Abstract
The proteasome is a major enzyme that cleaves proteins for antigen presentation. Cleaved peptides traffic to the cell surface, where they are presented in the context of MHC class I. Recognition of these complexes by cytotoxic T lymphocytes is crucial for elimination of cells bearing “non-self” proteins. Our previous studies revealed that ethanol suppresses proteasome function in ethanol-metabolizing liver cells. We hypothesized that proteasome suppression reduces the hydrolysis of antigenic peptides, thereby decreasing the presentation of the peptide-MHC class I-complexes on the cell surface. To test this, we used the mouse hepatocyte cell line (CYP2E1/ADH-transfected HepB5 cells) or primary mouse hepatocytes, both derived from livers of C57Bl/6 mice, which present the ovalbumin peptide, SIINFEKL, complexed with H2Kb. To induce H2Kb expression, HepB5 cells were treated with interferon gamma (IFNγ) and then exposed to ethanol. In these cells, ethanol metabolism decreased not only proteasome activity, but also hydrolysis of the C-extended peptide, SIINFEKL-TE and the presentation of SIINFEKL-H2Kb complexes measured after the delivery of SIINFEKL-TE to cytoplasm. The suppressive effects of ethanol were, in part, attributed to ethanol-elicited impairment of IFNγ signaling. However, in primary hepatocytes, even in the absence of IFNγ, we observed a similar decline in proteasome activity and antigen presentation after ethanol exposure. We conclude that proteasome function is directly suppressed by ethanol metabolism and indirectly, by preventing the activating effects of IFNγ. Ethanol-elicited reduction in proteasome activity contributes to the suppression of SIINFEKL-H2Kb presentation on the surface of liver cells.
Immune response to viral antigens plays a crucial role in the pathogenesis of hepatitis C or B viral infections (HCV and HBV, respectively). Professional antigen-presenting cells (dendritic cells and macrophages) are responsible for priming the immune response. HCV infection impairs the functioning of these cells (1, 2). However, when clonal expansion of cytotoxic T-lymphocytes (CTLs) is established, the next important restriction for elimination of infected cells is the availability of peptide-MHC class I complexes, which are recognized by CTLs on the surface of target cells (hepatocytes).
Keywords: ovalbumin peptides, hepatocytes, proteasome, ethanol, interferon gamma
The proteasome is a multicatalytic enzyme, which degrades the bulk of intracellular proteins and which generates peptides from intracellular proteins for MHC class I-restricted antigen presentation. In the cytosol, two proteasome particles, the 26S and 20S forms, catalyze ubiquitin-dependent and –independent protein cleavage, respectively. The proteasome is the first enzyme that initiates cleavage of antigenic peptides (3), while at the later stages of peptide degradation, other enzymes (like leucine aminopeptidase, etc.) also generate the peptides that fit into MHC class I groove (4, 5). The 20S proteasome particle ubiquitin-independently trims C-terminal extensions of antigenic peptides. Under inflammatory conditions, the release of IFNγ from T-lymphocytes stimulates the proteasome activator, PA28, to induce immunoproteasome formation, which, in turn, accelerates antigenic peptide cleavage (6). Generated peptides are transported to the endoplasmic reticulum (ER) by transporters associated with antigen processing (TAP) and then are assembled into a trimolecular complex with β2-microglobulin and the heavy chain of MHC class I molecules. Assembly is facilitated by TAP and a number of chaperones, to achieve the optimal MHC class I-peptide loading. All these steps of antigen processing/presentation are strongly IFN-dependent.
Ethanol metabolism induces oxidative stress in liver cells, disrupting the function of proteolytic systems, including the proteasome. Inhibition of proteasome function is CYP2E1- dependent (7-11) and correlates with enhanced generation of intracellular oxidants. Enhanced activity of CYP2E1 is a common feature of numerous pathologic events induced by ethanol-elicited oxidative stress in liver cells, both in the cytosolic and mitochondrial compartments (12, 13). Loss of proteasome function due to oxidative stress appears to occur from formation of adducts with carbonyls, 4-hydroxynonenal (4-HNE) and 3-nitrotyrosine derived from peroxynitrite (8, 14, 15). The 20S proteasome removes oxidized proteins even after 26S proteasome has been inhibited by oxidants, indicating differential resistance to oxidative insult (10, 16). Previously, by using ethanol-metabolizing recombinant VL-17A cells, we demonstrated that ethanol metabolism down-regulates proteasome function and the hydrolysis of C-extended 18-27 HBV core peptide (FLPSDFFPSVRDL) (16) and suppresses IFNγ signaling, which normally enhances the proteasome function (17). However, we did not examine whether ethanol treatment affected the presentation of antigenic peptide-MHC class I complex on the surface of ethanol-metabolizing liver cells as a FLPSDFFPSV-HLA-A2 complex-reactive antibody was unavailable.
To study antigen presentation in liver cells, we measured, by flow cytometry, the presentation of ovalbumin peptide, SIINFEKL, on the surface of H2Kb-expressing mouse hepatocyte-derived cells that stably express the ethanol-metabolizing enzymes, cytochrome P450 2E1 (CYP2E1) and alcohol dehydrogenase (ADH). The SIINFEKL-H2Kb complex is a well-known target for CTLs. We examined whether ethanol treatment affected the expression of SIINFEKL-H2Kb complex by its quantification with a complex-specific antibody. Here, we demonstrate that CYP2E1/ADH-transfected HepB5 cells serve as an appropriate model to study the effects of ethanol on the presentation of SIINFEKL-H2Kb complex. We show an ethanol-elicited reduction in the presentation of this complex on the cell surface, which corresponded to decreased hydrolysis of the precursor peptide, caused by decline in proteasome activity. The decline in antigen processing/presentation in HepB5 cells was, in part, attributed to ethanol-elicited suppression of IFNγ signaling.
MATERIALS AND METHODS
Reagents and Media
High glucose Dulbecco’s Modified Eagle Medium (DMEM), Ham’s F12 Medium and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA). Human recombinant interferon gamma (IFNγ) was from PeproTech, Inc. (Rocky Hill, NJ). The peptides, SIINFEKL and SIINFEKL-TE were purchased from SynPep (Menlo Park, CA). Chariot ™ and TransAM ™ DNA binding ELISA kit was purchased from Active Motive (Carlsbad, CA). Antibody to phosphorylated STAT1 (Tyr 701) was from Cell Signaling (Beverly, MA); antibody to the STAT1 protein was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Other reagents, all of analytical grade quality, were from Sigma (St. Louis, MO).
Cell Lines
HepB6 cells from C57Bl/6 mouse hepatocyte (18) were obtained from Drs I. Stroynowski and M. Chen, University of Texas Southwestern Medical Center, Dallas, TX. These cells were transfected with the plasmids, pIV-G2 and pIVL-2, as previously described for Hep G2 cells (9, 19). pIV-G2 was constructed by inserting the coding region of human CYP2E1 into the Hind III site of pcDNA 3.1 and PIVL-2 was constructed by inserting an eukaryotic expression plasmid containing cDNA encoding murine ADH into the corresponding sites of psDNA3.1/Zeo(+) (both plasmids were a kind gift from Dr. Dahn Clemens, VAMC, Omaha, NE). For transfection, Lipo TAXI (Invitrogen Corp., Carlsbad, CA) was used as described by the manufacturer. Recombinant cells, designated HepB5 cells, were selected in culture medium containing G418 and zeocin, each at 400 μg/ml. Clones were expanded and screened for ADH and CYP2E1 activity. Cells were cultured in a 1:1 mixture of DMEM and Ham’s F12 medium supplemented with 5 μg insulin/ml, 5 μg transferrin/ml, 5 ng selenium/ml, 40 ng dexamethasone /ml, 10% FBS, 100 U penicillin/ml, 100 μg streptomycin/ml and selective antibiotics, G418 and zeocin, each at 400 μg/ml.
Cell treatments
HepB5 cells were plated on 6-well plates in complete medium and exposed to 0 or 50 mM ethanol for 48 hr, with or without mouse IFNγ (10 ng/ml). In some experiments, ethanol treatment was in the presence or absence of 4 methyl pyrazole (4MP, 2 mM). Plates were covered with plastic film to prevent ethanol evaporation. After incubation, cells were detached using EDTA-based cell stripping solution (Cellstripper ™, Mediatech, Inc, Hamden, VA). To measure SIINFEKL-H2Kb complex expression, cells were stained with SIINFEKL-H2Kb antibody and further processed for flow cytometry. Total cell lysates were prepared by sonication in PBS and used to measure proteasome activity. Cytosolic fractions of cell lysates were obtained by a 1 hr centrifugation at 105,000 × g and glycerol was added to a final concentration of 20 % (w/v). This fraction was used as a source of proteasome, to study SIINFEKL-TE hydrolysis.
Hepatocytes were isolated from livers of C57Bl/6 mice by collagenase perfusion (20) and were plated on collagen- coated 6-well plates at 1×105 cell/well in Williams Medium supplemented with penicillin and streptomycin and 5% FBS. Cells were incubated overnight, in the presence or absence of 10 ng/ml IFNγ, 50 mM ethanol and in certain cases, with ethanol metabolism inhibitor, 4MP, 2 mM.
Detection of Peptide Cleavage
Crude cytosolic cell fractions (at a final concentration of 100 μg protein/ml) were mixed with 5 nM C-extended peptide (SIINFEKL-TE) in 50 mM Tris-HCl (pH 8.5), 5 mM MgCl2 in a total volume of 100 μl and incubated for 0, 15, 30 and 60 minutes at 37°C. The reaction was stopped by adding 20% trichloracetic acid and the supernatants were subjected to HPLC. The reverse-phase HPLC on a Vydac C18 monomeric column was performed as described before (Osna, 2007). The quantified peptide peak of SIINFEKL-TE peptide at 0 hr incubation with cytosols from control and ethanol-treated cells was marked as 100%. The percent of remaining (uncleaved) peptide was calculated after 30 min incubation of cell cytosols with the precursor peptide.
Proteasome activity
Proteasome chymotrypsin-like (Cht-L) activity was detected in vitro using the fluorogenic substrate Suc-LLVY-AMC as described (21).
Presentation of SIINFEKL-H2Kb complex on the cell surface
C-extended peptide, SIINFEKL-TE, was delivered to cells by Chariot™ (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. After 2 hr incubation of the precursor peptide-Chariot macromolecular complex with cells, the expression of a cleaved peptide, SIINFEKL, was measured in the context of H2Kb with antibody that recognizes SIINFEKL-H2Kb complex. This antibody was affinity-purified from the supernatants of 25D1.16 hybridoma cells (obtained from Dr. Germain, NIH, Bethesda, MD). After 30 min of exposure to SIINFEKL-H2Kb antibody, cells were washed and incubated with IgG-phycoerythrin (PE) for another 30 min on ice and then quantified by flow cytometry (BD FACSCalibur, Becton Dickenson, San Jose, CA). To control for spontaneous SIINFEKL-H2Kb expression, we incubated SIINFEKL-TE peptide with cells in the absence of Chariot ™. In addition, as a positive control for SIINFEKL-H2Kb staining, IFNγ-pretreated HepB5 cells were incubated with SIINFEKL peptide (without Chariot ™). To monitor the expression of MHC class I on the surface, the cells were double-stained with antibody to H2Kb-FITC.
Cytochrome P4502E1 (CYP2E1) Catalytic Activity
CYP2E1 activity was detected in microsome fractions of cell lysates by the formation of 4-nitrocatechol (4NC) detected spectrophotometrically, as previously described. (22). Specific activity is expressed as units (nmoles 4NC/hr) per mg protein.
Alcohol dehydrogenase (ADH) activity
ADH activity was measured in total cell lysates as previously described (11, 22)
Reactive oxygen species (ROS) production
ROS was measured by 2’7’dichlorodihydrofluorescein diacetate (DCDFA) (23). Data are expressed as DCFDA units (fluorescence detected at an excitation of 485 nm and an emission of 530 nm) per mg protein.
Ethanol and acetaldehyde levels were detected by head space gas chromatography in 300 μl medium taken from cells after 0, 24 and 48 hr incubation with ethanol as described (11), in the presence or absence of DAS, 20 μM. Acetaldehyde levels are expressed as μM acetaldehyde. Ethanol clearance determined after 48 hr cell incubation with ethanol, is expressed as nmoles ethanol/hr/10 6 cells.
IFNγ signaling
IFNγ signaling was measured by STAT1 phosphorylation on Western blots, as previously described (17). Nuclear extracts were obtained from the cells treated with or without IFNγ (10 ng/ml, 1 hr) (24). Attachment of activated STAT1 from nuclear extracts to DNA was detected by using Trans ™ DNA-binding ELISA (Active Motif, Carlsbad, CA), according to the manufacturer’s instructions.
Statistical analyses
Data are expressed as mean values ± standard deviation. Comparisons among multiple groups were determined by one-way ANOVA, using a Tukey post-hoc test. For comparisons between two groups, we used Student’s t-test. A probability value of 0.05 or less was considered significant.
RESULTS
Phenotypic characterization of HepB5 cells
To characterize the ethanol-metabolizing phenotype of HepB5 cells, we measured CYP2E1 and ADH activities as well as ethanol and acetaldehyde levels in the culture medium following exposure to ethanol. ADH activity, which ranged from 400 to 900 nmoles NADH/h/mgP was unchanged, while CYP2E1 activity was elevated up to 1.3-1.8 –fold by exposing HepB5 cells to ethanol for 48 hr (Fig.1A). Cells treated with 50 mM ethanol had 2.5 fold-higher ROS production (Fig.1B). Differential acetaldehyde (Ach) production was generated by exposure to various doses of ethanol (Fig.1C). Ach production appeared to be principally catalyzed by CYP2E1 because Ach levels were suppressed when cells were incubated with the specific CYP2E1 inhibitor, dyallyl sulfide (DAS). The rate of ethanol metabolism increased with the concentration of ethanol in the medium (Fig.1D).
Fig.1. HepB5 cells: ethanol-mediated CYP2E1 activity, ROS production, acetaldehyde generation and clearance of ethanol from the medium.
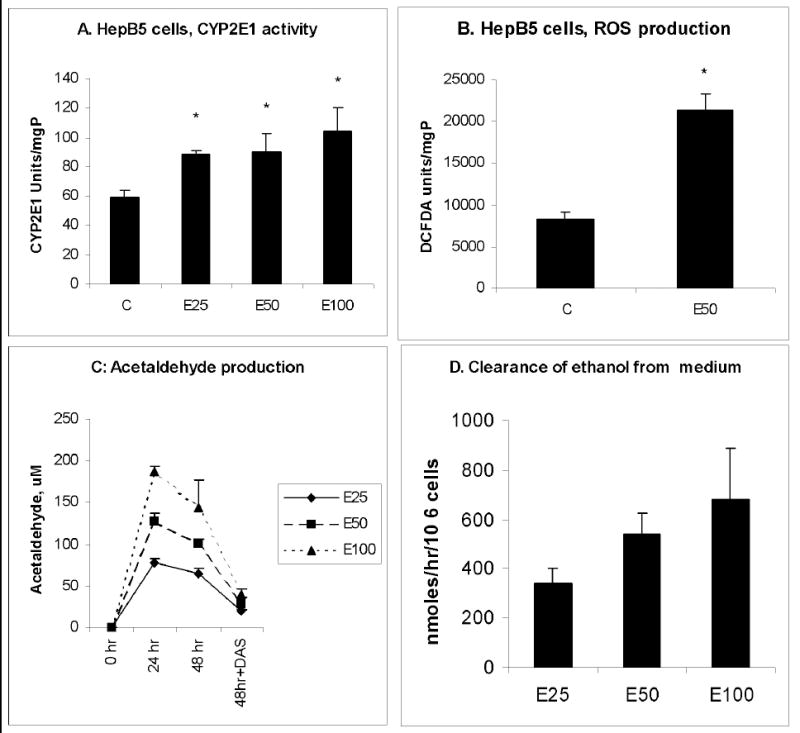
HepB5 cells were treated with various concentrations of ethanol for 48 hr and processed as indicated in Materials and Methods. A. CYP2E1 activity. B. ROS production. C. Acetaldehyde production. D. Ethanol clearance. Figures represent mean ± StDev from 3 experiments, * is p<0.05 between control and other treatments.
Proteasome activity in HepB5 cells
We measured the effects of ethanol on chymotrypsin-like (Cht-L) proteasome activity in HepB5 cells, because this activity plays a major role in cleavage of peptides for antigen presentation and because the proteasome is mainly responsible for the cleavage of C-extended peptides (3, 25). Ethanol treatment (50 mM, 48 hr) reduced Cht-L proteasome activity by 40%, but the activity was unaffected when cells were incubated with ethanol and 4MP (Fig. 2A). IFNγ induced proteasome activity, but ethanol exposure blocked this induction (Fig. 2A).
Fig.2. Ethanol down regulation of proteasome activity and peptide hydrolysis in HepB5 cells.
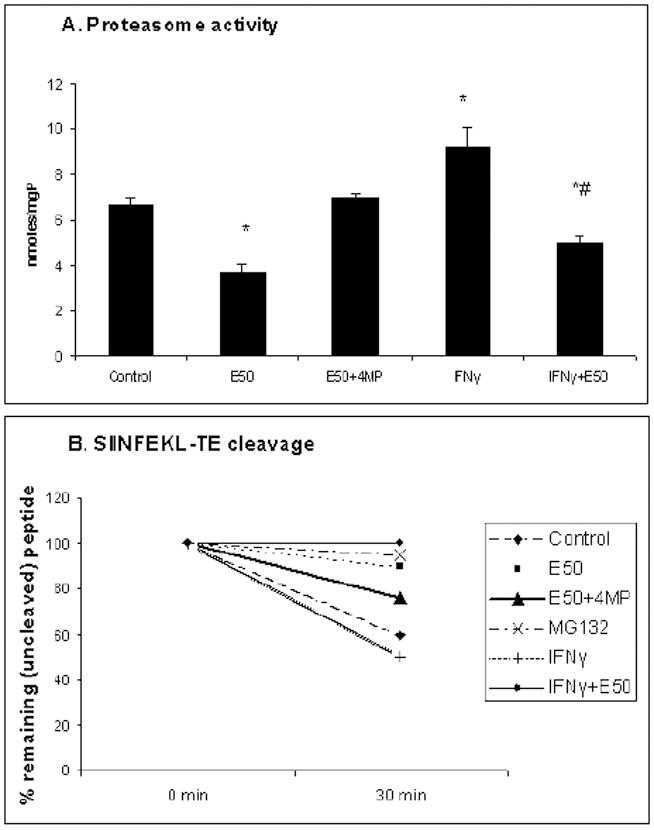
HepB5 cells were treated as described in Materials and Methods. A. Proteasome activity. Data from 3 experiments are presented as proteasome Cht-L activity, nmoles AMC/mgP, mean ± StDev. * is p<0.05 between control and treatments; # is p<0.05 between IFNγ and IFNγ+ethanol. B. Peptide hydrolysis. The percent of remaining (uncleaved) peptide was calculated after 30 min incubation of cell cytosols with the precursor peptide. The representative data from one out of two experiments with similar results are presented as % remaining SIINFEKL-TE peptide.
Peptide hydrolysis
Cytosols from control and 50 mM ethanol-exposed cells treated with or without IFNγ were incubated with SIINFEKL-TE as we described previously (16). We observed a 54% reduction in peptide cleavage by cytosols obtained from ethanol-treated cells compared with cytosols from untreated cells. The inhibitory effect of ethanol was partially reversed after cells were incubated with ethanol and 4MP (Fig.2B). Similarly, peptide hydrolysis was suppressed in IFNγ-treated cells, exposed to ethanol. MG132 completely blocked SIINFEKL-TE hydrolysis, confirming that the hydrolysis of C-extended precursor peptide is proteasome-dependent.
Presentation of SIINFEKL-H2Kb complex on HepB5 cells
To induce expression of H2Kb on the cell surface, HepB5 cells were treated with IFNγ (10 ng/ml, 48 hr). SIINFEKL-H2Kb processing/presentation were measured after exposure to 50 mM ethanol for 48 hr. The extended precursor peptide, SIINFEKL-TE, was delivered to cytoplasm using the Chariot ™ delivery vehicle. When the precursor peptide was incubated with HepB5 cells in the absence of Chariot ™, we observed no SIINFEKL-H2Kb complexes, quantified by using of SIINFEKL-H2Kb antibody. Delivery of the peptide into the cell was unaffected by ethanol treatment, as ethanol did not influenced intacellular β-galactosidase staining after its delivery into HepB5 cells (not shown). Prior exposure of cells to ethanol reduced the presentation of SIINFEKL-H2Kb complex on the surface of HepB5 cells by 30%; however, when 4 MP was included in the culture medium, the suppressive effects of ethanol on SIINFEKL-H2Kb presentation were blocked (Fig.3A,B). Similarly, co-treatment of cells with ethanol and catalase (which scavenges H2O2) restored SIINFEKL-H2Kb presentation up to 80% (Fig. 3B).
Fig.3. Ethanol exposure and inhibitors suppress presentation of SIINFEKL-H2Kb complex in HepB5 cells.
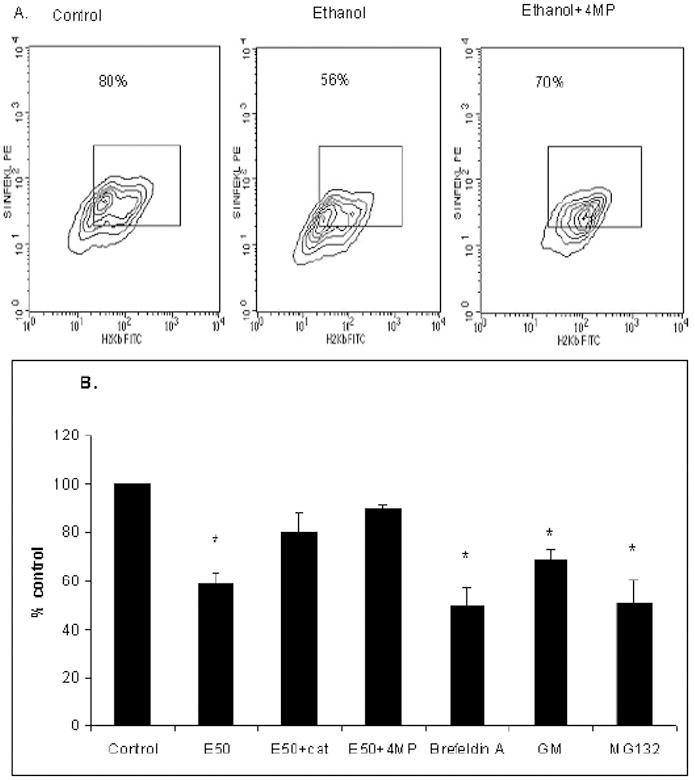
A. Effects of ethanol on SIINFEKL-H2Kb presentation in HepB5 cells. Cells were treated as indicated in Material and Methods. Data from one a representative experiment are expressed as percent anti-SIINFEKL-H2Kb positive cells out of H2Kb–positive HepB5 cells. B. Effects of various treatments on SIINFEKL-H2Kb presentation in HepB5 cells. HepB5 cells were pretreated with IFNγ for 48 hr in the presence or absence of ethanol, 4 MP and catalase and then exposed or not to either brefeldin A, geldanamycin and MG132 for 1 hr. Data from two to three independent experiments are presented as percent SIINFEKL-H2Kb-positive cells, mean ± StDev. * is p<0.05 between control and the treatments.
To confirm that a product of extended precursor peptide cleavage, SIINFEKL peptide, was delivered to cell surface via the endoplasmic reticulum (ER), the presentation of SIINFEKL –H2Kb complex was measured after the treatment of cells with brefeldin A (5 μg/ml), to block trafficking via the ER. In addition, chaperoning of the presented complex by heat-shock proteins was confirmed by treatment with geldanamycin (GM, 5 μM), which blocks HSP90. To demonstrate the involvement of proteasome into generation of SIINFEKL, the treatment with the proteasome inhibitor, MG132 (20 μM), has been used. Compared with controls, each of these individual treatments suppressed SIINFEKL-H2Kb presentation, indicating that proteasome cleaved the extended peptide to SIINFEKL size and this peptide traffics through ER, in a HSP90-mediated step, to be presented on the cell surface (Fig. 3B).
IFNγ signaling in HepB5 cells
HepB5 cells do not express H2Kb spontaneously. To present SIINFEKL-H2Kb complex, the cells required treatment with IFNγ to enhance H2Kb expression on the cell surface. Thus, the effects of ethanol on SIINFEKL-H2Kb presentation depend, in part, on ability of ethanol to regulate IFNγ signaling. Previously, we demonstrated that in VL-17A cells, ethanol suppresses IFNγ signaling (17). Therefore, we examined whether ethanol treatment influenced IFNγ-induced STAT1 phosphorylation and the attachment of activated (phosphorylated) STAT1 to DNA. Ethanol treatment caused no changes in STAT1 phosphorylation, nor did affect translocation of activated STAT1 to the nucleus (data not shown). However, ethanol exposure suppressed STAT1 attachment to DNA (Fig. 4A). Ethanol-elicited suppression in IFNγ signaling prevented the induction of the immunoproteasome subunit, LMP2, and 20S proteasome activator, PA28 expression by IFNγ (Fig.4B).
Fig.4. Effects of ethanol on IFNγ signaling in HepB5 cells.
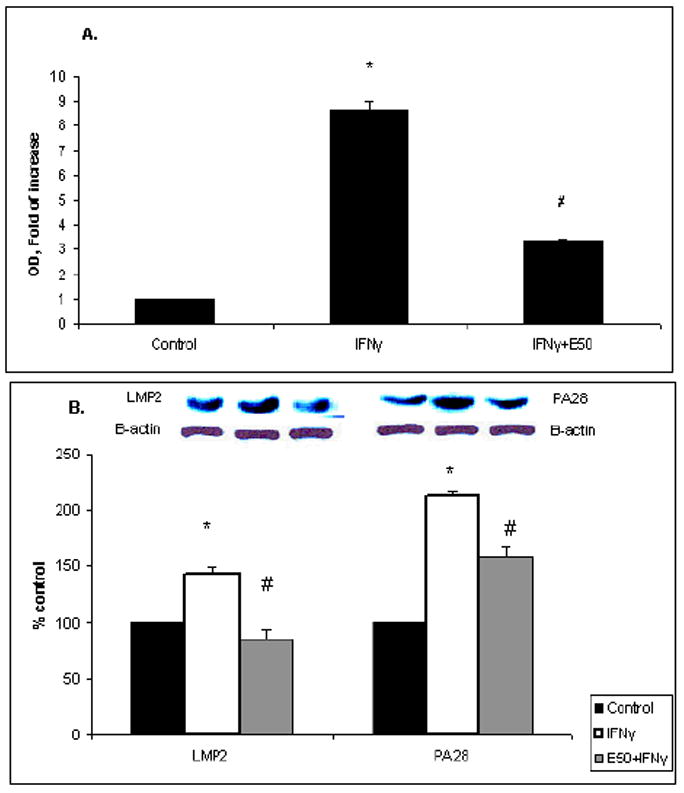
A. STAT1 attachment to DNA. HepB5 cells were treated in the presence or absence of ethanol for 48 hr and then exposed to IFNγ for 1 hr. Attachment of STAT1 to DNA was measured in nuclear extracts by Trans™ DNA-binding ELISA. Data from 3 experiments are presented as fold increase of absorbance. Fold increase is calculated as ratio of absorbance (OD) between treatments and control, mean ± StDev. * is p<0.05 between control and the treatments; # is p<0.05 between IFNγ and IFNγ+E50. B. Expression of LMP2 and PA28 in HepB5 cells. Cells were treated with or without IFNγ (10 ng/ml) and ethanol (50 mM) for 48 hr and then were lysed. LMP2 and PA28 were detected by Western blot with the specific antibodies and normalized to β-actin, to account for the equal protein load. Data from 3 experiments are presented as LMP2/β-actin and PA28/β-actin ratios, mean ± StDev. * is p<0.05 between control and the treatments; # is p<0.05 between IFNγ and IFNγ+E50.
SIINFEKL-H2Kb presentation and proteasome activity in hepatocytes
Because HepB5 cells require IFNγ-treatment for presentation of H2Kb and because IFNγ signaling is altered by ethanol exposure, we measured SIINFEKL-H2Kb presentation in primary cultures of hepatocytes of C57BL/6 mice, which express H2Kb constitutively, without IFNγ stimulation. Freshly isolated hepatocytes were attached to collagen and incubated with 50 mM ethanol for 18 hr, either with or without the inhibitor of ethanol metabolism, 4MP. SIINFEKL-TE peptide was delivered to the cells as described for HepB5 cells. Then, expression of the processed SIINFEKL-H2Kb complex was measured on the cell surface by flow cytometry and the proteasome activity-by in vitro assay. The proteasome inhibitor, MG132, was used as a positive control, to show the involvement of proteasome in processing of SIINFEKL from the precursor peptide. Additionally, we applied the trafficking inhibitor, brefeldin A, which showed the requirement of trafficking via the ER in the displaying of SIINFEKL-H2Kb. Presentation of the SIINFEKL-H2Kb complexes, as well as proteasome activity was decreased in ethanol-treated hepatocytes (Fig.5 A, B and 6). However, 4MP treatment partially prevented the ethanol-elicited reduction in SIINFEKL-H2Kb presentation and in proteasome activity. In addition, we measured SIINFEKL-H2Kb presentation and proteasome activity in IFNγ-pretreated hepatocytes and found that ethanol exposure also down-regulated both antigen presentation and proteasome function, in part, by preventing the activating effects of IFNγ (Fig. 5B and Fig. 6).
Fig.5. Effects of ethanol on SIINFEKL-H2Kb presentation and proteasome activity in hepatocytes.
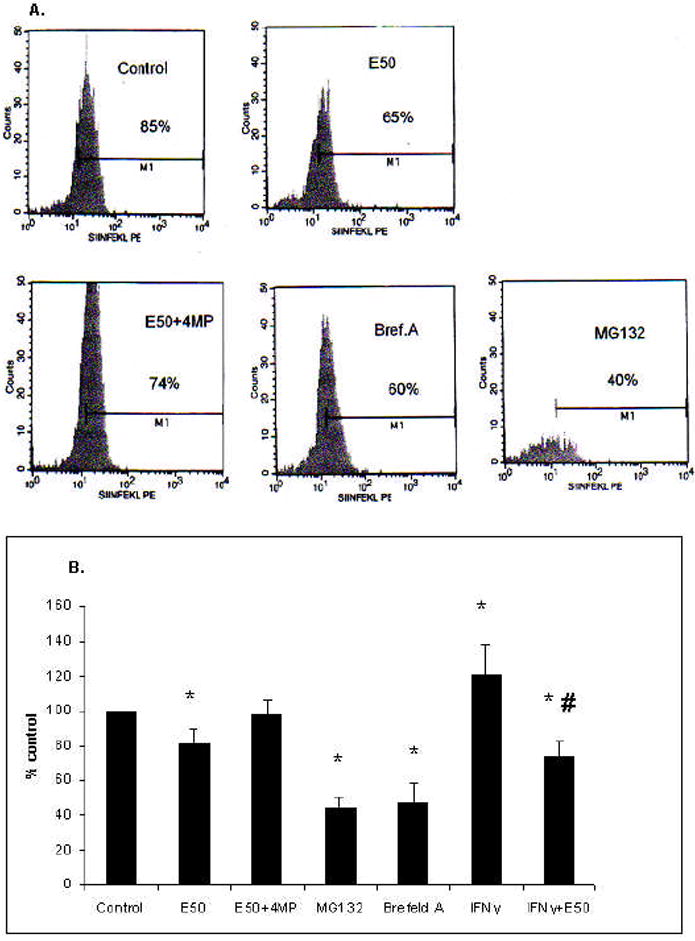
A. Presentation of SIINFEKL-H2Kb in hepatocytes. Hepatocytes obtained from the livers of C57BL/6 mice were plated on collagen-coated plates and treated overnight in the presence or absence of 50 mM ethanol and 2mM 4MP. Optionally, they were treated with brefeldin A or MG132 as described above. After delivery of SIINFEKL-TE, the presentation of SIINFEKL-H2Kb complex was measured using anti-SIINFEKL-H2Kb by flow cytometry. Data are from a representative experiment, percent SIINFEKL-H2Kb positive cells. B. Effects of various treatments on SIINFEKL-H2Kb presentation in hepatocytes. Cells were treated as indicated, in the presence or absence of IFNγ. Data from three experiments are presented as percent control. * is p<0.05 difference between control and treatments and #- between IFNγ and IFNγ+ethanol.
Fig. 6. Proteasome activity in hepatocytes.
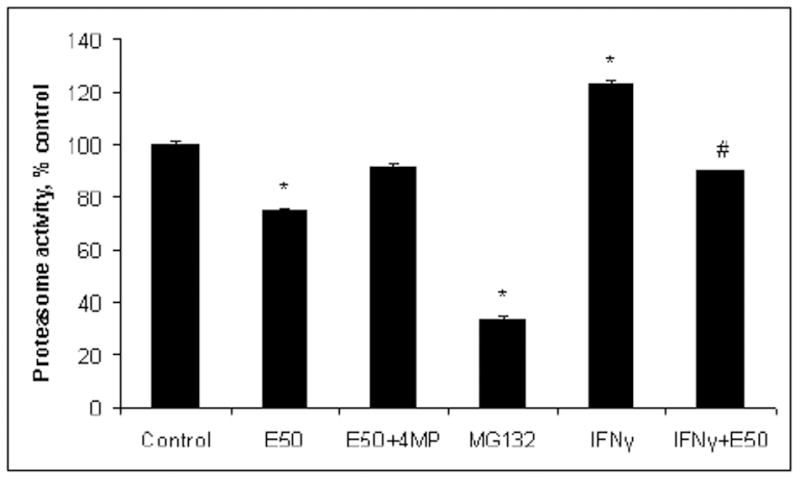
Hepatocytes were obtained and treated with IFNγ, ethanol, 4MP and MG132 as stated in A. Data from 3 experiments are presented as percent control, mean ± StDev. * is p<0.05 between control and the treatments and #- between IFNγ and IFNγ+ethanol.
DISCUSSION
The proteasome plays a pivotal role in antigen presentation by degrading antigenic proteins to peptides, which are incorporated into the MHC class I groove (25). While other proteases contribute to antigenic peptide cleavage, the proteasome is solely responsible for C-extended peptides trimming (26). To focus on proteasome-dependent processing of peptides for antigen presentation, we delivered C-extended peptide, SIINFEKL-TE, into liver cells and examined whether ethanol affected the presentation of the processed SIINFEKL peptide, a well-known CTL target.
In this study, we used HepB5 cells and primary hepatocytes derived from the livers of C57Bl/6 mice. HepB5 cells are a recently developed mouse hepatocyte-based cell line, which metabolize ethanol mainly via CYP2E1 and partially by ADH. Ethanol exposure to HepB5 cells increased CYP2E1 activity and induced acetaldehyde and ROS production, showing that these cells are suitable for investigation of ethanol-elicited effects on antigen processing and presentation. HepB5 cells do not express H2Kb spontaneously, but about 90% of them become H2Kb positive after 24-hr IFNγ exposure based upon flow cytometric analysis. For this reason, we used IFNγ-pre-treated cells for antigen processing/presentation experiments. The reduction in SIINFEKL-H2Kb presentation after MG132, the proteasome inhibitor, supports the involvement of proteasome in the intracellular generation of SIINFEKL peptide. The HSP90 inhibitor, geldanamycin, which inhibits the loading of the peptide to MHC class I groove (27) and the inhibitor of trafficking via ER, brefeldin A also decreased SIINFEKL-H2Kb presentation. Impairment of intacellular trafficking by ethanol has been shown in other studies (28-30). Thus, we recognize that in addition to impairment of proteasome function, ethanol may affect the trafficking of the cleaved SIINFEKL peptide to the ER. Nevertheless, we also observed reductions in both proteasome activity and precursor peptide hydrolysis, indicating that ethanol-mediated changes in peptide processing machinery plays a significant role in altered antigen presentation in hepatocytes. In fact, ethanol-elicited reduction of SIINFEKL-H2Kb presentation on HepB5 cell surface was dependent on ethanol metabolism and ROS production because the effects of ethanol were reversed by 4MP and catalase. The mechanisms, by which the proteasome function is affected by ethanol-generated oxidants, have been partially defined (8, 10, 14-16).
Because antigen presentation in HepB5 cells required IFNγ pretreatment, we could not completely exclude the suppressing effects of ethanol on IFNγ signaling, similar to those observed previously in VL-17A cells (17). In HepB5 cells, we detected reduced attachment of activated STAT1 to DNA in ethanol-exposed cells, while STAT1 phosphorylation was unaffected. Our previous studies reveled the reduction of STAT1 phosphorylation by ethanol treatment in VL-17A and WIF-B cells, dependent on ethanol metabolism. In that study, STAT1 phosphorylation was partially blocked due to ethanol-induced accumulation of a negative regulator of the Jak-STAT1 signaling, suppressor of cytokine signaling 1 (SOCS1). Here, in HepB5 cells, we observed no enhanced SOCS1 expression after IFNγ and ethanol treatment (not shown). This difference between VL-17A and HepB5 cells may be partially explained by human hepatoma origin of VL-17A cells, while HepB5 cells are derived from mouse hepatocytes. In addition, ADH activity is higher in VL-17A cells as compared with HepB5 cell, which may provide the higher magnitude of oxidant generation. The mechanism by which ethanol treatment reduces the attachment of STAT1 to DNA may be based on the ethanol-elicited enhancement of a complex formation between STAT1 and protein inhibitor of activated STAT1 (PIAS1), a negative regulator of the Jak-STAT1 signaling (31), which forms a complex with activated STAT1 to compete for STAT1 attachment to DNA (32). In preliminary experiments, we immunoprecipitated STAT1, from the cell lysates. These immunoprecipitates showed more intensive PIAS1 band in cells exposed to IFNγ and ethanol (not shown) than in untreated cells. However, PIAS1 mechanism requires further investigation. As a consequence of the ethanol-impaired IFNγ signaling, there was no induction of either the immunoproteasome subunit, LMP2, or of the 20S proteasome activator, PA28, which limits capacity of the proteasome to cleave peptides for antigen presentation in an IFNγ-dependent manner. Additionally to ethanol-induced suppression of IFNγ-controlled antigen processing machinery, we observed that in HepB5 cells, ethanol metabolism also affected proteasome activity and subsequent precursor peptide hydrolysis in the IFNγ-independent manner.
In contrast to HepB5 cells, mouse hepatocytes express H2Kb spontaneously and do not require IFNγ pretreatment before delivery of the precursor peptide to the cells. To further support the role of ethanol exposure in IFNγ-independent regulation of SIINFEKL-H2Kb presentation and proteasome activity, we used primary C57Bl/6 hepatocytes for antigen processing/presentation studies. Freshly isolated hepatocytes expressed H2Kb spontaneously (without IFNγ pretreatment), before the delivery of the precursor peptide into the cell. Similar to IFNγ-treated HepB5 cells and hepatocytes, in IFNγ non-treated hepatocytes SIINFEKL-H2Kb expression on the cell surface was suppressed after ethanol exposure and 4MP prevented this effect. The latter supports the hypothesis that ethanol treatment regulates antigen processing/presentation in both an IFNγ-dependent and independent manner.
We conclude that by impairing proteasome function, ethanol metabolism reduces the processing of precursor peptides, leading to suppression of antigen presentation in liver cells.
Acknowledgments
We thank Drs M. Chen and I. Stroynowski for providing of HepB6 cells, Dr. Germain-for providing of 25D1.16 hybridoma cells, Dr. Dahn Clemens for providing plasmids for CYP2E1 and ADH transfection in HepB5 cells and Dr. Carol Casey-for help with the perfusion of mouse livers. This study was supported by NIAAA, grant 5R21 AA015379-02.
Financial support: Supported by grant number 5R21 AA015379-02, from the National Institute on Alcohol Abuse and Alcoholism (NIAAA)
List of Abbreviations
- IFNγ
interferon gamma
- CTL
cytotoxic T-lymphocytes
- MHC
major histocompatibility complex
- CYP2E1
cytochrome P450 2E1
- DAS
diallyl sulfide
- 4MP
4-methylpyrazole
- Cht-L
chymotrypsin-like activity
- ER
endoplasmic reticulum
- GM
geldanamycin
- STAT1
signal transducer and activator of transcription 1
References
- 1.Aloman C, Gehring S, Wintermeyer P, Kuzushita N, Wands JR. Chronic ethanol consumption impairs cellular immune responses against HCV NS5 protein due to dendritic cell dysfunction. Gastroenterology. 2007;132:698–708. doi: 10.1053/j.gastro.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Szabo G, Dolganiuc A. Hepatitis C and innate immunity: recent advances. Clin Liver Dis. 2008;12:675–692. doi: 10.1016/j.cld.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg AL, Cascio P, Saric T, Rock KL. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol. 2002;39:147–164. doi: 10.1016/s0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 4.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 5.Saric T, Beninga J, Graef CI, Akopian TN, Rock KL, Goldberg AL. Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J Biol Chem. 2001;276:36474–36481. doi: 10.1074/jbc.M105517200. [DOI] [PubMed] [Google Scholar]
- 6.Del-Val M, Lopez D. Multiple proteases process viral antigens for presentation by MHC class I molecules to CD8(+) T lymphocytes. Mol Immunol. 2002;39:235–247. doi: 10.1016/s0161-5890(02)00104-9. [DOI] [PubMed] [Google Scholar]
- 7.Dey A, Cederbaum AI. Alcohol and oxidative liver injury. Hepatology. 2006;43:S63–74. doi: 10.1002/hep.20957. [DOI] [PubMed] [Google Scholar]
- 8.Kessova IG, Cederbaum AI. The effect of CYP2E1-dependent oxidant stress on activity of proteasomes in HepG2 cells. J Pharmacol Exp Ther. 2005;315:304–312. doi: 10.1124/jpet.105.088047. [DOI] [PubMed] [Google Scholar]
- 9.Osna N, Clemens D, Donohue TM., Jr Ethanol suppresses interferon gamma -mediated activation of the proteasome by preventing STAT1 phosphorylation in human recombinant Hep G2 cells. Hepatology. 2002;36:334A. Abstract No. 684. [Google Scholar]
- 10.Bardag-Gorce F, French BA, Nan L, Song H, Nguyen SK, Yong H, Dede J, et al. CYP2E1 induced by ethanol causes oxidative stress, proteasome inhibition and cytokeratin aggresome (Mallory body-like) formation. Exp Mol Pathol. 2006;81:191–201. doi: 10.1016/j.yexmp.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Donohue TM, Osna NA, Clemens DL. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. Int J Biochem Cell Biol. 2006;38:92–101. doi: 10.1016/j.biocel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Bai J, Cederbaum AI. Overexpression of CYP2E1 in mitochondria sensitizes HepG2 cells to the toxicity caused by depletion of glutathione. J Biol Chem. 2006;281:5128–5136. doi: 10.1074/jbc.M510484200. [DOI] [PubMed] [Google Scholar]
- 13.Bai J, Cederbaum AI. Overexpression of catalase in the mitochondrial or cytosolic compartment increases sensitivity of HepG2 cells to tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 2000;275:19241–19249. doi: 10.1074/jbc.M000438200. [DOI] [PubMed] [Google Scholar]
- 14.Bardag-Gorce F, Li J, French BA, French SW. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp Mol Pathol. 2005;78:109–115. doi: 10.1016/j.yexmp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Osna NA, Haorah J, Krutik VM, Donohue TM., Jr Peroxynitrite alters the catalytic activity of rodent liver proteasome in vitro and in vivo. Hepatology. 2004;40:574–582. doi: 10.1002/hep.20352. [DOI] [PubMed] [Google Scholar]
- 16.Osna NA, White RL, Todero S, McVicker BL, Thiele GM, Clemens DL, Tuma DJ, et al. Ethanol-induced oxidative stress suppresses generation of peptides for antigen presentation by hepatoma cells. Hepatology. 2007;45:53–61. doi: 10.1002/hep.21442. [DOI] [PubMed] [Google Scholar]
- 17.Osna NA, Clemens DL, Donohue TM., Jr Ethanol metabolism alters interferon gamma signaling in recombinant HepG2 cells. Hepatology. 2005;42:1109–1117. doi: 10.1002/hep.20909. [DOI] [PubMed] [Google Scholar]
- 18.Chen M, S I. Establishment of long-term cultured hepatocyte cell lines. The Henry M and Lillian Straton Basic Research Single Topic Conference: Human Liver Cells in Biomedical Research; 2002; June 7-9 2002; The Airlie cente, Warrenton, Virginia. 2002. p. 43. [Google Scholar]
- 19.Clemens DL, Forman A, Jerrells TR, Sorrell MF, Tuma DJ. Relationship between acetaldehyde levels and cell survival in ethanol-metabolizing hepatoma cells. Hepatology. 2002;35:1196–1204. doi: 10.1053/jhep.2002.32668. [DOI] [PubMed] [Google Scholar]
- 20.Casey CA, Kragskow SL, Sorrell MF, Tuma DJ. Chronic ethanol administration impairs the binding and endocytosis of asialo-orosomucoid in isolated hepatocytes. J Biol Chem. 1987;262:2704–2710. [PubMed] [Google Scholar]
- 21.Osna NA, White RL, Krutik VM, Wang T, Weinman SA, Donohue TM., Jr Proteasome activation by hepatitis C core protein is reversed by ethanol-induced oxidative stress. Gastroenterology. 2008;134:2144–2152. doi: 10.1053/j.gastro.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osna NA, Clemens DL, Donohue TM., Jr Interferon gamma enhances proteasome activity in recombinant Hep G2 cells that express cytochrome P4502E1: modulation by ethanol. Biochem Pharmacol. 2003;66:697–710. doi: 10.1016/s0006-2952(03)00252-1. [DOI] [PubMed] [Google Scholar]
- 23.Otani K, Korenaga M, Beard MR, Li K, Qian T, Showalter LA, Singh AK, et al. Hepatitis C virus core protein, cytochrome P450 2E1, and alcohol produce combined mitochondrial injury and cytotoxicity in hepatoma cells. Gastroenterology. 2005;128:96–107. doi: 10.1053/j.gastro.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 24.Pine R. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 1997;25:4346–4354. doi: 10.1093/nar/25.21.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 26.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 27.Callahan MK, Garg M, Srivastava PK. Heat-shock protein 90 associates with N-terminal extended peptides and is required for direct and indirect antigen presentation. Proc Natl Acad Sci U S A. 2008;105:1662–1667. doi: 10.1073/pnas.0711365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseph RA, Shepard BD, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Microtubule acetylation and stability may explain alcohol-induced alterations in hepatic protein trafficking. Hepatology. 2008;47:1745–1753. doi: 10.1002/hep.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuma DJ, Casey CA, Sorrell MF. Effects of ethanol on hepatic protein trafficking: impairment of receptor-mediated endocytosis. Alcohol Alcohol. 1990;25:117–125. doi: 10.1093/oxfordjournals.alcalc.a044986. [DOI] [PubMed] [Google Scholar]
- 30.Nagy LE, Lakshman MR, Casey CA, Bearer CF. Ethanol and membrane protein trafficking: diverse mechanisms of ethanol action. Alcohol Clin Exp Res. 2002;26:287–293. [PubMed] [Google Scholar]
- 31.Liang M, Melchior F, Feng XH, Lin X. Regulation of Smad4 sumoylation and transforming growth factor-beta signaling by protein inhibitor of activated STAT1. J Biol Chem. 2004;279:22857–22865. doi: 10.1074/jbc.M401554200. [DOI] [PubMed] [Google Scholar]
- 32.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]


