Abstract
We demonstrate that the ligand pocket of a lipocalin from Pieris brassicae, the bilin-binding protein (BBP), can be reshaped by combinatorial protein design such that it recognizes fluorescein, an established immunological hapten. For this purpose 16 residues at the center of the binding site, which is formed by four loops on top of an eight-stranded β-barrel, were subjected to random mutagenesis. Fluorescein-binding BBP variants were then selected from the mutant library by bacterial phage display. Three variants were identified that complex fluorescein with high affinity, exhibiting dissociation constants as low as 35.2 nM. Notably, one of these variants effects almost complete quenching of the ligand fluorescence, similarly as an anti-fluorescein antibody. Detailed ligand-binding studies and site-directed mutagenesis experiments indicated (i) that the molecular recognition of fluorescein is specific and (ii) that charged residues at the center of the pocket are responsible for tight complex formation. Sequence comparison of the BBP variants directed against fluorescein with the wild-type protein and with further variants that were selected against several other ligands revealed that all of the randomized amino acid positions are variable. Hence, a lipocalin can be used for generating molecular pockets with a diversity of shapes. We term this class of engineered proteins “anticalins.” Their one-domain scaffold makes them a promising alternative to antibodies to create a stable receptor protein for a ligand of choice.
For almost one century the immunoglobulins have been considered to be the universal type of ligand-binding proteins that can be employed for a variety of purposes in the fields of biochemistry, medicine, and biotechnology. After the establishment of methods for the production of functional antibody fragments in bacteria (for review, see ref. 1) many attempts were made to engineer antibodies with improved affinities or specificities or even to select artificial antibodies by means of genetic library techniques (for review, see refs. 2 and 3). However, from the practical point of view, antibodies have certain disadvantages: (i) they are rather large molecules, and even the smallest antigen-binding fragment, Fv, consists of ≈250 amino acids; (ii) they are composed of two different polypeptide chains, which necessitates complicated cloning steps for the pair of genes and, in the case of Fv, may lead to unstable domain association; (iii) they carry six hypervariable loops, which are difficult to manipulate all at once if the generation of synthetic antibodies is desired. Therefore, the question arises whether simpler molecular architectures exist that offer the potential for creating ligand-binding molecules with the help of combinatorial protein design.
The lipocalins, a family of diverse proteins that normally serve for the storage or transport of physiologically important compounds (for review, see ref. 4), seemed to be promising in this respect. Despite low mutual sequence homology they share a conserved barrel of eight antiparallel β-strands as their central folding motif. At one end of this supersecondary structure four loops connect each pair of β-strands and form the entrance to the binding pocket. In contrast to the β-barrel, which can be well superimposed between individual lipocalins, the loop regions are remarkably dissimilar in length and conformation (Fig. 1). This localized structural diversity among the members of the lipocalin family reflects the differing shapes and chemical properties of their cognate ligands. Thus, although being composed of a single polypeptide chain and being much smaller, the lipocalins resemble immunoglobulins, which owe their vast potential of antigen specificities to the six hypervariable loops on top of a rigid framework.
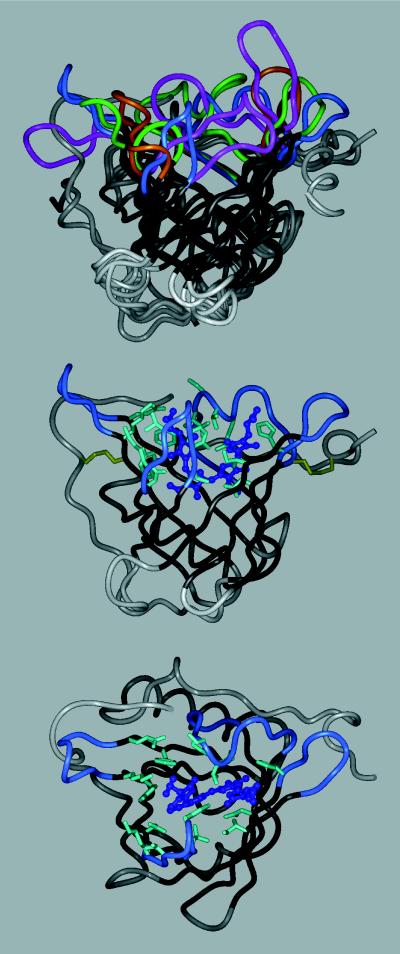
Figure 1.
Tertiary structure of the bilin-binding protein (5), either alone (Middle), with the randomized amino acids depicted, or superimposed (Top) with three other lipocalins: retinol-binding protein, RBP (6); mouse major urinary protein, MUP (7); and epididymal retinoic acid-binding protein, EPA (8). The polypeptide backbone is shown in a ribbon representation (InsightII molecular modeling software), with the β-barrel in black, the four loops surrounding the ligand pocket colored (BBP, blue; RBP, magenta; MUP, green; EPA, amber), and the remainder of the structure in gray. BBP (Middle) is shown with its natural ligand, biliverdin IXγ, in a ball and stick representation (dark blue). The 16 amino acid positions at the center of the binding site that were subjected to random mutagenesis are depicted with their original side chains (light blue). The two disulfide bonds are indicated as well (yellow). (Bottom) The same molecule is shown in an orientation that permits viewing into the binding pocket.
However, contrasting with the hundreds of millions of different antibodies that are permanently created by each individual’s immune system, there exist only a small number of lipocalins in nature and these, moreover, appear to be highly optimized for their particular task. To investigate to what extent the structure of the binding site in a given lipocalin can be deliberately modified we chose the BBP as a model system. BBP is a blue pigment protein of 174 amino acids (9), which complexes biliverdin IXγ and serves for the coloration and photoprotection of the butterfly Pieris brassicae. Its x-ray structure was determined to 0.2-nm resolution (5) and revealed the characteristic β-barrel architecture (Fig. 1). Compared with other representatives of this fold, the natural ligand pocket of BBP appeared rather wide and shallow, thus offering a promising starting point for the engineering of novel binding functions.
MATERIALS AND METHODS
Synthesis of Fluorescein Conjugates.
4-Glutarylamidofluorescein was synthesized as a mono-isomeric fluorescein derivative with an aliphatic carboxylate group linked by a spacer to a defined ring position by reacting fluoresceinamine, isomer I, with glutaric anhydride (10). The recrystallized reaction product, whose composition was confirmed by 1H NMR and fast atom bombardment MS, was activated with N-hydroxysuccinimide in the presence of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and coupled either to bovine serum albumin (BSA) or to bovine RNase A. After removal of excess reagent by gel filtration, a stoichiometry of 8 fluorescein groups per BSA molecule or of 3.3 fluorescein groups per RNase molecule was determined by using fluorescein’s extinction coefficient of 72,000 M−1⋅cm−1 at 495 nm.
Oligodeoxynucleotides.
For the construction of the BBP random library the oligodeoxynucleotides GB-1, 5′-CTT CGA CTG GTC CCA GTA CCA TGG TAA ATG GTG GGA-3′; GB-3, 5′-CCA TGG TAA ATG GTG GGA AGT CGC CAA ATA CCC CNN KNM SNN SNN KAA GTA CGG AAA GTG CGG A-3′; FS-15, 5′-GGG TAG GCG GTA CCT TCS NNA AAG TAT TCC TTG CCG TGG ATT ACM NNG TAS NNC GAA ACT TTG ACA CTC TT-3′; FS-18, 5′-AGA TCT TTC CAA TCT TGG AGT CAC CAA CTG GGT AGG CGG TAC CTT C-3′; GB-11, 5′-CCA AGA TTG GAA AGA TCT ACC ACA GCN NSA CTN NKG GAG GTN NSA CCV VSG AGN NKG TAT TCA ACG TAC TCT CC-3′; GB-4, 5′-TCT GGA GAG CAC CCA GAC MNN GTC SNN GTG TCC CTT CTT GTC CTC GTC GTA SNN GCA MNN GTA TCC GAT GAT GTA GTT-3′; and GB-2, 5′-CAC CAG TAA GGA CCA TGC TTC TGG AGA GCA CCC AGA C-3′, were synthesized by using phosphoramidite chemistry. One-letter abbreviations according to the International Union of Biochemistry nomenclature indicate the application of equimolar mixtures of the corresponding activated bases during synthesis. Thus, most of the 16 randomized base triplets code for all 20 amino acids, including TAG (amber) as the sole stop codon.
Genetic Library Construction.
The random library encoding the BBP mutants was constructed on the basis of the phasmid vector pBBP20, which was derived from the generic tetp/o expression vector pASK75 (11). pBBP20 codes for a fusion protein composed of the OmpA signal peptide, BBP (9), Strep-tag II (12) followed by an amber codon, and the C-terminal fragment of the gene III coat protein (amino acids 217–406) of the filamentous bacteriophage M13 (13). The structural gene for BBP carried four mutations: Asn-1 → Asp and Lys-87 → Ser for the grounds of monomeric structure (9) and increased protein stability, and Asn-21 → Gln and Lys-135 → Met to introduce two unique BstXI restriction sites with noncompatible overhangs permitting efficient cloning of the mutagenized gene cassette. For the random mutagenesis two PCRs (20 cycles) were performed in parallel with Taq DNA polymerase (Promega) according to the supplier’s recommendations, using pBBP20 phasmid DNA as template and GB-3/FS-15 or GB-11/GB-4, respectively, as primers. The corresponding amplification products of 159 and 164 bp were isolated by agarose gel electrophoresis, mixed, and, after addition of FS-18, used as template in the next PCR (20 cycles) with GB-1 and GB-2 as primers. The assembled mutagenized DNA fragment (371 bp) was isolated, digested with BstXI, and ligated with the similarly cut vector fragment of pBBP20. The ligation mixture containing 0.83 μg of the insert and 9.9 μg of the vector fragment was used for electroporation of competent Escherichia coli XL1-Blue cells (14). After plating on Luria–Bertani (LB) agar with 100 μg/ml ampicillin, 3.7⋅108 transformants were obtained.
Selection of Lipocalin Variants.
Phagemid particles were prepared from a mixed culture of these XL1-Blue transformants after superinfection with VCS-M13 helper phage, following published protocols (13). Synthesis of the fusion protein was induced by adding 25 μg/liter anhydrotetracycline (11) during this step with the internal amber codon being partially suppressed in the XL1-Blue background. Approximately 1011 of the isolated phagemids displaying the mutated BBP were subjected to panning with Nunc-Immuno Sticks that had been coated with the fluorescein-BSA conjugate. After 10 (in the first round 8) washing steps, bound phagemids were eluted with 0.1 M glycine⋅HCl, pH 2.2. The solution was immediately neutralized and used for infection of fresh XL1-Blue cells. From the mixture of these cells either phagemids were reamplified or the phasmid DNA was prepared.
Bacterial Protein Production.
Production of the soluble lipocalin variants was performed with the vector pBBP21, which was similar to pBBP20 but had the M13 gene III fragment deleted. In addition, it carried the gene for the bacterial protein disulfide isomerase, dsbC, as a second cistron (15). Gene expression was induced with anhydrotetracycline (11) after culturing of transformed JM83 or TG1/F− cells, and the recombinant protein was purified from the periplasmic cell fraction by means of the Strep-tag II on an affinity column with engineered streptavidin (16). The protein concentration was determined on the basis of calculated ɛ280 coefficients (17). SDS/PAGE was performed in the buffer system of Fling and Gregerson (18), and CD spectra were recorded on a JASCO J-710 instrument.
Introduction of Amino Acid Substitutions into the BBP Variants.
Site-directed mutagenesis was carried out according to standard methodology (19) with single-stranded DNA prepared from the pBBP21 derivative encoding FluA and appropriate oligodeoxynucleotides. Similarly, the amber stop codon in FluC was replaced by a Gln codon (CAG) for some of the binding experiments.
Binding Studies.
ELISAs were performed and evaluated essentially as described (16), using the fluorescein-BSA or the fluorescein-RNase conjugate for coating the microtiter plate. The BBP variant was applied in a dilution series. Bound protein was subsequently detected by means of the Strep-tag II with a streptavidin-alkaline phosphatase conjugate by following the hydrolysis of p-nitrophenyl phosphate in a SpectraMax 250 reader (Molecular Devices). Fluorescence titration was also carried out as described (16) with an LS 50 B (Perkin–Elmer) or an MSIII (Photon Technology International) instrument. Aliquots of a buffered ligand stock solution were added to 2 ml of the lipocalin at 1 μM in phosphate-buffered saline (PBS)/1 mM EDTA, and the protein fluorescence intensity was measured at 25°C. An inner filter effect due to the ligand absorption at 280 nm was corrected on the basis of analogous titrations with N-acetyl-l-tryptophanamide. In another experiment a set of solutions containing 1 μM fluorescein and various concentrations of the protein was prepared and the emission of fluorescein was detected. The data were fitted by nonlinear least-squares regression according to the law of mass action with KaleidaGraph software.
RESULTS
Construction of the BBP Random Library.
On the basis of structural considerations a set of amino acid positions at the center of BBP’s ligand pocket was chosen for targeted random mutagenesis according to two criteria. First, the residues should make contact to the natural ligand of BBP and they should in principle tolerate both small and large side-chain substitutions. Second, they should reach as deeply as possible into the ligand pocket without affecting the side-chain packing in the hydrophobic core of the protein, which is formed by the lower part of the β-barrel. As a result, altogether 16 amino acid positions, spread across the four loops and adjoining parts of the β-barrel, were used for the generation of the random library. Their location in the three-dimensional structure of BBP is illustrated in Fig. 1, and the corresponding sequence positions are listed in Table 1.
Table 1.
Sequence characteristics and dissociation constants of the fluorescein-binding lipocalin variants
| Residue no. | Residue present
|
||||
|---|---|---|---|---|---|
| BBP | FluA | FluB | FluC | FluD | |
| 34 | Asn | Ser | Gln | Ser | Glu |
| 35 | Ser | Pro | His | Lys | Ala |
| 36 | Val | Asn | Trp | Asn | Gly |
| 37 | Glu | Gly | Asp | Gly | Asn |
| 58 | Asn | Arg | Arg | Arg | Arg |
| 60 | His | Asp | Arg | Thr | Ser |
| 69 | Ile | Met | His | Gln* | Ser |
| 88 | Leu | Arg | Val | Arg | Ser |
| 90 | Tyr | Val | Arg | Val | Val |
| 93 | Val | Tyr | Arg | Lys | Arg |
| 95 | Lys | Arg | Arg | Arg | Arg |
| 97 | Asn | Thr | Gly | Gly | Val |
| 114 | Tyr | Ser | Arg | Arg | Arg |
| 116 | Lys | Arg | Arg | Arg | Leu |
| 125 | Gln | Trp | Trp | Leu | Arg |
| 127 | Phe | His | His | His | Leu |
| 40 | Gly | Arg† | |||
| 68 | Phe | Val† | |||
| 70 | Glu | Lys† | |||
| 96 | Glu | Lys† | |||
| Kd,‡ nM | >5·103¶ | 536 ± 174 | 55 ± 8 | 87 ± 11 | |
| Kd,§ nM | >105¶ | 427 ± 10 | — ∥ | 61.8 ± 3.1 | |
*Arising from an amber termination codon in a supE background.
Accidental mutation position not included in the random mutagenesis.
Determined as the protein concentration giving rise to half-maximal binding in an ELISA with the immobilized fluorescein-RNase conjugate.
Determined for 4-glutarylamidofluorescein by fluorescence titration of the protein.
Conservative estimate (cf. footnote to Table 2).
∥ Insufficient fluorescence quenching effect.
The 16 positions were mutagenized in a polymerase chain reaction (PCR) with degenerate oligodeoxynucleotide primers and the cloned BBP cDNA (9) as template. The mutations were simultaneously introduced into the four loop segments by means of a two-step approach. In the first step two separate PCRs were carried out such that in both reactions a pair of neighboring loop regions were mutagenized with two primers carrying degenerate codons at the relevant amino acid positions. In the second step the two isolated DNA fragments, each comprising the sublibrary for one pair of loops, were assembled by PCR.
The resulting collection of DNA molecules, which represented the BBP random library, was inserted into an appropriate phasmid vector (see Materials and Methods) and used for electroporation of E. coli. Thus, a genetic library with 3.7⋅108 independent transformants was obtained. After collection of the colonies and coinfection with a helper phage a pool of ≈1012 phagemids displaying the BBP variants (fused with a C-terminal fragment of the M13 gene III coat protein) was finally prepared (13).
Selection and Analysis of BBP Variants with Affinity for Fluorescein.
The BBP phagemid library was selected for binding to fluorescein, a well known immunological hapten (20). For this purpose the ligand was covalently attached by means of a flexible spacer to BSA and adsorbed onto a plastic surface. After six cycles of panning and phagemid reamplification the pooled phasmid DNA was isolated and the mutated BBP gene cassette was subcloned into a vector for expression of individual lipocalin variants in the periplasm of E. coli. DNA sequence analysis of 10 randomly selected clones revealed that just four different variants were encoded, which were subsequently named FluA (2 clones), FluB (4 clones), FluC (3 clones), and FluD (1 clone). Their nucleotide sequences were translated and the amino acid differences compared with BBP are summarized in Table 1.
The four variants as well as the recombinant wild-type BBP were produced in E. coli and purified (Fig. 2) by means of the Strep-tag II (12, 16). The affinities of the soluble proteins for their new ligand were first investigated in an ELISA with the fluorescein-BSA conjugate. The lipocalin variants FluA, FluB, and FluC gave rise to strong signals with a typical saturation behavior, whereas FluD and BBP just exhibited background activity. To confirm that the binding activity was not dependent on BSA as a carrier, another ELISA was performed with fluorescein coupled to RNase, and a similar result was obtained (for the apparent Kd values see Table 1).
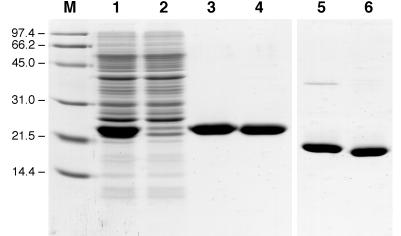
Figure 2.
A 0.1% SDS/15% polyacrylamide gel illustrating the purification of the bacterially produced FluA as a Strep-tag II fusion protein. Lane M, molecular size standard (masses labeled in kDa); lane 1, periplasmic protein extract of E. coli JM83 harboring pBBP21-FluA after overnight expression at 22°C; lane 2, flow-through of the streptavidin affinity column; lane 3, FluA eluted from the column; lane 4, BBP prepared in the same way after 3-h induction; lanes 5 and 6, as lanes 3 and 4 but without reduction of the disulfide bonds.
Affinities for glutarylamidofluorescein—i.e., the hapten derivative carrying the spacer group—were then determined by titrating the BBP variants with this soluble ligand and monitoring the fluorescence intensity of the protein’s Tyr and Trp residues. Quenching effects were observed for the variants FluA (QmaxProtein = 88.8% ± 0.3%) and, less pronounced, for FluC (QmaxProtein = 44% ± 2%) so that the corresponding dissociation constants (Kd) could be derived. According to these numbers, which were consistent with the Kd values from the solid-phase assays mentioned above, all three selected BBP variants bound to the hapten with submicromolar dissociation constants (Table 1).
Biochemical Characterization of a Soluble “Anticalin.”
Because of its favorable properties—i.e., high expression yield (≈5 mg/liter of shaker flask culture vs. ≈100 μg/liter for FluB and FluC), lack of an internal amber stop codon, and strong fluorescence quenching effect—the variant FluA was chosen for further analysis. The CD spectrum in the far-UV region was typical for a β-sheet protein and revealed close similarity in secondary structure content compared with BBP (not shown). In addition, the gel-electrophoretic mobilities were almost indistinguishable for the two proteins and, notably, were significantly enhanced when the disulfide bonds had not been reduced (see Fig. 2). Obviously, the gross conformation of the polypeptide chain was not changed in the engineered lipocalin and the two disulfide bridges were present.
The ligand-binding properties of FluA were investigated in solution by fluorescence titration of the purified protein with fluorescein and related compounds (Fig. 3; Table 2). Interestingly, it turned out that underivatized fluorescein was even bound tighter than 4-glutarylamidofluorescein, which had been used in its immobilized form for the selection, thus indicating an adverse influence of the spacer group on the molecular recognition by this lipocalin variant. In contrast, the triphenylmethane compounds pyrogallol red, phenolphthalein, and rhodamine B were poorly bound. Hence, FluA specifically recognizes fluorescein and discriminates between chemically quite similar substances.
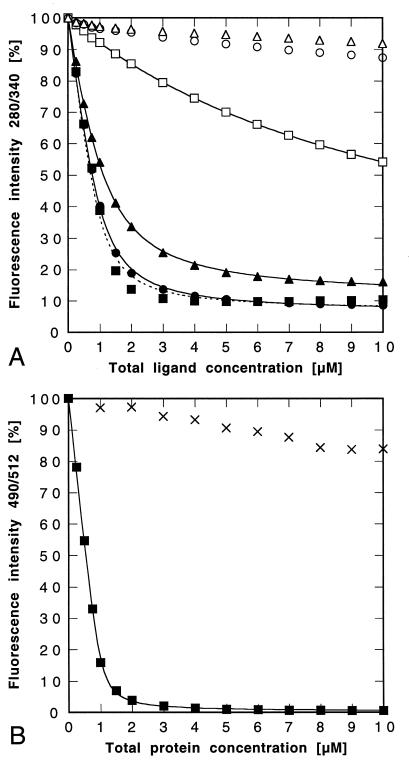
Figure 3.
Ligand binding studies with the purified lipocalin variant FluA. (A) Fluorescence titration (λEx = 280 nm; λEm = 340 nm) of a 1 μM protein solution with fluorescein (■), 4-aminofluorescein (●), 4-glutarylamidofluorescein (▴), pyrogallol red (□), phenolphthalein (○), and rhodamine B (▵). The data were corrected and fitted as described in Materials and Methods. The resulting Kd values are given in Table 2. In the case of fluorescein the curve (dotted line) could not be fitted as perfectly as with the other ligands. (B) Fluorescence titration (λEx = 490 nm; λEm = 512 nm) of a 1 μM fluorescein solution with FluA (■) or with BBP (×). The data for FluA were fitted without correction, yielding Kd = 35.2 ± 3.2 nM.
Table 2.
Dissociation constants for the interaction between FluA, FluC, orBBP and fluorescein as well as related compounds
| Ligand |
Kd, μM
|
||
|---|---|---|---|
| FluA | FluC | BBP | |
| Fluorescein | 0.152 ± 0.024 | 0.418 ± 0.029 | >50* |
| 4-Aminofluorescein | 0.201 ± 0.009 | 0.278 ± 0.044 | >100* |
| 4-Glutarylamidofluorescein | 0.427 ± 0.010 | 0.062 ± 0.003 | >100* |
| Pyrogallol red | 10.07 ± 0.14 | >100* | |
| Phenolphthalein | >50* | >100* | |
| Rhodamine B | >100* | 3.75 ± 0.09 | |
Kd values were determined by fluorescence titration of the purified protein with the ligand according to Materials and Methods.
Binding effects were so weak that lower estimates for the Kd were derived by comparing the data with simulated binding isotherms.
FluC, on the other hand, exhibited weaker affinity for fluorescein than for its glutarylamido derivative, as could be anticipated. In control experiments no binding effects were observed with the recombinant BBP and the fluorescein compounds. The same was the case when FluA was titrated with fluorescein after denaturation in the presence of 6 M Gdn⋅HCl (not shown). Surprisingly, some weak affinity was detected between BBP and rhodamine B (see Table 2), which carries positively charged groups and might in this respect resemble biliverdin IXγ, the natural tetrapyrrole ligand.
Finally, because of the known quenching effect by the anti-fluorescein antibody 4-4-20 (21) on the fluorescence emission of fluorescein (QmaxLigand = 83.6%), we investigated whether FluA has a similar influence on the spectroscopic properties of this hapten. A solution of the underivatized compound was therefore titrated with FluA, and its characteristic fluorescence was followed (Fig. 3B). Almost complete quenching of the fluorescence emission was observed in this case (QmaxLigand = 99.7% ± 0.3%), whereas a control titration with BBP did not reveal a comparable effect. Since the experimental data for the lipocalin variant FluA could be directly fitted according to the theory of bimolecular complex formation, the resulting dissociation constant of 35.2 ± 3.2 nM seems to be a reliable number for this protein–fluorescein complex.
A Model for the Molecular Interaction with Fluorescein.
From a sequence comparison between FluA and the two other fluorescein-binding variants, FluB and FluC, four identical amino acids became apparent among the randomized positions (cf. Table 1): Arg-58, Arg-95, Arg-116, and His-127. This preponderance of positively charged side chains was again reminiscent of the antibody 4-4-20 (21, 22). A similar tendency had been noted in an attempt to select antibody fragments with specificity for fluorescein from a semisynthetic combinatorial library (23).
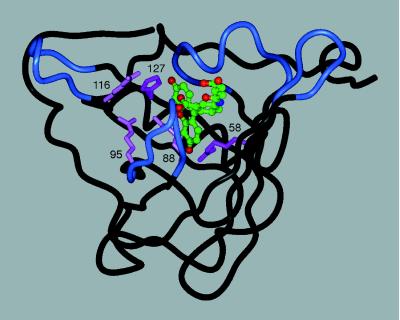
In the 4-4-20/fluorescein complex the xanthenolone moiety of the hapten is held in an extended orientation between a His side chain and an Arg side chain, whose Cα positions are 14.1 Å apart (24). On the basis of a similar distance between the Cα atoms of the residues in the crystal structure of BBP (14.8 Å) corresponding to His-127 and Arg-58 in FluA we modeled the complex between this BBP variant and 4-glutarylamidofluorescein (Fig. 4). The predicted interactions were supported by the following data from site-directed mutagenesis studies with FluA (Table 3).
Figure 4.
Tentative three-dimensional model of FluA complexed with 4-glutarylamidofluorescein (for coloring cf. Fig. 1). The backbone conformation corresponds to the crystal structure of BBP, whereas the five basic residues were introduced with their preferred side-chain rotamers (25) by using InsightII modeling software. The fluorescein structure was taken from the coordinates of its complex with the antibody 4-4-20 (24) and equipped with the aliphatic substituent in an extended standard conformation. The ligand (C, green; O, red; N, blue) was manually placed such that its fluorescein moiety can form hydrogen bonds with the two critical His and Arg residues (magenta), similarly as in the crystal structure of the antibody complex. Another three Arg residues (pink) can participate in electrostatic interactions with the negatively charged hapten.
Table 3.
Effects of substituting positively charged side chains in the ligand pocket of the fluorescein-binding BBP variant FluA
| Residue | Substitution | Kd, μM | Substitution | Kd, μM |
|---|---|---|---|---|
| FluA wild type | 0.427 ± 0.010 | |||
| His-127 | Phe | 5.56 ± 0.05 | ||
| Arg-58 | Lys | 25.1 ± 2.3 | ||
| Arg-88 | Lys | 0.356 ± 0.008 | Gln | 2.23 ± 0.03 |
| Arg-95 | Lys | 0.443 ± 0.012 | Gln | 0.918 ± 0.023 |
| Arg-116 | Lys | 0.597 ± 0.019 | Gln | 1.54 ± 0.04 |
Kd values were measured for 4-glutarylamidofluorescein by fluorescence titration of the protein.
Both the exchange His-127 → Phe and the exchange Arg-58 → Lys led to a strong drop in the affinity (by factors of 13 and 59, respectively), suggesting direct interactions of the imidazole and guanidinium groups with the ligand. In contrast, singular replacement of the Arg residues 95 and 116 and also of Arg-88 by Lys had virtually no effect, whereas substitution with Gln resulted in clearly increased Kd values. These experiments indicated that the fluorescein derivative is bound at the center of the ligand pocket and that electrostatic attractions contribute to the affinity, apart from specific interactions with some of the side chains.
DISCUSSION
Using a combinatorial protein design strategy, we identified three variants of BBP that recognize fluorescein, a compound with a rather different chemical structure compared with BBP’s natural ligand, biliverdin IXγ. Up to now lipocalins have mainly been known for their ability to complex hydrophobic organic molecules in a presumably promiscuous manner (4). However, a cluster of positively charged residues was found common to the three fluorescein-binding variants, which can be explained by the fact that the new ligand (especially the glutarylamido derivative) possesses several negative charge centers. Therefore, polar interactions play a major role in the molecular recognition. Closer investigation of the variant FluA revealed that the complexation of fluorescein is specific, and even compounds with just minor alterations in the chemical structure are much less well bound.
The described lipocalin variants complex a hapten with high affinity and specificity in a way that has so far been considered as typical for immunoglobulins. The observed dissociation constants are comparable to those for antibodies from the second immune response. Consequently, we denominate this class of small receptor proteins “anticalins.” Especially the anticalin FluA could be of practical value—e.g., for the detection of biomolecules coupled with fluorescein as a nonradioactive label (26) or because of its strong quenching effect (27) on this widely used fluorescent reagent. If desired it should furthermore be possible to raise the affinity by additional rounds of limited mutagenesis and selection, similar to what was demonstrated for engineered antibody fragments (28, 29).
The search for alternate protein scaffolds that are suitable for the engineering of novel binding sites has attracted recent attention (for review, see ref. 30). Single immunoglobulin domains (especially of the “camel VH” type, see ref. 31), helix-bundle structures, small protease inhibitors, and even disulfide-knotted peptides (32) were recruited for the generation of molecular repertoires and subsequent selection of artificial binding proteins. However, successful reports remained restricted to biological macromolecules as targets. In an attempt to select mutants of cytochrome b562 against an organic compound, for example, the hapten was recognized only when attached to the BSA carrier (33).
Hence it seems that the anticalins provide the first framework that permits the specific complexation of small molecules. Compared with conventional immunoglobulins, the use of the lipocalin scaffold should constitute an advantage, given, e.g., the well known difficulties in raising antibodies against metabolic compounds. Anticalins with specificities for ligands different from fluorescein have already been generated as well. A compilation of preliminary sequence data for BBP variants that were selected against several steroids and peptides, starting from the random library described here, revealed that none of the 16 mutagenized amino acid positions is apparently restricted for maintaining the structural integrity of the protein (S. Schlehuber, M. Weichel, T. Keller, T.S., and A.S., unpublished results).
In the present library a set of side chains at the center of BBP’s ligand pocket was chosen for random mutagenesis not only to create pockets that are likely to accommodate small ligands. Concomitantly it was sought to prove that the β-barrel tolerates amino acid exchanges even close to its hydrophobic core. Our observations indicate that the β-barrel of the lipocalins indeed represents a stable scaffold that can support binding sites with diverse shapes. The selection strategy described here should therefore be applicable to a variety of haptens.
Moreover, it seems reasonable that amino acid positions at more extreme positions of the four loops surrounding the pocket can be randomized as well, thus generating extended interfaces for the complexation of larger, antigen-like ligands. In this respect it should be noted that several lipocalins—such as the retinol-binding protein (RBP), which circulates through human serum in a complex with transthyretin (34)—can naturally interact with other proteins. Lipocalins of human origin could also be useful for the development of anticalins with therapeutic potential, apart from bioanalytical applications.
From a structural point of view it is remarkable how the lipocalins can furnish the substantial plasticity that is necessary for the specific recognition, especially of small ligands with a single and rather short polypeptide chain. One could thus wonder why the lipocalin architecture had not been selected during the evolution of the immune system. An explanation might be found on the genetic level because the combination of the independently folding heavy and light chains of immunoglobulins provides an efficient means for creating structural diversity by somatic gene shuffling. Since modern biochemistry offers other approaches for the combinatorial variation of amino acid residues on a given framework, as we could demonstrate here, the anticalins might have a future for replacing antibodies in several areas.
Acknowledgments
This work was supported by the Fonds der Chemischen Industrie, the Boehringer Ingelheim Fonds, and Grant 0310915 from the Bundesministerium für Forschung und Technologie, Germany.
ABBREVIATION
- BBP
bilin-binding protein from Pieris brassicae
References
- 1.Skerra A. Curr Opin Immunol. 1993;5:256–262. doi: 10.1016/0952-7915(93)90014-j. [DOI] [PubMed] [Google Scholar]
- 2.Winter G, Milstein C. Nature (London) 1991;349:293–299. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 3.Hoogenboom H R. Trends Biotechnol. 1997;15:62–70. doi: 10.1016/S0167-7799(97)84205-9. [DOI] [PubMed] [Google Scholar]
- 4.Flower D R. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huber R, Schneider M, Mayr I, Müller R, Deutzmann R, Suter F, Zuber H, Falk H, Kayser H. J Mol Biol. 1987;198:499–513. doi: 10.1016/0022-2836(87)90296-8. [DOI] [PubMed] [Google Scholar]
- 6.Cowan S W, Newcomer M E, Jones T A. Proteins Struct Funct Genet. 1990;8:44–61. doi: 10.1002/prot.340080108. [DOI] [PubMed] [Google Scholar]
- 7.Böcskei Z, Groom C R, Flower D R, Wright C E, Phillips S E V, Cavaggioni A, Findlay J B C, North A C T. Nature (London) 1992;360:186–188. doi: 10.1038/360186a0. [DOI] [PubMed] [Google Scholar]
- 8.Newcomer M E. Structure. 1993;1:7–18. doi: 10.1016/0969-2126(93)90004-z. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt F S, Skerra A. Eur J Biochem. 1994;219:855–863. doi: 10.1111/j.1432-1033.1994.tb18567.x. [DOI] [PubMed] [Google Scholar]
- 10.Ogamo A, Matsuzaki K, Uchiyama H, Nagasawa K. Carbohydrate Res. 1982;105:69–85. doi: 10.1016/s0008-6215(00)81855-8. [DOI] [PubMed] [Google Scholar]
- 11.Skerra A. Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt T G M, Koepke J, Frank R, Skerra A. J Mol Biol. 1996;255:753–766. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- 13.Kay B K, Winter J, McCafferty J. Phage Display of Peptides and Proteins: A Laboratory Manual. San Diego: Academic; 1996. [Google Scholar]
- 14.Tung W L, Chow K C. Trends Genet. 1995;11:128–129. doi: 10.1016/s0168-9525(00)89022-8. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt A M, Bloß I, Skerra A. Protein Eng. 1998;11:601–607. doi: 10.1093/protein/11.7.601. [DOI] [PubMed] [Google Scholar]
- 16.Voss S, Skerra A. Protein Eng. 1997;10:975–982. doi: 10.1093/protein/10.8.975. [DOI] [PubMed] [Google Scholar]
- 17.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 18.Fling S P, Gregerson D S. Anal Biochem. 1986;155:83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- 19.Geisselsoder J, Witney F, Yuckenberg P. BioTechniques. 1987;5:786–791. [Google Scholar]
- 20.Voss E W., Jr . Fluorescein Hapten: An Immunological Probe. Boca Raton, FL: CRC; 1984. [Google Scholar]
- 21.Denzin L K, Whitlow M, Voss E W., Jr J Biol Chem. 1991;266:14095–14103. [PubMed] [Google Scholar]
- 22.Herron J N, He X, Mason M L, Voss E W, Jr, Edmundson A B. Proteins Struct Funct Genet. 1989;5:271–280. doi: 10.1002/prot.340050404. [DOI] [PubMed] [Google Scholar]
- 23.Barbas C F, III, Bain J D, Hoekstra D M, Lerner R A. Proc Natl Acad Sci USA. 1992;89:4457–4461. doi: 10.1073/pnas.89.10.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitlow M, Howard A J, Wood J F, Voss E W, Jr, Hardman K D. Protein Eng. 1995;8:749–761. doi: 10.1093/protein/8.8.749. [DOI] [PubMed] [Google Scholar]
- 25.Ponder J W, Richards F M. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 26.Harmer I J, Samuel D. J Immunol Methods. 1989;122:115–121. doi: 10.1016/0022-1759(89)90341-4. [DOI] [PubMed] [Google Scholar]
- 27.Bar-Noy S, Darmon A, Ginsberg H, Cabantchik Z I. Biochim Biophys Acta. 1984;778:612–614. doi: 10.1016/0005-2736(84)90413-9. [DOI] [PubMed] [Google Scholar]
- 28.Low N M, Holliger P, Winter G. J Mol Biol. 1996;260:359–368. doi: 10.1006/jmbi.1996.0406. [DOI] [PubMed] [Google Scholar]
- 29.Barbas C F, III, Burton D R. Trends Biotechnol. 1996;14:230–234. doi: 10.1016/0167-7799(96)10029-9. [DOI] [PubMed] [Google Scholar]
- 30.Nygren P-Å, Uhlén M. Curr Opin Struct Biol. 1997;7:463–469. doi: 10.1016/s0959-440x(97)80108-x. [DOI] [PubMed] [Google Scholar]
- 31.Davies J, Riechmann L. Biotechnology. 1994;13:475–479. doi: 10.1038/nbt0595-475. [DOI] [PubMed] [Google Scholar]
- 32.Smith G P, Patel S U, Windass J D, Thornton J M, Winter G, Griffiths A D. J Mol Biol. 1998;277:317–332. doi: 10.1006/jmbi.1997.1621. [DOI] [PubMed] [Google Scholar]
- 33.Ku J, Schultz P G. Proc Natl Acad Sci USA. 1995;92:6552–6556. doi: 10.1073/pnas.92.14.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monaco H L, Rizzi M, Coda A. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]