Abstract
Sand fly-parasite and sand fly-host interactions play an important role in the transmission of leishmaniasis. Vector molecules relevant for such interactions include midgut and salivary proteins. These potential targets for interruption of propagation of Leishmania parasites have been poorly characterized. Transcriptomic analysis has proven to be an effective tool for identification of new sand fly molecules, providing exciting new insights into vector-based control strategies against leishmaniasis.
Keywords: Saliva, Midgut, Vector-based vaccine, Transmission-blocking vaccine, Salivary gland
1. Introduction
Leishmaniasis is a vector-borne neglected infectious disease that afflicts 88 countries with an estimated incidence of two million new cases each year [1]. With expanding endemicity, an estimated 350 million people at risk and 2,357,000 disability-adjusted life years lost, leishmaniasis is becoming a worldwide re-emerging public health problem. One intriguing aspect of leishmaniasis is the wide spectrum of distinct clinical manifestations that include visceral, cutaneous, mucocutaneous, and diffuse cutaneous leishmaniasis.
Leishmaniasis is sustained through a triad of complex interactions between Leishmania parasites, the sand fly, and the mammalian host. In vector sand fly species, Leishmania parasites undergo a complex developmental cycle within the midgut that is necessary for generation of infectious metacyclics (vector-parasite interface). In addition, the natural mode of transmission to the mammalian host is by the bite of an infective sand fly. At the bite site, sand flies release an array of pharmacologic, immunomodulatory, and immunogenic molecules that have immediate and long-lasting effects on the host (the vector-host interface). The availability of high-throughput approaches, mainly tissue-specific transcriptomes, has facilitated the identification of pertinent vector molecules that affect the development of the Leishmania parasite, its transmission, and its establishment in the mammalian host. This information can lead to novel strategies for the control of leishmaniasis.
2. Midgut
2.1. The sand fly-Leishmania molecular interface
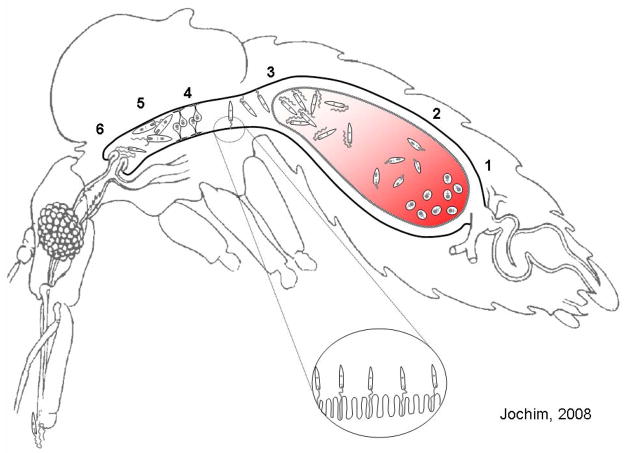
The Phlebotomus (Old World) and Lutzomyia (New World) genera include the majority of anthropophilic sand flies and are the most important vectors of leishmaniasis. Establishment of a transmissible Leishmania infection within the vector sand fly occurs solely within the lumen of the midgut. Once a sand fly feeds on an infectious host, it ingests a blood meal containing Leishmania-infected macrophages, beginning the life cycle in the sand fly (Fig. 1). Amastigotes are released after rupture of the macrophage and differentiate into several developmental stages, from flagellated procyclics to infectious-stage metacyclic promastigotes (Fig 1).
Fig. 1.
Lifecycle of Leishmania parasites within the sand fly. Promastigote forms: 1, amastigote; 2, procyclic; 3, nectomonad; 4, haptomonad; 5, leptomonad; 6, metacyclic. (Adapted from Schlein Y., Leishmania and sandflies: interactions in the life cycle and transmission. Parasitol Today 1993;9:255–8.)
Within the sand fly midgut are numerous natural barriers to parasite development, including resistance to digestive enzymes, escaping the peritrophic matrix (PM), and binding to the midgut epithelium. The midgut of a sand fly is therefore a fundamental organ representing a key target for interruption of Leishmania development and transmission. Despite the importance of this organ, very few molecules in the midgut of sand flies have been characterized to date.
2.2. Transcriptomics meets biology
Transcriptomics is a powerful tool for rapid identification of molecules expressed in a whole organism or particular tissue. Dillon et al. [2] generated 10,203 transcripts using whole Lutzomyia longipalpis sand flies that combined unfed, blood-fed, and flies infected with a variety of pathogens including Leishmania, providing a global descriptive repertoire of sand fly molecules. This was followed by more refined midgut-specific analysis of 2,934 transcripts from Lu. longipalpis [3] and 1,382 transcripts from Phlebotomus papatasi [4], offering a better characterization of midgut molecules and revealing for the first time the ability of Leishmania parasites to modulate vector midgut transcripts.
Following is an account of molecules identified through tissue-specific transcriptomic analysis that refine our understanding of key biologic processes within the sand fly midgut.
2.2.1. Midgut proteases
Midgut proteases facilitate blood-meal digestion and are likely to confer some defense against ingested organisms. The presence of Leishmania promastigotes in the midgut lumen of sand flies has been shown to inhibit proteolytic activity [5,6]. Infections initiated with Leishmania amastigotes, a more natural mode of infection, also caused a delay in trypsin and aminopeptidase activity [7]. Until recently, it has been unclear which specific proteolytic enzymes are regulated by the presence of the parasite, and knowledge of the full repertoire of sand fly midgut proteases was not available. An expressed sequence tag (EST) library using whole flies of Lu. longipalpis identified families of proteases such as trypsins, chymotrypsins, aminopeptidases, and carboxypeptidases [2]. Midgut-specific full-length cDNA libraries of the sand flies P. papatasi and Lu. longipalpis combined with customized bioinformatic analysis confirmed that these molecules are midgut proteases [3,4]. They also identified novel trypsins, chymotrypsins, carboxypeptidases, a serine protease, and an astacin-like metalloprotease present in the midgut of these vectors [3,4].
2.2.2. Midgut proteases modulated by blood
Comparison of unfed and blood-fed cDNA libraries demonstrated that most of the transcripts coding for proteases are upregulated by blood feeding, including one trypsin (PpTryp4), a chymotrypsin (Ppchym2 and LuloChym3), and two carboxypeptidases (LuloCpepA1 and LuloCpepB) [3,4]. Conversely, another trypsin (PpTryp1) and a chymotrypsin (LuloChym4) were downregulated by the blood meal, indicating that not all trypsins and chymotrypsins function in the same manner.
2.2.3. Midgut proteases modulated by Leishmania
Further comparison of blood-fed and Leishmania-infected cDNA libraries identified midgut molecules modulated by the presence of Leishmania parasites [3,4]. The presence of Leishmania in the sand fly midgut was shown to decrease the abundance (possibly a result of downregulation) of a transcript coding for a chymotrypsin molecule (Ppchym2 in P. papatasi and LuloChym1A in Lu. longipalpis) and to increase the abundance (which may be due to upregulation) of a trypsin molecule (PpTryp1 in P. papatasi and Lltryp2 in Lu. longipalpis) [3,4]. The presence of Leishmania also appears to modulate the abundance of transcripts expressed after the blood meal has been digested (5–7 days post-infection). A trypsin-like molecule Lltryp2 was more abundant, while LuloTryp3 transcripts were decreased by the presence of Leishmania [3]. This was the first report of the identity of the proteases specifically regulated by the presence of Leishmania parasites.
2.2.4. Peritrophic matrix
The proliferation and differentiation of the first parasite stages occur within the PM, a proteo-chitin structure formed to encapsulate the blood meal after feeding. The PM offers a protected environment during the first hours following ingestion of a blood meal, as amastigotes are susceptible to killing by digestive enzymes during their transformation to promastigotes [8]. Promastigotes are released into the lumen of the midgut following degradation of the PM. Schlein et al. [9] reported the absence of chitinolytic activity in uninfected P. papatasi midguts and attributed the breakdown of the PM solely to Leishmania chitinases. This was contested by the demonstration of an active chitinolytic system from the midgut of blood-fed P. papatasi [10]. The identity of the sand fly chitinase was validated by transcriptomic analysis. This will permit future studies of its effect on parasite development. Inhibition of the activity of the sand fly chitinase may prevent degradation of the PM and escape of the parasites into the midgut lumen. If this is the case, it may represent another attractive target for a vector-based transmission-blocking strategy.
Similar to chitinase, it is prudent to theorize that the Leishmania parasite may influence other sand fly molecules such as peritrophins, protein components of the PM, to ensure its escape to the midgut lumen. Two types of peritrophin molecules have been identified in the midgut transcriptomes of P. papatasi and Lu. longipalpis: multi-peritrophin domain proteins (PpPer1, 4 domains; PpPer3, 3 domains; and LuloPer1, 4 domains; likely necessary for crosslinking of chitin fibrils), and single-peritrophin domain proteins (PpPer2, LuloPer2, and LuloPer3) [3,4].
2.2.5. PM molecules modulated by Leishmania
P. papatasi infected with L. major downregulated the multi-domain peritrophin (PpPer1), whereas Lu. longipalpis infected with L. infantum chagasi upregulated the orthologous peritrophin (LuloPer1) [3,4]. The duality of the PM in Leishmania colonization (early protection from enzymes and the necessity for escape at a later time point) may provide a third target for transmission-blocking vaccines, either by disrupting early PM formation or by preventing PM dissociation and parasite escape.
2.2.6. Epithelial-parasite attachment
Once free of the PM, the procyclic promastigotes must adhere to the midgut epithelium to prevent its expulsion during defecation of digested blood. The outer surface of procyclic promastigotes is covered by a dense layer of lipophosphoglycans (LPGs), glycoconjugates with multiple functions [11]. LPG has been shown to restrict vector competence of certain sand fly species such as P. sergenti and P. papatasi as the ligand necessary for parasite attachment to the midgut epithelium [12–14]. A galactose binding protein, PpGalec, was identified as a relatively abundant transcript from the unfed midgut cDNA library of P. papatasi and was shown to be the midgut receptor for L. major [13]. PpGalec, a tandem repeat galectin on the luminal midgut epithelium, binds specifically to the LPG galactose residues of L. major, facilitating species-specific vector competence [13]. Additionally, PpGalec was the first molecule identified as a vector-based Leishmania transmission-blocking vaccine and provided evidence that blocking parasite binding to the midgut epithelium abrogated development of a transmissible infection [13]. A number of putative galectin molecules were identified in whole-fly analysis of Lu. longipalpis ESTs [2]; however, in the analysis of the Lu. longipalpis midgut-specific transcriptome, only one low-abundance transcript was identified, which was homologous to a single-domain galectin (GenBank: ABV60341) [3]. It is unlikely that this galectin acts as a receptor for L. infantum chagasi in Lu. longipalpis. This sand fly species is considered a permissive vector supporting mature infections of several different species of Leishmania under laboratory conditions [15]. Recent work using LPG-deficient L. major parasites demonstrated that LPG is not required for development of heavy promastigote infections in the permissive vectors Lu. longipalpis and P. arabicus [16]. The authors hypothesized that N-acetyl galactosamine-containing glycoproteins on the midgut epithelia of permissive vectors are ligands to which an as-yet-unidentified parasite lectin receptor binds, allowing full development of several different Leishmania species [16].
2.2.7. Other midgut proteins modulated by Leishmania
Transcripts coding for microvilli protein-like molecules from Lu. longipalpis (LuloMVP1, 2, 4, and 5) and P. papatasi (PpMVP1, PpMVP2) were downregulated in the presence of L. infantum chagasi and L. major infections, respectively [3,4]. This could be a reflection of their importance in parasite development. Of interest, these microvilli protein-like molecules represented the most abundant transcripts from the midgut of these sand fly species [3,4]. These proteins are approximately 20 kDa with a predicted signal secretory peptide, and their function remains unknown. Other proteins of unknown function were also modulated by the presence of Leishmania parasites. These include a protein of 29 kDa (EU124578) from the sand fly Lu. longipalpis that was more abundant in the presence of L. infantum chagasi [3] and two proteins from P. papatasi with a predicted molecular weight of 14.5 kDa (EU045347) and 50 kDa (EU045345) that were less abundant in the presence of L. major parasites [4].
2.3. Vector-based transmission-blocking vaccines
The above shows the power of transcriptomics in identifying midgut molecules pertinent to sand fly-Leishmania interactions. Functional studies will provide further insight into their relevance in sand fly biology and as potential targets for use as novel vector-based transmission-blocking vaccines to control leishmaniasis. The validity of this strategy was demonstrated by the disruption of L. major transmission through blocking PpGalec, its midgut receptor in P. papatasi [13].
3. Salivary glands
3.1. The sand fly-host molecular interface
In addition to the midgut, salivary glands represent another tissue of significance in the biology of sand flies as vectors of leishmaniasis. A fact often overlooked is the obligatory co-inoculation of Leishmania parasites with saliva. This initial brief encounter within the skin of the vertebrate host is the fundamental reason why saliva is relevant to every transmission event. In this instance, salivary glands impact the vertebrate host through secretion of a complex array of pharmacologic compounds that have evolved to facilitate blood feeding [17] but were shown to be both immunomodulatory (acting on the innate immune system) [18,19] and immunogenic (inducing an adaptive immune response) [20–23]. This alteration of the host immune status has important repercussions on survival of the Leishmania parasite and establishment of disease.
3.2. The first encounter—sand fly saliva and the naïve host
It has been shown that infection with L. major is significantly enhanced by saliva of the vector sand fly species P. papatasi and Lu. longipalpis [20,24–26]. The ability of saliva to enhance Leishmania infection has been attributed to modulation of the host immune system, potentially through anti-inflammatory properties described for Lu. longipalpis, P. duboscqi, and P. papatasi [18,19,27,28]. Such activities include downregulation of antigen presentation, co-stimulatory molecule expression, and nitric oxide production [29–32]. Disease enhancement by saliva is especially pronounced in the first encounter with a naïve animal, where immunomodulation is not diluted or ablated by an adaptive immune response to salivary proteins of a previously exposed host.
3.3 Memories—the host mounts an immune response to salivary proteins
Apart from their inherent pharmacologic, immunomodulatory, and anti-inflammatory activities, salivary proteins are immunogenic in several different species including humans [20,23,33–35]. It is important to note that immunity to sand fly saliva induced by salivary gland homogenate (SGH) injection or by bites of uninfected sand flies was shown to be protective against Leishmania infection in murine models [20,36,37]. Furthermore, immunization with two sand fly salivary proteins, maxadilan from Lu. longipalpis and PpSP15 from P. papatasi, has been shown to protect against leishmaniasis in mice [25,38].
3.3.1. Anti-saliva antibodies—do they play a role?
Interpretation of the significance of anti-saliva antibodies in leishmaniasis remains troublesome. A positive correlation was observed between protection from visceral leishmaniasis and intensity of Lu. longipalpis salivary antibodies [23,35]. Conversely, patients with cutaneous leishmaniasis had higher titers of anti-saliva antibodies associating them with disease [39,40]. Evidence from murine models indicates that anti-saliva antibodies are not required for protection, at least against L. major infection [38]. In that study, B cell-deficient C57BL/6 mice immunized with PpSP15, the salivary protein from P. papatasi, were protected from L. major infection, suggesting that cellular immunity observed in the form of a delayed-type hypersensitivity (DTH) response is sufficient to confer full protection [38].
3.3.2. Cellular immunity—a necessity?
In humans, the presence of a DTH response to bites of sand flies has been well documented [41]. The significance of this DTH response in protection from leishmaniasis was first demonstrated in murine models of cutaneous leishmaniasis and was correlated with the production of IL-12 and IFN-γ [36,38]. Recently, a subset of human volunteers repeatedly exposed to Lu. longipalpis produced a DTH response at the bite site. Peripheral blood mononuclear cells isolated from these individuals induced IFN-γ upon stimulation with sand fly SGH and controlled parasite growth in vitro [42]. This suggests that the correlates of protection from Leishmania infection demonstrated for rodent models may apply to humans as well. Nevertheless, outbred populations including humans probably recognize and mount immunity to different proteins within the saliva. Therefore, identification of immunodominant salivary proteins that can elicit a Th1-type DTH response should lead to the discovery of a protective salivary molecule to control Leishmania infection.
3.3.3. How does anti-saliva immunity control Leishmania infection?
Challenged in the absence of saliva, animals immunized with sand fly salivary proteins do not control Leishmania infection (Oliveira, unpublished results). These data suggest that the anti-saliva immune response is not directed against Leishmania parasites. We hypothesize that a DTH response to saliva affects the initial steps in establishment of Leishmania infection in the mammalian host. This anti-saliva immune response may alter the type and activation of macrophages or other host cells that otherwise would silently maintain the parasites. This could result in direct killing of Leishmania parasites, thus reducing the infective load. Additionally, a Th1 anti-saliva immunity may create an environment that accelerates priming of a protective Th1 anti-Leishmania immunity. Under these circumstances, any protein that induces a Th1 response in the dermis would affect Leishmania infection. The significance of anti-saliva immunity lies in the fact that, in nature, these sand fly salivary proteins will always be present at the site of Leishmania deposition during transmission. Indeed, salivary proteins can be considered ‘non-classical natural adjuvants’.
3.4. Sand fly salivary gland transcriptomics
Transcriptomics represent a rapid and efficient method to identify the most abundant secreted proteins from salivary glands of pertinent vectors of disease. Use of sand fly salivary gland transcriptomics resulted in the identification of complete sets of secreted salivary proteins from glands of several relevant vectors of cutaneous (P. papatasi, P. duboscqi) [38,43] and visceral (P. argentipes, P. ariasi, P. perniciosus, and Lu. longipalpis) leishmaniasis [44–46]. This is of particular significance since the sequence of the sand fly genome is not yet available.
3.4. Transcriptomics and anti-saliva immunity
The potential of anti-saliva immunity in protecting against leishmaniasis represents an untapped approach that may result in production of better vaccines. Through transcriptomic analysis, customized bioinformatics, and high-throughput DNA vaccination, we were able to screen complete repertoires of highly abundant salivary proteins in search of Th1 DTH-inducing molecules [45,47,48].
The salivary gland transcriptome of P. papatasi identified two DTH-inducing molecules that produced contrasting protective (PpSP15) and exacerbative (PpSP44) outcomes of L. major infection [47]. This study demonstrated that not all DTH-inducing molecules are protective and that some produce a Th2 profile that is exacerbative [47]. It also validated the transcriptomic approach for identification of protective molecules by corroborating the protective nature of PpSP15 against L. major infection in mice [47]. The contrasting immune responses to PpSP15 and PpSP44 provided the first evidence that anti-saliva immunity alters the environment in the skin hours following sand fly bites. This could favor or hinder the establishment of Leishmania parasites, depending on the nature of the salivary protein [47]. Another testament to the value of transcriptomics is the demonstration that immunity to a defined salivary protein (LJM19), identified from the salivary transcriptome of Lu. longipalpis [46], protected from the fatal outcome of visceral leishmaniasis in hamsters [48]. The systemic protection from L. infantum chagasi conferred by immunization with LJM19 further alludes to the effect of anti-saliva immunity on priming a Th1 anti-Leishmania immune response.
Despite the powerful protection observed in rodents immunized with salivary proteins, their mode of infection (injection of SGH and parasites) challenges their efficacy under field conditions. It is prudent to test these promising vaccine candidates by a more natural route of transmission (bites of experimentally infected sand flies).
3.6. Comparative salivary gland transcriptomics
When considering sand fly salivary proteins as potential anti-Leishmania vaccines, further information is needed regarding the diversity or similarity of these proteins among different sand fly species and populations. Comparative transcriptomic analysis of salivary glands from different sand fly species revealed the presence of both common proteins and genus-specific salivary molecules [44]. Among the salivary proteins shared by at least by five different sand fly species, including two different genera (Phlebotomus and Lutzomyia), are the PpSP15-like proteins, apyrases, yellow-related proteins, antigen 5-related proteins, PpSP32-like proteins, 33-kDa proteins, D7-related proteins, and an endonuclease. The level of similarity between these proteins among different species indicates that salivary vaccines may work at the species level or even within a single genus [44]. This is further supported by the high level of conservation observed in salivary proteins from P. duboscqi sand flies at the ends of its geographic distribution (Mali to the west and Kenya to the east). Conserved regions included the predicted MHC class II T cell epitopes of PpSP15-like, D7-related, PpSP32-like, antigen 5-related, apyrase, and yellow-related salivary proteins [43].
4. Overall conclusion
The genomes of P. papatasi and Lu. longipalpis are currently in their infancy. This underlines the value of tissue-specific transcriptomics as a powerful approach for identification of vector-based, salivary gland- and midgut-specific, vaccine candidates against leishmaniasis.
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank NIAID intramural editor Brenda Rae Marshall for assistance.
Because the authors are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Abbreviations used
- DTH
delayed-type hypersensitivity
- EST
expressed sequence tag
- LPG
lipophosphoglycans
- PM
peritrophic matrix
- SGH
salivary gland homogenate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27:305–18. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Dillon RJ, Ivens AC, Churcher C, Holroyd N, Quail MA, Rogers ME, Soares MB, Bonaldo MF, Casavant TL, Lehane MJ, Bates PA. Analysis of ESTs from Lutzomyia longipalpis sand flies and their contribution toward understanding the insect-parasite relationship. Genomics. 2006;88:831–40. doi: 10.1016/j.ygeno.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jochim RC, Teixeira CR, Laughinghouse A, Mu J, Oliveira F, Gomes RB, Elnaiem DE, Valenzuela JG. The midgut transcriptome of Lutzomyia longipalpis: comparative analysis of cDNA libraries from sugar-fed, blood-fed, post-digested and Leishmania infantum chagasi-infected sand flies. BMC Genomics. 2008;9:15. doi: 10.1186/1471-2164-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramalho-Ortigão M, Jochim RC, Anderson JM, Lawyer PG, Pham VM, Kamhawi S, Valenzuela JG. Exploring the midgut transcriptome of Phlebotomus papatasi: comparative analysis of expression profiles of sugarfed, blood-fed and Leishmania-major-infected sandflies. BMC Genomics. 2007;8:300. doi: 10.1186/1471-2164-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlein Y, Romano H. Leishmania major and L. donovani: effects on proteolytic enzymes of Phlebotomus papatasi (Diptera, Psychodidae) Exp Parasitol. 1986;62:376–80. doi: 10.1016/0014-4894(86)90045-7. [DOI] [PubMed] [Google Scholar]
- 6.Borovsky D, Schlein Y. Trypsin and chymotrypsin-like enzymes of the sandfly Phlebotomus papatasi infected with Leishmania and their possible role in vector competence. Med Vet Entomol. 1987;1:235–42. doi: 10.1111/j.1365-2915.1987.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 7.Dillon RJ, Lane RP. Influence of Leishmania infection on blood-meal digestion in the sandflies Phlebotomus papatasi and P. langeroni. Parasitol Res. 1993;79:492–6. doi: 10.1007/BF00931590. [DOI] [PubMed] [Google Scholar]
- 8.Pimenta PF, Modi GB, Pereira ST, Shahabuddin M, Sacks DL. A novel role for the peritrophic matrix in protecting Leishmania from the hydrolytic activities of the sand fly midgut. Parasitology. 1997;115:359–69. doi: 10.1017/s0031182097001510. [DOI] [PubMed] [Google Scholar]
- 9.Schlein Y, Jacobson RL, Shlomai J. Chitinase secreted by Leishmania functions in the sandfly vector. Proc Biol Sci. 1991;245:121–6. doi: 10.1098/rspb.1991.0097. [DOI] [PubMed] [Google Scholar]
- 10.Ramalho-Ortigão JM, Kamhawi S, Joshi MB, Reynoso D, Lawyer PG, Dwyer DM, Sacks DL, Valenzuela JG. Characterization of a blood activated chitinolytic system in the midgut of the sand fly vectors Lutzomyia longipalpis and Phlebotomus papatasi. Insect Mol Biol. 2005;14:703–12. doi: 10.1111/j.1365-2583.2005.00601.x. [DOI] [PubMed] [Google Scholar]
- 11.Descoteaux A, Turco SJ. Glycoconjugates in Leishmania infectivity. Biochim Biophys Acta. 1999;1455:341–52. doi: 10.1016/s0925-4439(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 12.Kamhawi S, Modi GB, Pimenta PF, Rowton E, Sacks DL. The vectorial competence of Phlebotomus sergenti is specific for Leishmania tropica and is controlled by species-specific, lipophosphoglycan-mediated midgut attachment. Parasitology. 2000;121:25–33. doi: 10.1017/s0031182099006125. [DOI] [PubMed] [Google Scholar]
- 13.Kamhawi S, Ramalho-Ortigão M, Pham VM, Kumar S, Lawyer PG, Turco SJ, Barillas-Mury C, Sacks DL, Valenzuela JG. A role for insect galectins in parasite survival. Cell. 2004;119:329–41. doi: 10.1016/j.cell.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Kamhawi S. Phlebotomine sand flies and Leishmania parasites: friends or foes? Trends Parasitol. 2006;22:439–45. doi: 10.1016/j.pt.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Sacks D, Kamhawi S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu Rev Microbiol. 2001;55:453–83. doi: 10.1146/annurev.micro.55.1.453. [DOI] [PubMed] [Google Scholar]
- 16.Myskova J, Svobodova M, Beverley SM, Volf P. A lipophosphoglycan-independent development of Leishmania in permissive sand flies. Microbes Infect. 2007;9:317–24. doi: 10.1016/j.micinf.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 18.Titus RG, Bishop JV, Mejia JS. The immunomodulatory factors of arthropod saliva and the potential for these factors to serve as vaccine targets to prevent pathogen transmission. Parasite Immunol. 2006;28:131–41. doi: 10.1111/j.1365-3024.2006.00807.x. [DOI] [PubMed] [Google Scholar]
- 19.Gillespie RD, Mbow ML, Titus RG. The immunomodulatory factors of bloodfeeding arthropod saliva. Parasite Immunol. 2000;22:319–31. doi: 10.1046/j.1365-3024.2000.00309.x. [DOI] [PubMed] [Google Scholar]
- 20.Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, Rowton E, Ribeiro J, Sacks DL. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med. 1998;188:1941–53. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belkaid Y, Valenzuela JG, Kamhawi S, Rowton E, Sacks DL, Ribeiro JM. Delayed-type hypersensitivity to Phlebotomus papatasi sand fly bite: an adaptive response induced by the fly? Proc Natl Acad Sci USA. 2000;97:6704–9. doi: 10.1073/pnas.97.12.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva F, Gomes R, Prates D, Miranda JC, Andrade B, Barral-Netto M, Barral A. Inflammatory cell infiltration and high antibody production in BALB/c mice caused by natural exposure to Lutzomyia longipalpis bites. Am J Trop Med Hyg. 2005;72:94–8. [PubMed] [Google Scholar]
- 23.Barral A, Honda E, Caldas A, Costa J, Vinhas V, Rowton ED, Valenzuela JG, Charlab R, Barral-Netto M, Ribeiro JM. Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am J Trop Med Hyg. 2000;62:740–5. doi: 10.4269/ajtmh.2000.62.740. [DOI] [PubMed] [Google Scholar]
- 24.Titus RG, Ribeiro JM. Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science. 1988;239:1306–8. doi: 10.1126/science.3344436. [DOI] [PubMed] [Google Scholar]
- 25.Morris RV, Shoemaker CB, David JR, Lanzaro GC, Titus RG. Sandfly maxadilan exacerbates infection with Leishmania major and vaccinating against it protects against L. major infection. J Immunol. 2001;167:5226–30. doi: 10.4049/jimmunol.167.9.5226. [DOI] [PubMed] [Google Scholar]
- 26.Norsworthy NB, Sun J, Elnaiem D, Lanzaro G, Soong L. Sand fly saliva enhances Leishmania amazonensis infection by modulating interleukin-10 production. Infect Immun. 2004;72:1240–7. doi: 10.1128/IAI.72.3.1240-1247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteiro MC, Lima HC, Souza AA, Titus RG, Romão PR, Cunha FQ. (Effect of Lutzomyia longipalpis salivary gland extracts on leukocyte migration induced by Leishmania major. Am J Trop Med Hyg. 2007;76:88–94. [PubMed] [Google Scholar]
- 28.Carregaro V, Valenzuela JG, Cunha TM, Verri WA, Jr, Grespan R, Matsumura G, Ribeiro JM, Elnaiem DE, Silva JS, Cunha FQ. Phlebotomine salivas inhibit immune inflammation-induced neutrophil migration via an autocrine DC-derived PGE2/IL-10 sequential pathway. J Leukoc Biol. 2008;84:104–14. doi: 10.1189/jlb.1107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall LR, Titus RG. Sand fly vector saliva selectively modulates macrophage functions that inhibit killing of Leishmania major and nitric oxide production. J Immunol. 1995;155:3501–6. [PubMed] [Google Scholar]
- 30.Theodos CM, Titus RG. Salivary gland material from the sand fly Lutzomyia longipalpis has an inhibitory effect on macrophage function in vitro. Parasite Immunol. 1993;15:481–7. doi: 10.1111/j.1365-3024.1993.tb00634.x. [DOI] [PubMed] [Google Scholar]
- 31.Costa DJ, Favali C, Clarêncio J, Afonso L, Conceição V, Miranda JC, Titus RG, Valenzuela J, Barral-Netto M, Barral A, Brodskyn CI. Lutzomyia longipalpis salivary gland homogenate impairs cytokine production and costimulatory molecule expression on human monocytes and dendritic cells. Infect Immun. 2004;72:1298–305. doi: 10.1128/IAI.72.3.1298-1305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers KA, Titus RG. Immunomodulatory effects of Maxadilan and Phlebotomus papatasi sand fly salivary gland lysates on human primary in vitro immune responses. Parasite Immunol. 2003;25:127–34. doi: 10.1046/j.1365-3024.2003.00623.x. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh KN, Mukhopadhyay J. The effect of anti-sandfly saliva antibodies on Phlebotomus argentipes and Leishmania donovani. Int J Parasitol. 1998;28:275–81. doi: 10.1016/s0020-7519(97)00152-5. [DOI] [PubMed] [Google Scholar]
- 34.Gomes RB, Mendonça IL, Silva VC, Ruas J, Silva MB, Cruz MS, Barral A, Costa CH. Antibodies against Lutzomyia longipalpis saliva in the fox Cerdocyon thous and the sylvatic cycle of Leishmania chagasi. Trans R Soc Trop Med Hyg. 2007;101:127–33. doi: 10.1016/j.trstmh.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Gomes RB, Brodskyn C, de Oliveira CI, Costa J, Miranda JC, Caldas A, Valenzuela JG, Barral-Netto M, Barral A. Seroconversion against Lutzomyia longipalpis saliva concurrent with the development of anti-Leishmania chagasi delayed-type hypersensitivity. J Infect Dis. 2002;186:1530–4. doi: 10.1086/344733. [DOI] [PubMed] [Google Scholar]
- 36.Kamhawi S, Belkaid Y, Modi G, Rowton E, Sacks D. Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science. 2000;290:1351–4. doi: 10.1126/science.290.5495.1351. [DOI] [PubMed] [Google Scholar]
- 37.Thiakaki M, Rohousova I, Volfova V, Volf P, Chang KP, Soteriadou K. Sand fly specificity of saliva-mediated protective immunity in Leishmania amazonensis-BALB/c mouse model. Microbes Infect. 2005;7:760–6. doi: 10.1016/j.micinf.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JM. (Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med. 2001;194:331–42. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Moura TR, Oliveira F, Novais FO, Miranda JC, Clarêncio J, Follador I, Carvalho EM, Valenzuela JG, Barral-Netto M, Barral A, Brodskyn C, de Oliveira CI. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl Trop Dis. 2007;1:e84. doi: 10.1371/journal.pntd.0000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohousova I, Ozensoy S, Ozbel Y, Volf P. Detection of species-specific antibody response of humans and mice bitten by sand flies. Parasitology. 2005;130:493–9. doi: 10.1017/s003118200400681x. [DOI] [PubMed] [Google Scholar]
- 41.Theodor O. A study of the reaction to Phlebotomus bites with some remarks on ‘Harara’. Trans Roy Soc Trop Med Hyg. 1935;29:273–84. [Google Scholar]
- 42.Vinhas V, Andrade BB, Paes F, Bomura A, Clarêncio J, Miranda JC, Báfica A, Barral A, Barral-Netto M. Human anti-saliva immune response following experimental exposure to the visceral leishmaniasis vector, Lutzomyia longipalpis. Eur J Immunol. 2007;37:3111–21. doi: 10.1002/eji.200737431. [DOI] [PubMed] [Google Scholar]
- 43.Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, Sangare CS, Samake S, Sissoko I, Garfield M, Sigutova L, Volf P, Doumbia S, Valenzuela JG. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya) BMC Genomics. 2006;7:226. doi: 10.1186/1471-2164-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson JM, Oliveira F, Kamhawi S, Mans BJ, Reynoso D, Seitz AE, Lawyer P, Garfield M, Pham M, Valenzuela JG. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics. 2006;7:52. doi: 10.1186/1471-2164-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliveira F, Kamhawi S, Seitz AE, Pham VM, Guigal PM, Fischer L, Ward J, Valenzuela JG. (From transcriptome to immunome: identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2006;24:374–90. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- 46.Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J Exp Biol. 2004;207:3717–29. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- 47.Oliveira F, Lawyer PG, Kamhawi S, Valenzuela JG. Immunity to distinct sand fly salivary proteins primes the anti-Leishmania immune response towards protection or exacerbation of disease. PLoS Negl Trop Dis. 2008;2:e226. doi: 10.1371/journal.pntd.0000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes R, Teixeira C, Teixeira MJ, Oliveira F, Menezes MJ, Silva C, de Oliveira CI, Miranda JC, Elnaiem DE, Kamhawi S, Valenzuela JG, Brodskyn CI. Immunity to a salivary protein of a sand fly vector protects against the fatal outcome of visceral leishmaniasis in a hamster model. Proc Natl Acad Sci U S A. 2008;105:7845–50. doi: 10.1073/pnas.0712153105. [DOI] [PMC free article] [PubMed] [Google Scholar]