Abstract
A family of proteins conserved throughout the eukaryotic lineage is characterized by the presence of a common sequence motif – the methyl CpG binding domain, or MBD. This sequence motif corresponds to a structural domain which, in some but not all cases, confers the ability to bind methylated cytosine residues in the context of the dinucleotide 5’ CG 3’. Mammals have five well characterized members of this family, each with unique biological characteristics. Recently, much progress has been made in defining the biochemical properties of one member of this family, MeCP2. This protein has a very high affinity for chromatin and considerable insight has been gained into its interactions with naked DNA and with chromatin fibers. Previous models have proposed that several members of the MBD family contribute to establishment and/or maintenance of transcriptional repression by recruiting enzymes that locally modify histones. Surprisingly, recent data indicate that MeCP2 is likely to contribute to chromatin properties through an architectural role, participating in higher order chromatin structures that facilitate both gene repression as well as gene activation. These observations suggest that existing models probably do not explain the entire gamut of biological functions performed by this very interesting protein family.
Keywords: epigenetics, DNA methylation, methyl CpG binding protein, transcriptional repression, histone deacetylase
1. Introduction
In the late 1980’s and early 1990’s, Adrian Bird and colleagues established reliable biochemical assays for the detection and eventual biochemical isolation from mammalian cells of proteins with the capacity to selectively recognize methylated DNA [1,2]. The founding protein, termed MeCP2, was demonstrated to possess a sequence motif both necessary and sufficient for methylated DNA binding [3]. Approximately 10 years ago, Hendrich and Bird published a landmark paper enlarging the family of mammalian proteins containing the methyl CpG binding domain to its current size [4]. Subsequent work has established that this protein motif is conserved across the eukaryotic lineage [5]. The intervening decade has witnessed considerable interest in these proteins, with characterization of their biochemistry and genetics. Here we highlight recent findings that suggest an unanticipated level of complexity to the biology of MBD proteins.
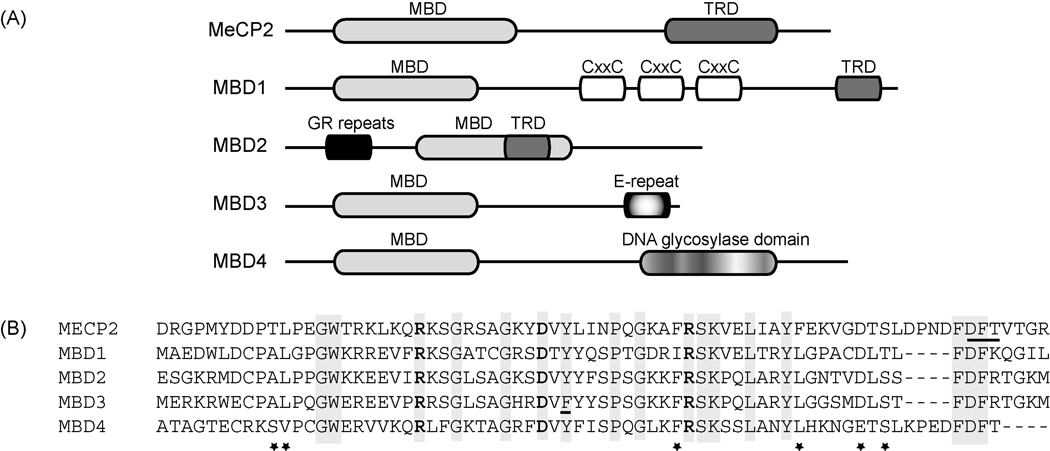
The MBD sequence motif was defined by molecular analysis of the prototype MBD protein, MeCP2 [3]. This sequence motif was found to be both necessary and sufficient to direct specific interaction with methyl CpG containing DNA fragments. It consists of approximately 70 amino acids and represents the sole sequence feature found in common in all MBD family members (Figure 1A). The sole exception to this rule is the case of MBD2 and MBD3, which are nearly identical in amino acid sequence, have common gene structures and are believed to have arisen from an ancient duplication in the evolution of the vertebrate lineage [5]. The primary amino acid sequence of the motif is remarkably conserved across the different family members within mammals (Figure 1B).
Figure 1. The primary amino acid sequence of the MBD motif is conserved across the various members of the family.
A. The general structural domains of MeCP2 and the MBD family are depicted. MBD=methyl CpG binding domain; MeCP2=methyl CpG binding protein 2; TRD=transcriptional repression domain; CxxC=cysteine rich domain; GR=glycine and arginine repeats; E-repeat=glutamate repeat
B. The MBD motif from humans is depicted in the sequence alignment. Identical residues are indicated by gray shading. Conserved amino acids are indicated by asterisks (*). The residues in MeCP2 making direct contact with DNA in the crystal structure are highlighted in bold type, the Asx-ST motif in the MeCP2 MBD motif, implicated in a phosphate backbone contact, is underlined.
The MBD sequence motif corresponds very closely with a structural domain now characterized at atomic level resolution. Solution structures of the MBD from multiple different MBD proteins as well as from different species adopt very similar folds [6–8], forming a wedge-shaped structure. The front face of the wedge consists of beta strands (which vary in number depending on the protein), the other face of the wedge contains a short alpha helix. The NMR structure of the MBD domain from MBD1 in complex with a short methylated duplex oligonucleotide [9] demonstrated a conformational change consistent with an induced fit mechanism on interaction with DNA. This finding has been confirmed recently by solution of the crystal structure of MeCP2 in complex with methylated DNA [10]. A pair arginine residues and an aspartic acid residue, all three being absolutely conserved across the MBD family, make direct contact with DNA bases (Figure 1). Further contacts between the protein and DNA recognition site are mediated by several highly structured water molecules, including a hydrogen bond made by a highly conserved tyrosine hydroxyl group (Figure 1). Finally, the MeCP2 MBD domain contains an unusual Asx-ST motif that makes contact with a DNA backbone phosphate placed in an atypical structure (for standard B form DNA) by a local AT run [10], a sequence feature found associated with high affinity MeCP2 binding sites in vitro and in vivo [11].
2. MeCP2 as a chromatin component
In the late 1990’s a seminal set of observations established a biochemical link between the MBD family and enzymes that modify chromatin components. MeCP2 [12,13], MBD3 [14,15], MBD2 [15,16], and MBD1 [17] were linked by biochemical and/or molecular analysis to histone deacetylase enzymes. These studies led to the propagation of a model suggesting that members of the MBD family bind specifically to methylated DNA through the action of the MBD motif where they recruit enzymatic activities, such as histone deacetylases, to establish and/or maintain a locally repressive chromatin environment [18]. As is usually the case, subsequent findings have indicated that this model cannot account for all the biological properties of the MBD proteins.
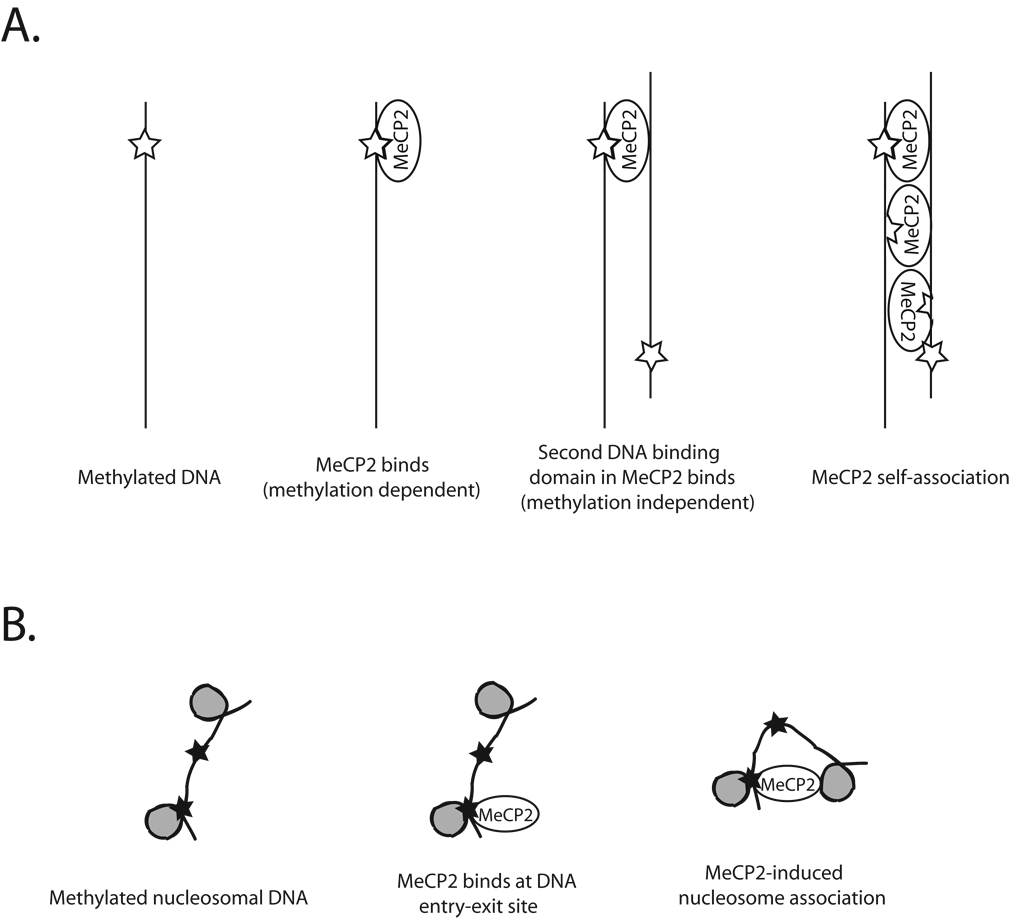
Recent studies by Hansen, Woodcock and colleagues have provided a wealth of data describing solution properties of MeCP2, its interactions with DNA, and its interactions with biochemically defined chromatin fibers. Somewhat surprisingly, the full length protein is largely devoid of secondary structure (approximately 2/3 unstructured by circular dichroism spectroscopy). Analysis of structure by protease digestion confirmed the presence of large regions lacking detectable structural features punctuated by short regions of protease resistance, including the previously defined MBD and transcriptional repression domains [19]. Consistent with early descriptions of native MeCP2 [20], recombinant MeCP2 was shown to have the capacity to bind both unmethylated and methylated DNA, with a preference for methyl CpG containing substrates [19,21]. Electron micrographs of MeCP2 in complex with naked DNA reveal complicated structures, including DNA loops, regions of close contact between 2 DNA molecules and extensive regions in which 2 DNA segments are in close juxtaposition [21]. These MeCP2-DNA complexes are consistent with binding of a single MeCP2 molecule to 2 different DNA molecules (presumably utilizing two distinct DNA binding surfaces), followed by cooperative formation of multiple side-by-side interactions of the same type (Figure 2).
Figure 2. Models for the interaction of MeCP2 with naked DNA and with chromatin fibers.
A. Generation of DNA-MeCP2-DNA ‘sandwich’ structures. The model depicts a possible scenario in which DNA methylation dependent binding by MeCP2 followed by non-methylation dependent interaction with a second DNA duplex and eventual MeCP2 self association leads to formation of unusual protein-DNA structures.
B. MeCP2-mediated chromatin compaction. The model depicts one potential mode of generating compact nucleosomal structures by MeCP2. The MeCP2 protein interacts with methylated CpG residues (denoted by filled black stars) on nucleosomal arrays. IN this case, binding occurs at the DNA entry-exit site. Association of nucleosomal DNA on a second surface of MeCP2 leads to local chromatin compaction with formation of ‘looped’ linker DNA.
Analysis of the interaction of recombinant MeCP2 with defined chromatin fibers demonstrated that the protein has high affinity for chromatin [22]. Full length forms of the protein form highly regular, compacted structures at low ionic strength [22], this binding is moderately enhanced if the underlying DNA is methylated [21]. While the compaction evident in these structures is impressive, linker DNA between individual nucleosomes is still accessible to enzymatic cleavage [22]. Surprisingly, both overall binding efficiency and enhancement of binding by DNA methylation is lost if the carboxyl terminus of the protein is truncated (note that the truncations do not impinge on the MBD domain), suggesting that the C terminus of MeCP2 harbors a chromatin interaction surface [21]. The mode of chromatin compaction seems to depend on nucleosome-nucleosome interactions (Figure 2), again suggesting a ‘sandwich’ consisting of a single MeCP2 molecule simultaneously bound to 2 different nucleosomes [21]. MeCP2 was observed in these structures to form a ‘stem-like’ structure at the DNA entry-exit site on the nucleosome surface, reminiscent of the interaction of histone H1 with nucleosomal DNA. Indeed, MeCP2 binding to nucleosomal DNA protects linker DNA from micrococcal nuclease digestion in a manner similar to H1 [23]. However, MeCP2 interaction with DNA at the nucleosome entry-exit site differs from H1 binding in that less DNA is protected (11 bp versus approximately 20), the protection frequently occurs on only one end of the nucleosome, and MeCP2 binding does not lead to the characteristic zig-zag structures formed by interaction of H1 with chromatin [23].
These rich structural data seem difficult to reconcile with models depicting transcriptional regulatory events mediated by MeCP2 being restricted to recruitment of histone modification enzymes. Rather, they imply that the protein has an intrinsic capacity to organize chromatin in a manner similar to, but distinct from, linker histones. The key question is whether these in vitro observations have predictive power for the biology of MeCP2 in vivo.
3. MeCP2 as an architectural component of chromatin
Mutations in the human MeCP2 locus are known to be causal for the autism spectrum disorder Rett Syndrome [24], prompting the creation of animal models. Initial characterization of the potential impact of MECP2 deficiency on the transcriptome in these models failed to yield substantial insights into genetic targets [25]. These studies were performed with whole brain tissue, potentially masking specific alterations of a cell type specific nature. In fact, convincing evidence emerged that MeCP2 serves, consistent with existing models, as a corepressor for the brain-derived neurotrophic factor (BDNF) gene [26,27].
The lack of striking defects in global transcriptional profiling experiments has led some groups to search for unique functions for the protein that might be refractory to such analysis. Of particular interest are imprinted genes. Genomic imprinting, in which one and only one allele of a given gene is expressed depending on parent of origin, is altered in human diseases with neurological defects [28]. Loss of imprinting should, in theory, lead to a modest alteration in transcript levels which might be difficult to detect by microarray.
Given these potential connections, Terumi Kohwi-Shigematsu and colleagues investigated whether MeCP2 might be involved in regulating the allele specific expression pattern of imprinted genes [29]. Their studies focused on an imprinted gene cluster located on mouse chromosome 6 which includes several genes known to be imprinted in either mice or humans [30]. Two high affinity MeCP2 binding sites were identified in this locus by chromatin immunoprecipitation – these sites are separated by megabases of genomic DNA. Importantly, several genes in this cluster, including the Distal-less homeobox 5 protein (DLX5), were observed to lose monoallelic expression patterns in either brain tissue of MeCP2 null mice or in lymphoblast cell lines isolated from Rett Syndrome patients [29]. High resolution analysis of MeCP2 localization to the imprinted gene cluster revealed several sites of occupancy, which coincided with local enrichment of histone deacetylase [29]. On the surface, these observations seem entirely consistent with models depicting local recruitment of histone modification enzymes to genes by MeCP2 bound to methylated DNA. However, the sites involved were distant from the promoters of imprinted genes within the cluster. Further, while high affinity MeCP2 binding sites contained methylated CpG residues, the methylation density was not high and there were no apparent differences in methylation pattern between the two alleles. These observations suggested that the mode of regulation at this imprinted gene cluster was dependent on MeCP2 function, but might not fit with current paradigms.
Analysis of higher order chromatin structure revealed the presence of unusual chromatin organization at this imprinted cluster. Several long range interactions of distant sequences, separated by kilobases, were detected. A subset of these were detected in brain tissue from wild type, but not MeCP2 deficient mice [29]. On the surface, these observations appear to implicate long range chromatin interactions as integral to the appropriate regulation of an imprinted gene cluster and suggest that MeCP2 might regulate genes through architectural functions. These implications are entirely consistent with in vitro data indicating that MeCP2 has the capacity to induce formation of highly compact, ordered chromatin structures in a seemingly cooperative fashion. However, it should be noted that these observations are not universally accepted. In fact, a recent publication concludes that DLX5 is not imprinted at all in either mouse or human samples [31]. Obviously, further study is required to resolve the controversy regarding this particular gene cluster. However, the suggestion that MeCP2 might nucleate formation of higher order chromatin architecture remains attractive.
4. Association of MeCP2 with active genes
The emergence of the genomic era has provided a wealth of new research tools for studying the function of components of chromatin. One particularly valuable tool has been the application of chromatin immunoprecipitation as a tool to generate DNA probes for high density microarrays that tile regions of mammalian genomes. Such experiments have the potential to map, at high resolution, the association of proteins with genomic chromatin in living cells [32].
The application of this ChIP-chip technology is still in its initial stages and we do not yet have comprehensive data on the full MBD family in a broad range of biological samples. However, Janine Lasalle and colleagues have provided a glimpse of the potential surprises to come in a landmark paper describing the localization of MeCP2 across approximately 25 megabases of genomic chromatin in a cellular model of neuronal differentiation [33]. ChIP-chip analysis was performed using antibodies against MeCP2 in the human neuronal cell line SH-SY5Y. A modest number of MeCP2 binding sites of high confidence were observed, approximately 170. Surprisingly, a majority (approximately 60%) were located between genes with many sites being more than 10 kilobases distant from the nearest known gene [33]. In fact, this distribution was not significantly different from the distribution expected from random chance. A very small number were associated with CpG islands.
Perhaps the most surprising result of this study was the association of MeCP2 with transcriptionally active genes. Comparison of the ChIP-chip location analysis of MeCP2 binding with expression analysis indicated that roughly 2/3 of strongly MeCP2 bound promoters were transcriptionally active [33]. Clearly this intriguing result is not consistent with models depicting MeCP2 as a methylation dependent transcriptional repressor. Furthermore, DNA methylation analysis performed in the very same cell line indicated that only a very small percentage of highly methylated genes (2.2%) were also bound by MeCP2 [33]. Once again, these data seem to directly contradict the current paradigm for MeCP2 function stipulating that DNA methylation recruits MeCP2, which in turn recruits enzymatic activities that create locally repressive chromatin.
5. Summary and future perspectives
How do we rationalize the seemingly disparate stories describing MeCP2 biology? The in vitro biochemical properties of the protein seem to indicate that MeCP2 is highly unusual. It binds methylated DNA, but prefers a local sequence context [11]. It is also a general DNA binding protein and the difference in affinity for any DNA compared to methylated DNA is not huge [19,21]. High resolution imaging of the protein bound to DNA indicates the presence of multiple DNA interaction surfaces and a seemingly cooperative self-assembly process [21]. The protein has unusual interactions with chromatin fibers in the test tube. It appears to utilize multiple surfaces to bring individual nucleosomes into intimate contact [21,22]. It directs dramatic changes in solution properties of nucleosomal arrays inducing major alterations in conformation with some similarities to linker histones [22,23].
In cells and tissues, MeCP2 has been associated with gene silencing that appears to be local and mediated through recruitment of histone modification enzymes [26,27]. However, some data indicate that the protein itself is not strongly associated with such activities in the mammalian brain [34]. There are reports of MeCP2-dependent higher-order chromatin structures associated with imprinted gene clusters [29], although these have also been questioned [31]. In addition, MeCP2 has been implicated as an RNA binding protein involved in splicing of mRNA [35]. Finally, the question of where the protein is bound in vivo and what genes it associates with casts doubts on long-held models [33].
Alexander Pope in An Essay on Criticism warned that "A little learning is a dangerous thing; drink deep, or taste not the Pierian spring: there shallow draughts intoxicate the brain, and drinking largely sobers us again." The seemingly conflicting data on MeCP2 regarding what proteins it associates with, how it interacts with DNA and chromatin, what genes it regulates, and in what manner it affects gene expression serve as a stark reminder that we know very little about MeCP2 and the MBD family of which it is a member.
Critical challenges remain to be addressed in this area. We do not fully understand how DNA methylation in mammalian cells is distributed within the genome, or whether these patterns change during development or as a consequence of normal physiology. The MBD protein family has been proposed as potential readers of the DNA methylation mark. Yet it is clear that the phenotypic consequences of mutation of the system for deposition of DNA methylation are far more profound than are the consequences of mutation of the MBD proteins themselves [36]. The different members of the DNA methyltransferase family are essential in mammals, null mutants in MBD family members in mice are largely viable and fertile. This discrepancy implies that either we have not yet uncovered the entire protein family that interacts with methylated DNA, or that the functions of this essential DNA modification extend beyond the biology of the MBD family. The questions of where MBD proteins are localized and how this pattern responds to the biology of DNA methylation are currently under active investigation in a number of laboratories. The resulting answers will no doubt bring new surprises as well as some expected results. The ensuing discussions promise to make the study of MBD proteins, where they are localized in the genome, and how they impact chromosomal biology a topic of intense interest for years to come.
Acknowledgements
This research was supported (Project number 1Z01ES101965) by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. We thank the members of the Wade laboratory for useful discussions throughout the course of preparation of this manuscript. We apologize to our colleagues whose work could not be cited here due to space considerations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 2.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 3.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 6.Ohki I, Shimotake N, Fujita N, Nakao M, Shirakawa M. Solution structure of the methyl-CpG-binding domain of the methylation- dependent transcriptional repressor MBD1. Embo J. 1999;18:6653–6661. doi: 10.1093/emboj/18.23.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakefield RI, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, Bird AP, Barlow PN. The solution structure of the domain from MeCP2 that binds to methylated DNA. J Mol Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- 8.Heitmann B, Maurer T, Weitzel JM, Stratling WH, Kalbitzer HR, Brunner E. Solution structure of the matrix attachment region-binding domain of chicken MeCP2. Eur J Biochem. 2003;270:3263–3270. doi: 10.1046/j.1432-1033.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 9.Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 10.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 13.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex [see comments] Nature. 1998:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 14.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation [see comments] Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeke J, Ammerpohl O, Kegel S, Moehren U, Renkawitz R. The minimal repression domain of MBD2b overlaps with the methyl-CpG- binding domain and binds directly to sin3A [In Process Citation] J Biol Chem. 2000;275:34963–34967. doi: 10.1074/jbc.M005929200. [DOI] [PubMed] [Google Scholar]
- 17.Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 19.Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J Biol Chem. 282;2007:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- 20.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 23.Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J Biol Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 24.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl- CpG-binding protein 2 [see comments] Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 25.Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 27.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 28.Lalande M. Parental imprinting and human disease. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 29.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 30.Ono R, Shiura H, Aburatani H, Kohda T, Kaneko-Ishino T, Ishino F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003;13:1696–1705. doi: 10.1101/gr.906803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schule B, Li HH, Fisch-Kohl C, Purmann C, Francke U. DLX5 and DLX6 expression is biallelic and not modulated by MeCP2 deficiency. Am J Hum Genet. 2007;81:492–506. doi: 10.1086/520063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TH, Barrera LO, Ren B. ChIP-chip for genome-wide analysis of protein binding in mammalian cells. Curr Protoc Mol Biol Chapter. 2007;21 doi: 10.1002/0471142727.mb2113s79. Unit 21 13. [DOI] [PubMed] [Google Scholar]
- 33.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu K, Nan X, Bird A, Wang W. Testing for association between MeCP2 and the brahma-associated SWI/SNF chromatin-remodeling complex. Nat Genet. 2006;38:962–964. doi: 10.1038/ng0906-962. author reply 964–967. [DOI] [PubMed] [Google Scholar]
- 35.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]