Summary
Microtubule based motility is often thought of as specifically referring to the directed stepping of microtubule based motors such as kinesin or dynein. However microtubule lattice diffusion (also known as diffusional motility) provides a second mode of transport that is shared by a much broader class of microtubule binding proteins. Microtubule lattice diffusion offers distinct advantages as a transport mechanism including speed, bi-directional microtubule end targeting, and no requirement for direct chemical energy (ie. ATP). It remains to be seen whether a universal binding mechanism for this interaction will be identified but electrostatic interactions appear to play a significant role. In the meantime, the well-studied subject of DNA binding proteins that diffuse along the DNA backbone provides an insightful analogue for understanding the nature of microtubule based diffusional motility.
Introduction
The microtubule cytoskeleton provides the scaffold for a multitude of protein-protein interactions. Proteins that are able to bind directly to microtubules form a diverse ensemble referred to as microtubule binding proteins. Microtubule binding proteins, when examined at the level of single molecules, tend not to remain statically bound to the microtubule, but instead travel along the surface of the microtubule. Perhaps the most familiar example of such translocation is the directed motility exhibited by kinesin molecules, which step in a constant direction along the microtubule lattice. However a second mechanism, referred to as diffusional motility (Figure 1), has generated a significant amount of interest as an increasing number of microtubule binding proteins have been found to exhibit this behavior. Diffusional motility may even contribute to the stepping mechanism in kinesins by means of a diffusional search process of the free motor domain head [1], (also see companion review in this issue from Gennerich and Vale).
Figure 1. Diffusive and directed modes of microtubule based motility.
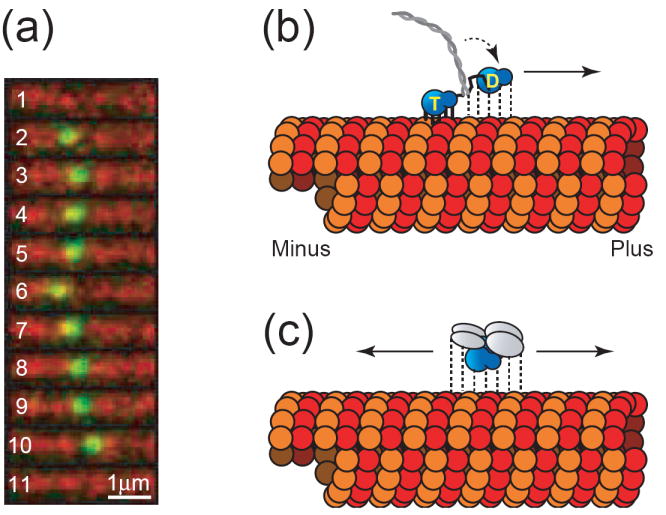
(a) Still frame series exhibiting unbiased diffusional motility of a single MCAK molecule. The molecule binds to the microtubule lattice (frame 2), diffuses 1-dimensionally along the microtubule lattice (frames 2-10), and then detaches (frame 10). Frames 1 and 11 show the microtubule immediately before and immediately after the binding event, respectively. Frame rate is 5 frames per second. GFP-labeled MCAK is shown in green and Cy5-labeled microtubules are shown in red (unpublished data). (b) Model of hand-over-hand directed motility of a kinesin motor. The two motor domain heads alternate between strongly bound (solid lines) and weakly bound states (dashed lines) while hydrolyzing ATP to advance toward the microtubule plus end. The strongly bound head possesses ATP (labeled “T”) in its nucleotide-binding pocket, while the weakly bound head has hydrolyzed its nucleotide to ADP (labeled “D”). During interludes where both motor domain heads possess ADP (not shown), the entire molecule would be weakly bound which can explain the diffusional component sometimes observed superimposed on top of directed motility [9]. (c) Model of unbiased diffusional motility of MCAK. The motor domain is shown in blue while the n-terminal and c-terminal dimerization domains are shown in gray. With either ADP or ATP in its nucleotide-binding pocket, MCAK maintains relatively weak binding to the microtubule lattice [3]. This allows for the molecule to slide randomly in either direction along the microtubule lattice while maintaining its attachment.
Diffusional motility is defined as a one-dimensional random walk along the microtubule lattice driven solely by thermal energy. The equations characterizing these motions are identical to that of classical Brownian motion, although the diffusion coefficients tend to be significantly lower [2-5]. Interestingly, such a reduction in diffusional speed has been predicted by models of DNA binding proteins [6]. This model assumes that 1-D diffusion is limited not only by the binding protein’s 3-dimensional translational diffusion coefficient, but also by its rotational diffusion coefficient. Similar to a key sliding into a lock, the protein must adopt the correct rotational orientation before advancing along the polymeric filament. Any rotation away from the correct orientation will stall its progress.
Survey of microtubule binding proteins that exhibit 1-D lattice diffusion
Some of the earliest single molecule observations of diffusional motion on the microtubule lattice were made while studying the directed motility of kinesin molecules [7,8]. Kinesin motor domains can bind strongly to the microtubule, which allows for the directed “hand-over-hand” motility characteristic of kinesin superfamily members. However, this process requires that the kinesin molecule’s trailing head disengage (calling for a weakly bound state) while the leading head remains strongly bound. Then the weakly bound head swivels around to become the new leading head. This weakly bound state characteristically occurs with ADP in the kinesin motor domain’s nucleotide binding pocket, while strongly bound states occur with either ATP or no nucleotide at all (Figure 1b). Typically, kinesin motility experiments are carried out with ATP in solution, which makes it likely for at least one of the kinesin molecule’s two heads to be strongly bound at any given time. However, kinesin single molecule motility experiments sometimes exhibit a non-directional diffusive component superimposed upon the otherwise directed path. This so-called “biased diffusion” has been observed in a variety of kinesins including conventional kinesin [8], KIF1A [7], CENP-E [9], Eg5 [10], and Ncd [11]. The diffusional component has been hypothesized to occur during brief interludes in which both motor domain heads are disengaged [9], resulting in relatively free movement of the molecule along the microtubule axis. Here the word “disengaged” is not meant to imply that the kinesin molecule is unbound (in which case it would be free to diffuse in 3 dimensions), but rather that the classical “lock-and-key” strong binding motif is not engaged. This hypothesis is supported by experiments in which ADP is substituted for ATP [10,12]. The kinesin molecules imaged in these experiments exhibit pure unbiased diffusion, presumably because the weakly bound motor domains spend a significant amount of time disengaged from the microtubule lattice.
Cytoplasmic dynein, like motile kinesins, also exhibits a combination of directed motility and 1-dimensional diffusion [13,14]. There are also a number of microtubule binding proteins that exhibit pure diffusional motility (ie. lacking the directed motility component) along the microtubule lattice including the kinesin-13 protein (MCAK) [3], the Dam1 complex [5], XMAP215 [15], the Ndc80 complex [16], and tau [17]. Finally, even one of the myosin family members, Myosin Va, has been shown to exhibit rapid unbiased diffusion along the microtubule lattice [4].
Advantages of microtubule lattice diffusion as a transport mechanism
Clearly, the phenomenon of microtubule lattice diffusion is a ubiquitous mechanism occurring across a diverse set of microtubule binding proteins. Importantly, diffusional motility holds several key advantages over classical directed motility. First, as pure diffusion is by definition unbiased, it allows the binding protein to reach either end of the microtubule [18]. Significantly many of the microtubule binding proteins that exhibit pure diffusion have a function that requires localization to microtubule ends and diffusional motility provides a mechanism for delivery to the ends. The potential to target either the −end or the +end (or both simultaneously) greatly enhances the utility of diffusion as a targeting mechanism. Second, it is likely that diffusion allows the microtubule binding protein to more effectively skirt around obstacles on the microtubule surface. This is based on the assumption that the weakly bound state corresponding to diffusive motion [7] permits more frequent transitions between protofilaments while the strongly bound state of directed motility does not [19-21]. Such a feature may be relevant not only to those proteins that exhibit pure diffusion but also to the directed motility of kinesins and dynein [21]. As a specific example, the microtubules in neuronal axons are heavily decorated with the protein tau [22]. In order to navigate through this “forest-like” layer of tau, it seems likely that a motor protein such as kinesin, which normally travels axially along the microtubule [20], would periodically need to make diffusive side-steps. In addition, diffusion of the tau itself might make the process more akin to moving through a forest of seaweed than a forest of pine trees. Third, over short distances (< 1 μm), the speed of diffusive motion allows binding proteins to reach microtubule ends more rapidly than the directed motility mechanism of delivery. This is potentially advantageous because such rapid delivery permits the microtubule lattice to remain minimally cluttered. However, since diffusion is unbiased, directed motility is the clear winner when longer distances must be traversed, e.g. in larger cells and axons. Fourth, diffusional motility demands no external chemical energy (ie ATP) to propel the binding protein, making it an economical mode of transport.
DNA binding proteins - an analogue for diffusion of microtubule binding proteins
The concept of microtubule binding proteins diffusing along the microtubule lattice has only recently evolved into a significant area of research. However, the similar phenomenon of DNA binding proteins diffusing along DNA strands has been intensely studied for decades [23-26]. Although the structures of DNA and microtubule biopolymers are radically different, the overall 1-D morphology and strong negative surface charge that they have in common makes this a potentially enlightening comparison. Interestingly, 1-D diffusion coefficients measured for both DNA binding proteins [27] and microtubule binding proteins [3-5] tend to be roughly in line with one another, ranging between 0.1 μm2/sec and 0.4 μm2/sec. Surprisingly, a recent study has shown that hOgg1 (a DNA binding protein involved in base excision repair) was found to have a 1-D diffusion coefficient that approaches the theoretical maximum [27]. The theoretical maximum diffusion coefficient is calculated assuming that the diffusing molecule encounters no barriers as it slides along the polymer surface [6]. In the case of hOgg1, the energy barrier (referred to as the activation energy) was calculated to be an almost nonexistent 0.9 kBT. Typically the mechanism of 1-D diffusive motion is modeled as if the binding protein makes discrete, albeit directionally un-biased, steps between binding positions (nucleotides in the case of DNA or tubulin subunits in the case of microtubules). In this model, the activation energy refers to the energy required for the binding protein to release from one binding position such that it can move to an adjacent position. The fact that hOgg1 exhibits such a minimal activation energy suggests that the entire concept of “steps” in diffusive motion may provide an inaccurate representation of the mechanism. Perhaps a more relevant concept is that of the binding protein associating with an elongated space around the DNA polymer, coined the “DNA domain” [28]. In this model, as the binding protein diffuses throughout the DNA domain, local variations in electrostatic potential would likely generate roughness in the energy profile, thus accounting for the observed non-zero activation energy. However, the barriers between local minima corresponding to this activation energy would be too small to legitimately refer to the minima as distinct chemical species. Instead, according to this model, the entire diffusive interaction should be thought of as a single chemical species [28].
A theoretical estimation for the maximum diffusion coefficient of barrierless diffusion on microtubules (equivalent to the theoretical treatment for DNA binding proteins [6]) is currently lacking. Thus, activation energies for the diffusing microtubule binding proteins have not yet been calculated. Consequently, it is unknown if the diffusion exhibited by these proteins is better described by an unbiased stepping behavior (from one tubulin subunit to the next) or instead by a concept in which it moves relatively freely throughout an extended “microtubule domain”, as has been described for DNA binding proteins (above) [28]. Indeed, since the 1-D diffusion coefficients measured for several microtubule binding proteins [3,4,15] approach that measured for hOgg1 [27], it is a strong possibility that these molecules may also diffuse with very little activation energy throughout a roughly isoenergetic microtubule domain. Future theoretical work is needed to estimate the barrier-free diffusion coefficients for microtubule binding proteins.
The diffusive binding interaction: flexible and non-specific
A question remains as to the nature of the interaction that would allow such freedom of motion without complete detachment from the microtubule lattice. Commonly, the binding interaction is thought to be purely electrostatic in nature due to the observation that the microtubule surface carries a strong negative surface charge and binding proteins tend to hold a net positive charge. This is additionally supported by studies that have shown that proteolytic removal of the microtubule’s negative surface charges [3,4] or mutational neutralization of a binding protein’s positive charges [29] can greatly reduce the interaction probability and duration. In addition, homology modeling suggests that there is little energy difference between the negatively charged C-terminal tails that project outward from the microtubule versus those that lie along the lattice surface, suggesting that the flexible C-termini are considerably unconstrained [30]. This feature is likely to be essential for microtubule function as deletion of either the c-terminus of alpha- or beta tubulin is lethal. Yet the c-termini are interchangeable in that cells are viable with either tail on both tubulins [31]. Such flexibility and interchangeability supports the idea that the tails promote the formation of an extended “microtubule domain” along the entire polymer. In addition, microtubule binding proteins often include highly disordered regions which might enhance the flexibility and non-specificity of the interaction. For example, crystallographic studies suggest that the distal two-thirds of MCAK’s positively charged neck region is quite flexible and disordered [32]. Importantly the neck is thought to be a critical component of MCAK’s ability to interact with microtubules [3,32,33].
Structural studies of the binding interaction are unfortunately limited by the fact that microtubules have not been able to be crystallized to date. Fortunately, small DNA oligomers can be crystallized [34-36] and we can once again use the DNA binding proteins as a model of what might be expected for the microtubule binding system. Curiously, a feature that is often found in the vicinity of DNA strands is structured hydration shells that surround the molecule, particularly in the minor groove and encircling the strongly negatively charged phosphate backbone [37-39]. Unlike stereotypical “lock-and-key” type protein-protein interactions, the non-specific, electrostatically-driven interactions of DNA binding proteins are greatly facilitated by semi-structured water molecules that often line the interfaces between the DNA polymer and binding-protein [37,38,40]. Conveniently, this provides an explanation for the extremely small activation energies observed for one-dimensional sliding. The interfacial water molecules can be thought of as a lubricant that mediates the electrostatic interaction while still allowing the binding molecule to slide almost freely along the axis of the polymer (Figure 2). Furthermore, the displacement of structured water (that had previously resided in the hydration shells) upon protein binding has been hypothesized to add an entropic component to the binding free energy [37,38].
Figure 2. Mechanistic model of microtubule based 1-dimensional diffusion.
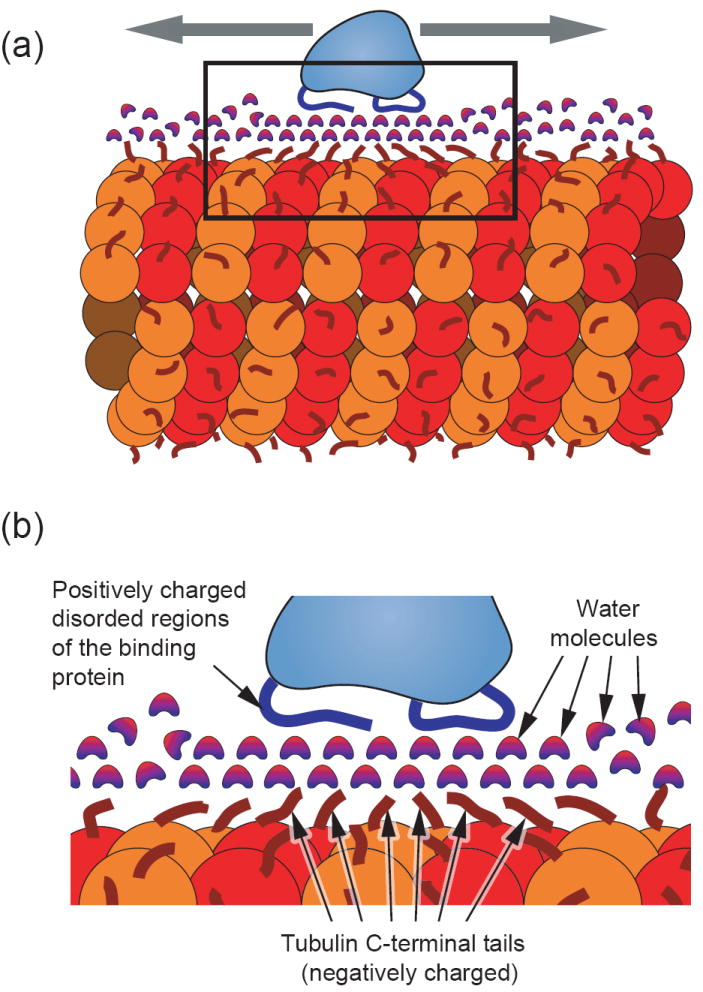
(a) Model of a microtubule binding protein (light blue) diffusing axially along the length of a microtubule (red). The microtubule surface is covered with flexible negatively-charged c-terminal tubulin ends (dark red) [30]. Complementing this, the microtubule binding protein is shown with flexible positively-charged regions (blue) that interact with the microtubule. (b) An expanded view of the boxed area from part (a) depicting a hypothetical interaction scheme in which a layer (or possibly multiple layers) of structured water molecules form an intermediary for the electrostatic interaction. This concept is drawn from the analogous situation of DNA binding proteins in which structural data indicates the existence of water “lubricating” the interfacial surface [40] [37] [38]. This scenario could provide low energy barriers for relatively free sliding along the length of the polymer. An alternative model would be one in which the charged disordered regions of both the microtubule and binding protein interact directly with very little interfacial water. In either scenario, the non-specificity of the interaction is a key feature for providing low energy barriers and the potential for fast sliding.
The lack of atomic level structural data for microtubules prevents us from being able to directly observe the potential role of water molecules in the weak interactions corresponding to 1-dimensional diffusion of microtubule binding proteins. However, the common electrostatic nature of the interactions and the fact that very similar diffusion dynamics are observed between the microtubule binding and DNA binding proteins, presents a compelling case that a very similar phenomenon might be occurring. Perhaps future studies exploring the thermodynamics (entropic vs. enthalpic) of the microtubule binding interactions may provide more evidence of such a mechanism.
Conclusions
Microtubule lattice diffusion is a widespread and effective mode of protein transport along microtubules. The fact that such a diversity of microtubule binding proteins exhibit this behavior has hindered attempts to develop a unifying description of the mechanism. However, microtubule-based diffusional motility shares important similarities with the well-studied subject of DNA-based diffusion (including comparable diffusion coefficients and a common electrostatic component of the interaction). This suggests that the latter may provide valuable insight into understanding the nature of this interaction. Based upon this analogy, we would expect extremely low activation energies for the diffusive motion of microtubule binding proteins (thus casting doubt on the idea that diffusive motion consists of discrete “steps”). Furthermore, we anticipate that the proximal water structure plays a significant role, both as a mediator of electrostatic interactions and as an entropic contributor to binding.
Finally, the diversity of molecules that diffuse on the surface of microtubules strongly implies that no obvious molecular structure (such as “two-headedness”) is required for robust diffusive motility. While motors and other microtubule binding proteins may have undergone small evolutionary refinements to capitalize on these properties, considerable responsibility also lies with the microtubule. Post-translational modifications of tubulin have the potential to define both subpopulations of microtubules and also heterogenous regions on the microtubule domain. Here, lessons can be inferred from the DNA methyltransferases whose diffusive motility and methyltransferase activity is enhanced upon encountering hemi-methylated DNA [41]. In the near future, we expect that increasingly sensitive methods to measure the activity and diffusive behavior of microtubule binding proteins will make it likely that the numerous post-translational modifications of the microtubule substrate will have a measurable and functionally relevant effect on these molecules both in vitro and in cells.
Acknowledgments
The authors acknowledge support from the National Institutes of Health and from the National Science Foundation (IGERT traineeship to J. C. and NIH grant GM69429 to L. W.). We thank Chip Asbury and Mike Wagenbach for insightful comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 2.Young ME, Carroad PA, Bell RL. Estimation of diffusion coefficents of proteins. Biotechnology and Bioengineering. 2004;22:947–955. [Google Scholar]
- 3.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 4••.Ali MY, Krementsova EB, Kennedy GG, Mahaffy R, Pollard TD, Trybus KM, Warshaw DM. Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc Natl Acad Sci U S A. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104.. Myosin Va is shown to diffuse on microtubules. The fact that myosin (classically known to be an actin based motor protein) exhibits microtubule based diffusion highlights the tremendous diversity of proteins that possess such functionality.
- 5•.Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702.. The yeast Dam1 complex diffuses as a monomer along the microtubule lattice. In addition, phosphorylation is shown to reduce the dwell time of the Dam1/microtubule interaction hinting that electrostatics may be involved.
- 6.Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA. Hydrodynamic considerations. Biophys Chem. 1979;9:413–414. [PubMed] [Google Scholar]
- 7.Okada Y, Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 8.Inoue Y, Iwane AH, Miyai T, Muto E, Yanagida T. Motility of single one-headed kinesin molecules along microtubules. Biophys J. 2001;81:2838–2850. doi: 10.1016/s0006-3495(01)75925-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–485. doi: 10.1038/nchembio812.. The kinesin Eg5 exhibits directed motility superimposed with a diffusive motility component (ie. biased diffusion) in the presence of ATP and pure, unbiased diffusion in the presence of ADP.
- 11.Furuta K, Toyoshima YY. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr Biol. 2008;18:152–157. doi: 10.1016/j.cub.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 12.Okada Y, Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci U S A. 2000;97:640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vale RD, Soll DR, Gibbons IR. One-dimensional diffusion of microtubules bound to flagellar dynein. Cell. 1989;59:915–925. doi: 10.1016/0092-8674(89)90614-4. [DOI] [PubMed] [Google Scholar]
- 14.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powers AF, Franck AD, Gestaut DR, Cooper J, Graczyk B, W RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex uses biased diffusion to couple chromosomes to dynamic microtubule tips. Cell. 2009 doi: 10.1016/j.cell.2008.12.045. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konzack S, Thies E, Marx A, Mandelkow EM, Mandelkow E. Swimming against the tide: mobility of the microtubule-associated protein tau in neurons. J Neurosci. 2007;27:9916–9927. doi: 10.1523/JNEUROSCI.0927-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Kamimura S, Mandelkow E. Tubulin protofilaments and kinesin-dependent motility. J Cell Biol. 1992;118:865–875. doi: 10.1083/jcb.118.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray S, Meyhofer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Khan S, Sheetz MP. Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophys J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay R, Kumar S, Hoh JH. Molecular mechanisms for organizing the neuronal cytoskeleton. Bioessays. 2004;26:1017–1025. doi: 10.1002/bies.20088. [DOI] [PubMed] [Google Scholar]
- 23.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 24.Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor--operator interaction: equilibrium measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 25.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 26•.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441.. A thorough review of the advances made in the study of the diffusion of DNA-binding proteins, with particular focus on direct visualization by single molecule TIRF imaging.
- 27•.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103.. The DNA-repair protein, hOgg1 is found to approach the theoretical maximum rate of diffusion.
- 28.Shimamoto N. One-dimensional diffusion of proteins along DNA. Its biological and chemical significance revealed by single-molecule measurements. J Biol Chem. 1999;274:15293–15296. doi: 10.1074/jbc.274.22.15293. [DOI] [PubMed] [Google Scholar]
- 29.Ali MY, Lu H, Bookwalter CS, Warshaw DM, Trybus KM. Myosin V and Kinesin act as tethers to enhance each others’ processivity. Proc Natl Acad Sci U S A. 2008;105:4691–4696. doi: 10.1073/pnas.0711531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuszynski JA, Carpenter EJ, Huzil JT, Malinski W, Luchko T, Luduena RF. The evolution of the structure of tubulin and its potential consequences for the role and function of microtubules in cells and embryos. Int J Dev Biol. 2006;50:341–358. doi: 10.1387/ijdb.052063jt. [DOI] [PubMed] [Google Scholar]
- 31.Duan J, Gorovsky MA. Both carboxy-terminal tails of alpha- and beta-tubulin are essential, but either one will suffice. Curr Biol. 2002;12:313–316. doi: 10.1016/s0960-9822(02)00651-6. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- 33.Ovechkina Y, Wagenbach M, Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J Cell Biol. 2002;159:557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drew HR, Dickerson RE. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981;151:535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- 36.Kopka ML, Fratini AV, Drew HR, Dickerson RE. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983;163:129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- 37.Jayaram B, Jain T. The role of water in protein-DNA recognition. Annu Rev Biophys Biomol Struct. 2004;33:343–361. doi: 10.1146/annurev.biophys.33.110502.140414. [DOI] [PubMed] [Google Scholar]
- 38.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 39•.Privalov PL, Dragan AI, Crane-Robinson C, Breslauer KJ, Remeta DP, Minetti CA. What drives proteins into the major or minor grooves of DNA? J Mol Biol. 2007;365:1–9. doi: 10.1016/j.jmb.2006.09.059.. Thermodynamic analysis of DNA-binding proteins reveals that there is a significant entropic component to binding within the minor groove of DNA, apparently due to release of structured water molecules from the DNA.
- 40.Schwabe JW. The role of water in protein-DNA interactions. Curr Opin Struct Biol. 1997;7:126–134. doi: 10.1016/s0959-440x(97)80016-4. [DOI] [PubMed] [Google Scholar]
- 41.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]


