavioral activity coincides with a specific phase of SCN rhythms. These results suggest that in the circadian clock, the overall pattern and organization of rhythmic neuronal activity that drives the temporal organization of locomotor behavior is encoded by the mean temporal population vector of SCN neuron rhythms.
Materials and Methods
Animals and housing.
Mice with a targeted VIP gene disruption (Colwell et al., 2003) were backcrossed four generations to mice (B6C3 hybrid from Jackson Laboratories) carrying the mPer1::d2EGFP transgene (Kuhlman et al., 2000) yielding Per1::GFP+/+, VIP+/− mice which were bred in a 12L:12D light/dark cycle (LD) to yield experimental mice. Male offspring 3–5 weeks of age were placed in litter-filled wheel cages (Coulbourn Instruments) with food and water ad libitum. All mice were entrained to 12L:12D light/dark cycles for 9–22 d, and then either killed and their SCN explanted to assay SCN molecular rhythms ex vivo as part of the LD experimental group, or maintained in constant darkness (DD) for an additional 20–60 d and then killed and their SCN explanted to assay SCN rhythms ex vivo as part of the DD group. Animals were killed by cervical dislocation between Zeitgeber time (ZT) 9–12 in LD (where ZT 12 is lights off), or Circadian time (CT) 9–13 in DD (where CT 12 is the onset of activity). Behavioral onset of mice behaviorally arrhythmic in DD could not be assigned. They were killed along with their rhythmic counterparts and their SCN rhythm phases are reported as time ex vivo. All animal care was conducted in accordance with Vanderbilt University Institutional Animal Care and Use Committe guidelines.
Behavioral analyses.
Wheel-running activity was monitored and recorded in 5-min bins using ClockLab software (Actimetrics). Behavior was analyzed using ClockLab Analysis software, and activity was quantified as the number of wheel revolutions occurring during 5-min bins. In LD, the proportions of activity during lights on and lights off, as well as the total amount of activity per day were determined for the last 9 d in LD using the “activity profile” function. In DD, activity was measured for the first 20 d in darkness. Chi-squared periodogram analysis yielded rhythmic power as a measure of the amplitude and coherence of behavioral rhythms (Colwell et al., 2003). Linear regression (Pittendrigh and Daan, 1976) was also used to project behavioral onset (CT12) in rhythmic animals to establish a consistent time of euthanization (CT 9–13).
Ex vivo culture.
After behavioral characterization, mouse brains were extracted and blocked in cold sampling media as described previously (Ohta et al., 2006). One or two 200 μm coronal hypothalamic sections per animal were made on a Vibroslicer (Campden Instruments). SCN were isolated by trimming the surrounding hypothalamic tissue and placed on a Millicell (Millipore) culture membrane insert in 37°C recording media (Ohta et al., 2006), conditioned with Invitrogen N-2 supplement (Invitrogen) and contained in 35 mm culture dishes (Falcon, BD). Dishes were then sealed with sterile high-vacuum grease and placed in a custom 6-well aluminum chamber heated to 35.5°C by a voltage-regulator.
Confocal imaging.
The 6-well chamber containing cultured slices was placed on an automated stage on a laser-scanning confocal microscope (LSM5 PASCAL, Zeiss) with GFP excitation at 488 nm, and bandpass emission recorded between 505 and 555 nm. Three Z-stack images totaling 40 μm in depth were taken of each slice every hour for 90 h. Focus was adjusted manually to account for slice swelling and flattening as needed.
Ex vivo analyses.
Raw LSM files of SCN time-lapse imaging were compiled in MetaMorph (Molecular Devices). The Z-stack for each time-point was compressed into a single maximum projection image from which a background subtraction was made (Close-Open morphological filter with a 75 pixel-round sequential average). Whole SCN or single cells were selected as regions of interest from the resultant time-series stack, and time-lapse intensity measurements were made and exported to an in-house program created in R (http://www.r-project.org/) that allowed selection of minima and maxima for each SCN or cell, and calculation of phase. The first 12 h ex vivo were omitted from the analyses. Peaks in fluorescence were visually scored as having a clear rising and falling phase above baseline fluorescence, and the number of circadian peaks per SCN or per cell was determined by 3-person independent visual scoring. For the purposes of this study, SCN or cells with two or more peaks were considered rhythmic. Overall slice fluorescence was determined by the total integrated fluorescence over hours 12–36 ex vivo divided by the area.
Statistical analysis.
Circular statistics and Rayleigh vector plots were performed in Oriana 2.0 (Rockware). Statistics were calculated for each animal for further comparison. Means were statistically compared in SPSS 14.0 (SPSS) with independent samples t tests (two-tailed) and one-way ANOVA followed by post hoc Fisher's least significant difference tests, except for when the variances were not homogeneous, as indicated by a significant Levene's test. In these cases, nonparametric Kruskal–Wallis or Median tests were used. For correlational analysis, Pearson's product moment correlations were used. Significance was ascribed at p < 0.05.
Results
VIP−/− Per1::GFP mice exhibit weakened, phase-advanced circadian behaviors
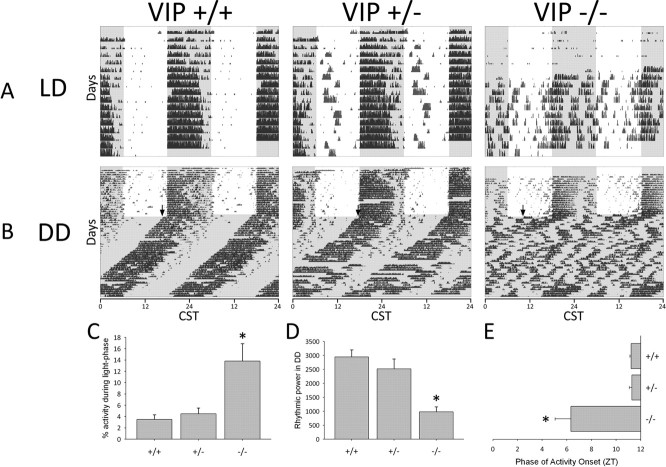
VIP knock-out mice have been previously shown to exhibit specific circadian behavioral deficits, including altered light entrainment in LD and disrupted free-running circadian rhythms in DD (Colwell et al., 2003). To confirm that these traits persisted following breeding onto the Per1::GFP circadian reporter background, we examined the circadian behavior of VIP knock-out (VIP−/−), heterozygous (VIP+/−) and wild-type (VIP+/+) Per1::GFP mice in LD and DD (Fig. 1). In light/dark cycles, all genotypes exhibited primarily nocturnal locomotor patterns, but VIP−/− mice had less robust partitioning of activity into the night (dark) portion of the cycle, performing a greater percentage of wheel running during the light phase than the other genotypes (Median test, χ2(2) = 3.76, p < 0.05) (Fig. 1A,C). In constant darkness, VIP−/− mice exhibited severely disrupted free-running circadian behavior compared with wild-type and heterozygous mice. They lacked clear, coherent circadian rhythms (Fig. 1B) and exhibited a significant reduction in rhythmic power in periodogram analysis (F(2,22) = 13.4, p < 0.01) (Fig. 1D). At the transition from LD into DD, many VIP−/− mice did exhibit an initial behavioral onset during the first day in DD that was phase advanced by 5–6 h compared with the onsets of VIP+/− and VIP+/+ mice (Kruskal–Wallis, χ2(2) = 8.8, p < 0.05) (Fig. 1E). These results demonstrate that the signature circadian deficits associated with the lack of VIP signaling in the SCN (weakened light entrainment, disrupted endogenous rhythms and advanced phase angle of activity onset) (Colwell et al., 2003) are preserved in VIP−/− Per1::GFP mice.
Figure 1.
VIP knock-out Per1::GFP mice exhibit altered circadian behavior. Left to right, Example VIP wild-type (+/+), heterozygous (+/−) and knock-out mice (−/−) on the Per1::GFP reporter background. A, Double-plotted actograms illustrate circadian behavior in a 12L:12D light/dark cycle. CST, Central Standard Time. B, Double-plotted actograms illustrate circadian behavior in DD. Black arrow represents the phase of activity onset on the first day in DD. A, B, Black ticks represent activity in 5-min bins; white background denotes lights on; gray background denotes lights off. C, Percentage of total wheel-running activity during the light phase in LD for VIP+/+ (N = 14), VIP+/− (N = 16) and VIP−/− (N = 16) mice. D, Rhythmic power in constant darkness as measured by χ2 periodogram amplitude for VIP+/+ (N = 8), VIP+/− (N = 9) and VIP−/− (N = 8) mice. E, Time of activity onset on the first day in constant darkness for VIP+/+ (N = 7), VIP+/− (N = 9) and VIP−/− (N = 8) mice. Error bars represent SEM; *p < 0.05.
VIP−/− Per1::GFP mice exhibit desynchronized, phase-advanced SCN neuronal rhythms
We next characterized the SCN organization of the mice behaviorally characterized in Figure 1 by measuring circadian rhythms in the expression of Per1 promoter-driven short half-life GFP in acute, ex vivo hypothalamic slice culture (Kuhlman et al., 2000; Quintero et al., 2003; Ohta et al., 2005, 2006). After behavioral characterization in LD or DD, coronal SCN slices from each mouse were acutely explanted into organotypic slice culture. Cultures were imaged hourly for 90 h on a confocal microscope, providing an assay of SCN network organization that could then be correlated with the animal's previous behavior, genotype and lighting condition.
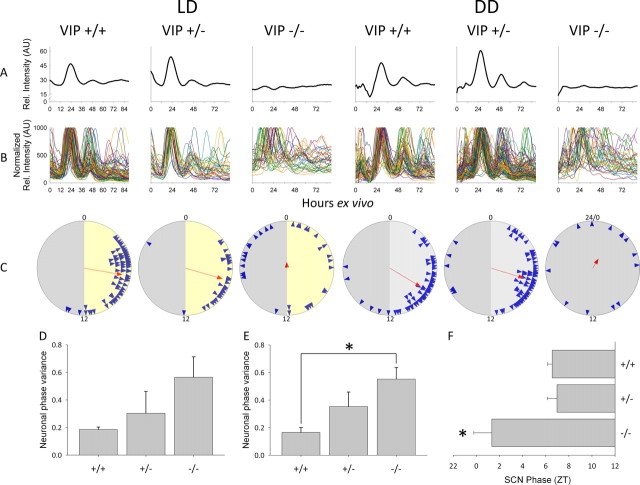
Figure 2, A–C (and supplemental Movies 1 and 2, available at www.jneurosci.org as supplemental material), shows the SCN tissue and the SCN neuron Per1::GFP rhythms from the individual mice that were behaviorally characterized in Figure 1. Each column shows imaging data from an individual mouse matching the corresponding genotype and condition in Figure 1 (i.e., the data in the LD, VIP+/+ column in Fig. 2A–C are imaging data from the SCN explant of the mouse for which the behavioral record is Fig. 1A LD, VIP+/+, etc.). Overall SCN rhythmicity, as measured by the integrated Per1::GFP fluorescence of the SCN, was robustly rhythmic in explants from VIP+/+ and VIP+/− mice in LD and DD, but was significantly disrupted in VIP−/− explants in both LD and DD (Fig. 2A). Like locomotor behavior of VIP−/− mice at the LD to DD transition, Per1::GFP gene expression in explants from LD VIP−/− mice often exhibited a detectable initial onset cycle, followed by damped or disrupted rhythmicity during subsequent cycles ex vivo (Fig. 2A), suggesting that deafferentation of the explant may mimic the LD to DD transition. SCN explants from VIP−/− mice maintained in DD exhibited disrupted tissue-level circadian rhythms (Fig. 2A) similar to their disrupted circadian behavior (Fig. 1A).
Figure 2.
Ex vivo circadian gene expression rhythms from SCN and clock neurons. Left to right, Example ex vivo SCN Per1::GFP imaging data from VIP wild-type (+/+), heterozygous (+/−) and knock-out mice (−/−). A, Representative relative fluorescence intensity plots of ex vivo SCN for each of the VIP genotypes from LD (left) and DD (right) over 90 h. B, Representative normalized relative fluorescence intensity plots of individual neurons over 90 h ex vivo. C, Rayleigh plots from LD (left) and DD (right). Blue arrowheads represent the 50% peak rising phases of individual rhythmic neurons from a representative mouse of a particular genotype. Red arrow indicates the mean phase vector of rhythmic neurons, where length is inversely proportional to the neuronal phase variance, and the direction indicates timing relative to the previous light cycle in LD or previous behavioral cycles in DD. For LD (left), numbers indicate projected ZT, where projected ZT 0–12 is represented with a yellow background and projected ZT 12–24(0) is represented with a gray background; for DD (right), numbers indicate CT, where CT 0–12 is represented with a light gray background and CT 12–24(0) is represented with a darker gray background. For the VIP−/− mouse in DD, in which CT could not be reliably assigned, the phases of neurons are plotted as time ex vivo. D, Neuronal phase variance in SCN from VIP+/+ (N = 4), VIP+/− (N = 5) and VIP−/− (N = 4) mice maintained in LD. E, Neuronal phase variance in SCN from VIP+/+ (N = 7), VIP+/− (N = 8) and VIP−/− (N = 7) mice maintained in DD. F, SCN phase from VIP+/+ (N = 9), VIP+/− (N = 9) and VIP−/− (N = 6) mice previously maintained in LD. Error bars represent SEM; *p < 0.05.
At the cellular level, individual SCN neurons exhibited robust circadian rhythms in reporter gene expression in explants from all genotypes maintained in both LD and DD, even in VIP−/− mice with disrupted behavioral and SCN rhythms. Figure 2B shows all the cellular rhythms from the corresponding SCN slices in Figure 2A, whereas supplemental Figure 1, available at www.jneurosci.org as supplemental material, shows a limited number of typical cellular rhythms from the same slices to better illustrate the properties of individual cellular rhythms. Rayleigh vector plots of the distribution of neuronal Per1::GFP rhythm phases revealed that the degree of synchrony in neuronal rhythms, as shown by vector arrow length, was reduced in VIP−/− mouse explants, particularly in DD (Fig. 2C). Further quantitative analysis of the imaging data showed that in explants from mice maintained in LD, there was a trend toward increased neuronal phase variance within each mouse as the number of functional VIP alleles decreased, with VIP−/− explants exhibiting the highest degree of neuronal phase variance (F(2,10) = 2.0, p = 0.19) (Fig. 2D). Similarly, in explants from mice in DD, the variance of the individual cell phases within each animal was significantly greater in VIP−/− explants than in VIP+/+ explants (Kruskal–Wallis, χ2(2) = 9.171, p = 0.01) (Fig. 2E). Finally, the onset phase (10% rise) of the first Per1::GFP cycle ex vivo occurred 5–6 h earlier in VIP−/− SCN compared with VIP+/− and VIP+/+ SCN (F(2,21) = 4.1, p < 0.05) (Fig. 2F), similar to the onset of behavioral rhythms at the LD to DD transition. The lack of detectable synchrony in SCN neuronal phases across slices from VIP−/− mice in DD (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), in which the time axis is hours ex vivo, indicated that dissection and transition to ex vivo conditions did not set neuronal phase.
SCN neuronal phase distribution sets behavioral coherence and timing
Given the striking correspondence of changes in the neuronal phase distribution with alterations in circadian behavior that we observed across mouse populations with altered VIP communication, we next tested if SCN neuronal phase organization was correlated with circadian behavioral characteristics within individual mice across VIP genotype and lighting condition. Indeed, the proportion of wheel-running activity that occurred during the light phase of LD cycles was significantly and positively correlated with increased neuronal phase variance within individual animals across all genotypes (i.e., increased daytime activity was associated with decreased neuronal synchrony in all mice; r(15) = 0.568, p < 0.05) (Fig. 3A). Similarly, behavioral rhythmic power was significantly and negatively correlated with neuronal phase variance within individual animals across VIP gene dosage (i.e., decreased behavioral rhythmic power was associated with increased neuronal desynchrony in all mice; r(18) = −0.641, p < 0.01) (Fig. 3B). In addition to the correlations of neuronal phase variance with the coherence of behavioral rhythms, the timing of molecular rhythms in the SCN neural network also correlated with the timing of behavioral activity. In SCN from mice maintained in LD, both the onset of the Per1::GFP rhythm (10% above baseline on the rising phase) during the first cycle ex vivo and the onset of behavioral activity during the first cycle in DD were phase advanced by 5–6 h in VIP−/− mice compared with other genotypes (Fig. 3C).
Figure 3.
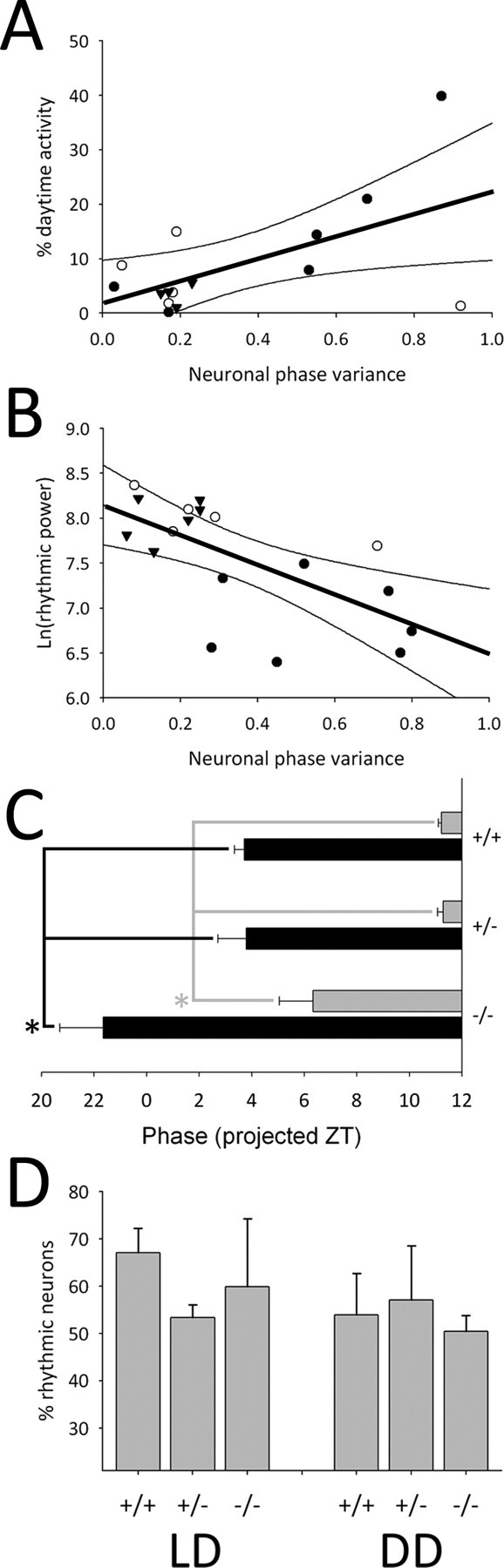
Correlation of SCN neuronal rhythms with behavioral characteristics. A, Plot of percentage of daytime wheel-running activity versus neuronal phase variance for individual mice (N = 15) by VIP genotype maintained in LD. B, Plot of rhythmic power versus neuronal phase variance for individual mice (N = 18) by VIP genotype maintained in DD. A, B, Closed circles represent VIP−/− mice, open circles represent VIP+/− mice and triangles represent VIP+/+ mice. Thick black line represents single order regression, and the thin black lines represent the 95% confidence interval. C, Phase of SCN Per1::GFP onset (10% rising phase) on the first day ex vivo from mice maintained in LD (black bars) and onset of behavioral activity on the first day in DD (gray bars). For number of animals, see Figure 1E and Figure 2F. D, Plot of the percentage of rhythmic neurons (2+ peaks) per SCN slice for mice maintained in LD (left) or in DD (right). LD: VIP+/+ (N = 5), VIP+/− (N = 5) and VIP−/− (N = 4); DD: VIP+/+ (N = 6), VIP+/− (N = 8) and VIP−/− (N = 7). Error bars represent SEM; *p < 0.05.
Whereas the characteristics of circadian behavior mirrored SCN neuronal phase distribution and onset time, there was no significant correlation between other aspects of SCN rhythms and circadian behavior. Overall levels of Per1::GFP expression, measured as fluorescence intensity, were similar across VIP genotypes (F(2,70) = 1.359, p = 0.26), and there was no correlation between the number or percentage of rhythmic neurons in an individual animal's SCN explant and the behavioral rhythmic power of that mouse in DD (Pearson's product moment correlation for: number, r(20) = 0.255, p = 0.28 and percent, r(20) = 0.238, p = 0.31). Furthermore, the percentage of rhythmic neurons in individual SCN explants was similar across VIP genotypes and across lighting conditions (LD: F(2,11) = 2.7, p = 0.11; DD: F(2,18) = 1.3, p = 0.29) (Fig. 3D; supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Discussion
In this study, we have disrupted SCN network organization by targeted deletion of a key communicating molecule, VIP, and then assayed the circadian behavior and the neural correlates of circadian behavior by ex vivo real-time gene expression imaging of circadian clock neurons in the SCN of individual mice. Our principal findings are that the timing (phase) and coherence (power) of circadian behavior is encoded by the timing and degree of synchrony of SCN neuronal rhythms. The timing and strength of circadian locomotor rhythms is encoded by the timing and strength of the SCN average neuronal temporal population vector.
Targeted knockout of the VIP gene disrupts the circadian neural network by leading to neuronal desynchronization and results in stereotypical behavioral deficits such as increased daytime wheel running and arrhythmicity in constant darkness. In this study, we found that the weakened behavioral rhythms and phase-advanced DD behavioral onset in adult VIP−/− Per1::GFP mice reflect the less synchronized and phase-advanced patterns of neural rhythms recorded ex vivo in the SCN of the same individual animals. Specifically, the degree of degradation of behavioral rhythmic coherence was correlated with the degree of degradation of rhythmic coherence in the population of SCN neurons within an individual (Fig. 3A,B). We also found that the first peak of SCN network Per1::GFP expression maintains a constant phase relationship to activity onset across VIP genotypes, with the initial rise in gene activity anticipating behavioral activity onset by 7–8 h and matching the 5–6 h phase advance of activity in the VIP−/− mice. Unexpectedly, we also found that while SCN neuronal synchrony was disrupted in VIP−/− mice, SCN neurons from VIP−/− mice exhibited robust rhythmicity in similar number and proportion to SCN neuronal molecular rhythms from VIP+/− and VIP+/+ mice.
There are both similarities and differences between our results and the results from previous circadian studies using VIP communication-deficient mice. Our study used acutely explanted, ex vivo SCN from adult VIP−/− mice to assay circadian behavior and establish circadian neural correlates within individual mice. In contrast, previous studies used reconstituted dispersed neonatal SCN networks or reorganized neonatal SCN explants that had been maintained chronically in vitro for 2 or more weeks before recording, and then inferred a relationship of in vitro findings to adult circadian behavior based on genotype (Aton et al., 2005; Maywood et al., 2006). Our finding that compromised VIP signaling leads to desynchronization of SCN clock neurons is in accordance with studies of neonatal SCN networks, and strengthens the conclusion that VIP acts as a synchronizing substance in the SCN (Aton et al., 2005; Maywood et al., 2006; Brown et al., 2007). In addition, we have now demonstrated directly the previous speculation that the desynchronization of SCN neurons is accompanied by loss of behavioral rhythmic coherence within individuals. Our results demonstrating preservation of the number and proportion of individual neuronal rhythms in explanted adult VIP−/− SCN stand in contrast to the neonatal VIP studies in which there was significant loss in the proportion of rhythmic neurons following loss of VIP communication in SCN networks (Aton et al., 2005; Maywood et al., 2006). A study of VIP receptor knock-out adult animals also revealed a surprisingly high proportion of rhythmic but desynchronized SCN neurons (Hughes et al., 2008). The reasons for these differences are unknown, but could be explained by a number of factors including a difference in the role of VIP in mature versus neonatal SCN networks, differences in culture time (4 d vs 2–3 weeks), access to a running wheel or compensation in the adult by upregulation of other signaling molecules in the SCN network such as GRP that could sustain cellular rhythms (Maywood et al., 2006). Whatever the precise cause, it is clear that one inference drawn from the neonatal SCN studies, that cellular desynchronization in the SCN necessarily leads to reduced cellular rhythmicity, does not hold in the case of VIP communication, or in the case of disruption of SCN neuronal synchrony by constant light, where cellular rhythm robustness is also preserved in the face of cellular desynchrony (Ohta et al., 2005, 2006).
Our findings have significant implications for understanding how the SCN neural network encodes and drives circadian behavior. The present study of VIP gene-dosage effects, in combination with work on mice exposed to constant light (Ohta et al., 2005, 2006), demonstrates a strong correlation between the timing and coherence of circadian locomotor behavior, and the timing and coherence of neuronal phases within the SCN, represented as the temporal direction and magnitude of the population vector of SCN neuronal rhythms. Work on neonatal SCN and SCN neurons from Tau-mutant hamsters and ClockΔ19-mutant mice, has suggested that behavioral circadian period is approximated by the population average period of the individual neurons that comprise the SCN (Liu et al., 1997; Herzog et al., 1998). Similarly, chimeric SCN composed of wild-type and ClockΔ19-mutant SCN neurons drive circadian behavior with the intrinsic period of the majority of neurons (Low-Zeddies and Takahashi, 2001). The present data suggest that, similar to period, the timing (phase) of behavioral onset is encoded by the average phase of the neural population represented as a temporal population vector with the temporal direction of the vector determining the time of activity onset and the magnitude of the vector determining the coherence of the behavioral rhythms. In this way, the manner in which the circadian network encodes behavioral phase is not unlike the population coding observed in the voluntary motor system, in which the spatial direction of targeted limb movements is coded by the average population vector of directional motor neurons in the cortex (Georgopoulos et al., 1986).
This concept of population encoding can be extended to the paired SCN and to multiple neuronal populations within each SCN to account for exceptional circumstances in which SCN neuronal subpopulations can encode bimodal circadian behavioral output. For example, “splitting” of behavioral circadian rhythms in response to constant light correlates with antiphase oscillation of the gene expression rhythms in the SCN neurons of the paired SCN (de la Iglesia et al., 2000; Ohta et al., 2005). Similarly, seasonal light cycles induce changes in the distribution of neuronal phases within the SCN (Inagaki et al., 2007; VanderLeest et al., 2007; Naito et al., 2008) that likely encode for photoperiodic changes in circadian behaviors. Population coding in the SCN has these potential advantages: (1) the summation of neuronal phases may lead to greater accuracy and precision of circadian behavioral activity onset similar to the increase in precision of period gained by population encoding (Liu et al., 1997; Herzog et al., 1998); (2) a network composed of neurons with variable phase relationship and a population encoded output to behavior can flexibly encode the amplitude and phase of changing environmental stimuli (such as seasonal light cycles) and through the phase organization of the neuronal population, drive circadian behavior appropriately. VIP is a signal for light stimuli within the SCN network as well as a synchronizing agent (Piggins et al., 1995; Aton et al., 2005; Maywood et al., 2006). Thus, the overall role of VIP communication in the SCN appears to be to modulate the timing and coherence of population vector encoding by SCN neurons to adjust circadian behavior to environmental light input.
Footnotes
This work was supported by National Institutes of Health Grants RO1 MH63341 (D.G.M.), F31 MH080547 (C.M.C.), NS043169 (C.S.C.), P30 EY008126 (J. D. Schall), and T32 MH64913 (E. Sanders-Bush). We thank Terry Page for his helpful comments and Tongrong Zhou for technical assistance.
References
- Albers HE, Stopa EG, Zoeller RT, Kauer JS, King JC, Fink JS, Mobtaker H, Wolfe H. Day-night variation in prepro vasoactive intestinal peptide/peptide histidine isoleucine mRNA within the rat suprachiasmatic nucleus. Brain Res Mol Brain Res. 1990;7:85–89. doi: 10.1016/0169-328x(90)90077-q. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Shigeyoshi Y, Okamura H. Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J Neurosci. 1997;17:3920–3931. doi: 10.1523/JNEUROSCI.17-10-03920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Colwell CS, Waschek JA, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophysiol. 2007;97:2553–2558. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Meyer J, Carpino A, Jr, Schwartz WJ. Antiphase oscillation of the left and right suprachiasmatic nuclei. Science. 2000;290:799–801. doi: 10.1126/science.290.5492.799. [DOI] [PubMed] [Google Scholar]
- Georgopoulos AP, Schwartz AB, Kettner RE. Neuronal population coding of movement direction. Science. 1986;233:1416–1419. doi: 10.1126/science.3749885. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- Hughes AT, Guilding C, Lennox L, Samuels RE, McMahon DG, Piggins HD. Live imaging of altered period1 expression in the suprachiasmatic nuclei of Vipr2(-/-) Mice. J Neurochem. 2008;106:1646–1657. doi: 10.1111/j.1471-4159.2008.05520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, McMahon DG. GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport. 2000;11:1479–1482. [PubMed] [Google Scholar]
- Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, Bittman EL. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7:1626–1638. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O'Neill JS, O'Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Moore RY, Eichler VB. Central neural mechanisms in diurnal rhythm regulation and neuroendocrine responses to light. Psychoneuroendocrinology. 1976;1:265–279. doi: 10.1016/0306-4530(76)90016-0. [DOI] [PubMed] [Google Scholar]
- Naito E, Watanabe T, Tei H, Yoshimura T, Ebihara S. Reorganization of the suprachiasmatic nucleus coding for day length. J Biol Rhythms. 2008;23:140–149. doi: 10.1177/0748730408314572. [DOI] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- Ohta H, Mitchell AC, McMahon DG. Constant light disrupts the developing mouse biological clock. Pediatr Res. 2006;60:304–308. doi: 10.1203/01.pdr.0000233114.18403.66. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh C, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents: V. Pacemaker structure: a clock for all seasons. J Comp Physiol A. 1976;106:333–355. [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusak B. Role of suprachiasmatic nuclei in generation of circadian-rhythms in golden-hamster, Mesocricetus-Auratus. J Comp Physiol A. 1977;118:145–164. [Google Scholar]
- Shinohara K, Tominaga K, Isobe Y, Inouye ST. Photic regulation of peptides located in the ventrolateral subdivision of the suprachiasmatic nucleus of the rat: daily variations of vasoactive intestinal polypeptide, gastrin-releasing peptide, and neuropeptide Y. J Neurosci. 1993;13:793–800. doi: 10.1523/JNEUROSCI.13-02-00793.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Okamura H, Yanaihara N, Hamada S, Fujita S, Ibata Y. Vasoactive intestinal peptide immunoreactive neurons in the rat suprachiasmatic nucleus demonstrate diurnal variation. Brain Res. 1989;497:374–377. doi: 10.1016/0006-8993(89)90283-7. [DOI] [PubMed] [Google Scholar]
- VanderLeest HT, Houben T, Michel S, Deboer T, Albus H, Vansteensel MJ, Block GD, Meijer JH. Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol. 2007;17:468–473. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Weaver DR. The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms. 1998;13:100–112. doi: 10.1177/074873098128999952. [DOI] [PubMed] [Google Scholar]
- Yang J, Cagampang FR, Nakayama Y, Inouye SI. Vasoactive intestinal polypeptide precursor mRNA exhibits diurnal variation in the rat suprachiasmatic nuclei. Brain Res Mol Brain Res. 1993;20:259–262. doi: 10.1016/0169-328x(93)90049-u. [DOI] [PubMed] [Google Scholar]