Abstract
Serine/threonine kinases secreted from rhoptry organelles constitute important virulence factors of Toxoplasma gondii. Rhoptry kinases are highly divergent and their structures and regulatory mechanism are hitherto unknown. Here, we report the X-ray crystal structures of two related pseudokinases named ROP2 and ROP8, which differ primarily in their substrate-binding site. ROP kinases contain a typical bilobate kinase fold and a novel N-terminal extension that both stabilizes the N-lobe and provides a unique means of regulation. Although ROP2 and ROP8 were catalytically inactive, they provided a template for homology modelling of the active kinase ROP18, a major virulence determinant of T. gondii. Autophosphorylation of key residues in the N-terminal extension resulted in ROP18 activation, which in turn phosphorylated ROP2 and ROP8. Mutagenesis and mass spectrometry experiments revealed that ROP18 was maximally activated when this phosphorylated N-terminus relieved autoinhibition resulting from extension of aliphatic side chains into the ATP-binding pocket. This novel means of regulation governs ROP kinases implicated in parasite virulence.
Keywords: autophosphorylation, serine/threonine kinase, signaling, virulence factor
Introduction
Toxoplasma gondii is an obligate intracellular parasite that actively invades a wide range of cell types where it survives and replicates within a non-fusigenic vacuole (Sibley et al, 2007). During host invasion, the contents of apical secretory organelles called rhoptries are discharged into the forming vacuole (Carruthers and Sibley, 1997). In the process, some rhoptry proteins (ROPs) are also released directly into the host cell cytosol at the point of cell invasion (Håkansson et al, 2001). The mechanism by which ROP proteins cross the host membrane is unclear, but it may involve a momentary breach in the cell wall followed by resealing, as suggested by early electron microscopy observations (Nichols and O'Connor, 1981) and more recent electrophysiology measurements (Suss-Toby et al, 1996). A recent proteomic study of the contents of T. gondii rhoptries characterized ∼40 constitutive proteins from this organelle, including those with conserved domains for kinases, proteases and phosphatases (Bradley et al, 2005). The functions of ROP proteins are largely undefined and limited to suggestions of a potential role of ROP2 in modulation of the parasite-containing vacuole. Although this compartment resists fusion with lysosomes and endosomes, it is enshrouded with host endoplasmic reticulum and mitochondria (Sibley et al, 2007). This association may be mediated by interaction with the N-terminus of the ROP2 protein (Sinai and Joiner, 2001), which is characterized by a series of low complexity regions. The original model for ROP2 suggested it was an integral membrane protein that spanned the parasite-containing vacuole membrane (Beckers et al, 1994). However, recent analysis of the related rhoptry protein ROP5 suggests that ROPs may adopt a kinase fold, thus burying the putative transmembrane domain within the core of the protein (El Hajj et al, 2006).
Recent genetic studies have demonstrated that ROP proteins are the major mediators of acute virulence in laboratory mice (Bradley and Sibley, 2007; Boothroyd and Dubremetz, 2008), which provide a model for toxoplasmosis in humans. ROP16 was identified based on its influence on host gene transcription and consistent with this, it has been observed to accumulate in the host nucleus following parasite infection (Saeij et al, 2007). ROP18 was identified in two parallel genetic crosses that mapped genes affecting acute virulence of T. gondii (Saeij et al, 2006; Taylor et al, 2006). ROP18 is highly expressed in type I strains where it accounts for more than 90% of the extreme virulence phenotype characteristic of this lineage (Taylor et al, 2006). ROP18 also contributes to the intermediate level of virulence seen in type II strains (Saeij et al, 2006). The virulence enhancing potential of ROP18 relies on its kinase activity (Taylor et al, 2006; El Hajj et al, 2007), although the precise target(s) of this kinase are unknown. Rhoptries also contain a large number of related proteins that contain kinase domains, with a number of these likely not active in phosphorylation and can therefore be classified as pseudokinases (Boothroyd and Dubremetz, 2008).
Serine/threonine (S/T) kinases are highly conserved in eukaryotes and have been grouped into several major families based on sequence conservation and domain structure (Hanks and Hunter, 1995). In general, S/T kinases contain 12 subdomains encapsulated in structural features that include an N-terminal lobe, hinge region and a C-terminal lobe. The N-lobe has a predominantly antiparallel β-mesh primarily involved in binding and orienting the nucleotide ATP. The C-lobe is typically α-helical and largely responsible for peptide substrate binding and mediating phosphotransfer (Hanks and Hunter, 1995). The two lobes are joined at a hinge that includes a long helix known as αC. Binding to ATP typically results in flexing of the hinge to bring the two lobes closer together, a motion that is often accompanied by re-orientation of the αC helix (Jeffrey et al, 1995; Kannan et al, 2008).
A set of conserved motifs within the core of each kinase are essential for catalysis (Hanks and Hunter, 1995). Those deficient in these motifs but featuring the bilobed fold are known as pseudokinases, which can nevertheless have important roles in regulating diverse cellular processes (Boudeau et al, 2006). For example, some pseudokinases have been found to be active under specific physiological conditions in vivo (Mukherjee et al, 2008). In JAK kinases, a conventional active tyrosine kinase domain (JH1) is adjoined by a catalytically inactive pseudokinase domain (JH2), which regulates the activity of the fusion protein (Saharinen and Silvennoinen, 2002). In another example, the activity of an active kinase, LKB1, was increased 100-fold upon binding with the pseudokinase domain of STRAD (Baas et al, 2003).
Apicomplexans contain a diverse array of kinases, including those belonging to the CMGC, CaMK and AGC families, as well as a few novel families seen only in parasites (Ward et al, 2004). For example, ROP kinases are not highly conserved with those found in mammalian cells or yeast, suggesting that they have diverged for specialized functions. Notably, although Plasmodium, the causative agent of malaria, is related to T. gondii, it does not share the ROP kinases, but rather has its own distinct family of secretory kinases called FIKKs (Ward et al, 2004). Named for a conserved stretch of amino acids they contain, FIKK kinases are over-represented in Plasmodium falciparum, where they are exported from the vacuole of infected red blood cells through a PEXEL motif (Schneider and Mercereau-Puijalon, 2005; Nunes et al, 2006).
Previous studies modelled the catalytic core of ROP18 on the mammalian kinase Tao2, a homologue of the yeast Ste20 MAP kinase family member; however, this model was largely incomplete due to sequence divergence of ROP18 (Taylor et al, 2006). To provide a framework for exploring the functions of ROP kinases, we expressed ROP2 and ROP8—two canonical but inactive members of the ROP2 family—as recombinant proteins, obtained their crystallographic structures and generated a homology model of ROP18 that revealed insight into its mechanism of regulation.
Results and discussion
Phylogenetic comparison of ROP kinases
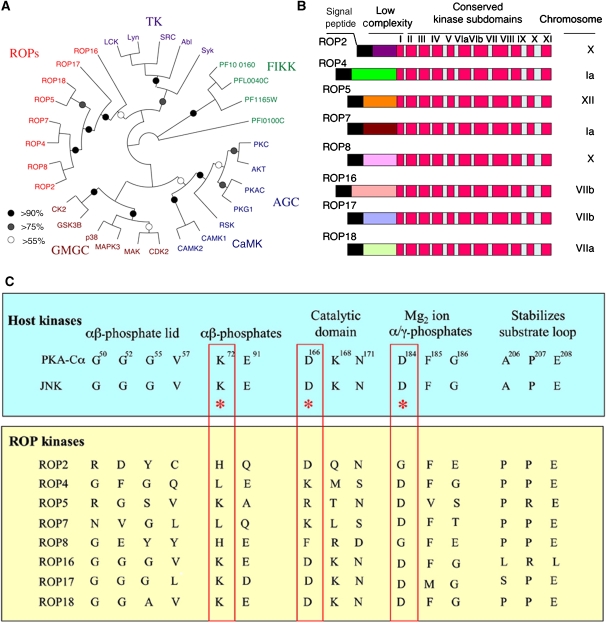
To provide insight into their functional and evolutionary relationships, ROP2 family members were compared with a set of eukaryotic kinases representing the major families of S/T and tyrosine kinases, as described previously (Hanks and Hunter, 1995). Phylogenetic analysis revealed that the ROP2 family was divergent from AGC, CaMK, CMGC and TK kinases and instead formed its own deep-branching group (Figure 1A). Several groups of ROP paralogues were supported from this analysis, including ROP2–ROP8, ROP4–ROP7 and ROP5–ROP18, suggesting that this gene family has undergone recent divergence due to gene duplication (Figure 1A). Included for comparison were ROP16 and ROP17, two other related parasite ROP kinases that branch separately from the ROP2 clade and define other major subfamilies of the ROP kinase superfamily (data not shown). ROP kinases also branched separately from and were not orthologues of FIKK kinases found in P. falciparum (Figure 1A), despite the fact that both are unique to apicomplexan organisms and share a S/T kinase domain. A larger analysis of additional ROP kinases from T. gondii and with other divergent eukaryotic kinases was consistent with the conclusion that the ROP family of kinases constitutes a highly divergent group (data not shown).
Figure 1.
Relationship of ROP kinases to conventional S/T kinases. (A) Phylogenetic comparison of Toxoplasma ROP kinases and Plasmodium FIKK kinases with typical eukaryotic kinases from the AGC, CaMK, GMGC and tyrosine kinase (TK) families. Neighbour-joining distance analysis, bootstrap values are indicated by %. (B) Conserved domain structure of ROPs containing N-terminal signal peptide (black) followed by a low-complexity region and a conserved C-terminal domain including the 12 subdomains typical of S/T kinases (Hanks and Hunter, 1995). (C) Conservation of key ATP-binding and catalytic residues in host and ROP kinases. For comparison, human protein kinase A (PKA-Cα, numbering of residues) and JNK are shown. ROP2 and ROP8 lack the conserved residues in the catalytic triad (KDD in boxed segments) and also contain degenerate residues involved in ATP binding. Only ROP17 and ROP18 contained conserved ATP-binding residues and a highly conserved catalytic triad.
Domain structures of ROP kinases
The domain structure of ROPs was analysed using InterProScan to define conserved domains. All of the ROPs examined here contain an N-terminal signal peptide recognized by SignalP (probability of >0.9) (Figure 1B). Internal transmembrane domains were not predicted by this analysis. Additionally, most ROPs contained the signature ROP2 domain (IPR016815) and a core S/T kinase domain (IPR000719) (Figure 1B). The N-terminal domains of ROPs were not highly conserved but contained several stretches of low complexity defined by an abundance of basic residues, which are predicted to form alpha helical regions.
Previous studies on eukaryotic kinases have revealed 12 key subdomains involved in ATP binding and γ-phosphate transfer (Hanks and Hunter, 1995) (Figure 1B). However, most ROPs are degenerate in key positions normally invariant in active eukaryotic kinases (Figure 1C). For example, ROP2 lacks the conserved G-rich loop implicated in binding to the αβ-phosphates of ATP (Figure 1C). Also missing are the lysine residue corresponding to K72 in subdomain II of PKA and part of the catalytic triad, as well as the aspartic acid of the DFG motif corresponding to D184 in subdomain VII of PKA (numbering of amino acids based on the Protein Data Bank record 1ATP) and involved in Mg2+ ion binding. Both residues are thought to be essential for optimal kinase activity (Hanks and Hunter, 1995), and their absence predicts ROP2 to be inactive. ROP4, ROP5, ROP7, and ROP8 were similarly degenerate in one or more of these three regions and therefore are also unlikely to be catalytically active (Figure 1C). In contrast, ROP17 and ROP18 contained a conserved catalytic triad (red boxes in Figure 1C) as well as showing excellent conservation of other key residues. ROP16 was intermediate in that it lacks K72 (E instead), but otherwise retains the other key catalytic residues (Figure 1C). Although K72 is often essential, there are exceptions, for example the human kinase WNK1 lacks the equivalent of K72 yet is active; substitution of a K residue from subdomain I fulfills this critical function (Boudeau et al, 2006). Finally, ROPs contained a highly conserved segment in subdomain VII involved in substrate binding where the APE triad commonly found in other kinases was converted to PPE (Figure 1C).
ROP18 actively phosphorylates other ROPs
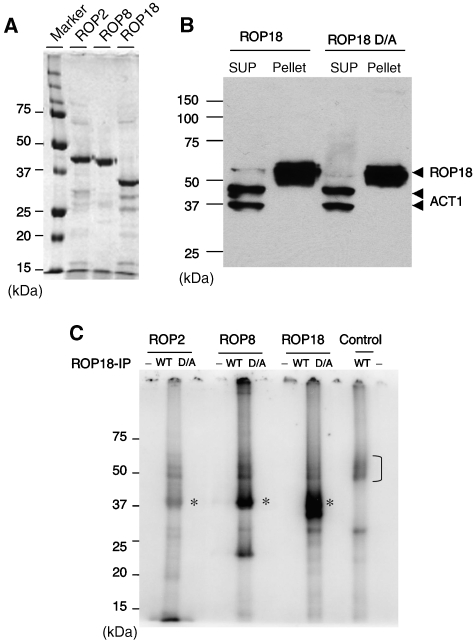
To examine the functional activity of ROPs, we tested the ability of ROP18 to phosphorylate other recombinant ROPs purified from Escherichia coli (Figure 2A). Active ROP18 was obtained by immunoprecipitation from transgenic parasites that overexpress the type I allele of ROP18, or an inactive version (D394A) from parasites expressing a mutant kinase (Figure 2B), as described previously (Taylor et al, 2006). Addition of ROP18 that was immunoprecipitated from wild-type cells to recombinant ROP2, ROP8 or a truncated form of ROP18, resulted in phosphorylation of these substrates in a 32P-ATP kinase assay (* in Figure 2C). Labelling of ROP2 was relatively weak, whereas ROP8 and ROP18 were strongly labelled (Figure 2C). Recombinant ROP2, ROP8 and ROP18 were not labelled when a catalytically inactive form of ROP18 (D/A) was added, indicating that phosphorylation relies on the kinase activity of ROP18 (Figure 2C). This result is consistent with ROP2 and ROP8 being inactive, as confirmed when they were incubated alone in the kinase reaction (Figure 2C), although it remains formally possible that they might be active on different substrates not tested here. ROP18, which was immunoprecipitated from parasites, underwent autophosphorylation in the presence and absence of ROPs as substrates (bracket in Figure 2C). We conclude from these experiments that ROP18 is able to actively phosphorylate recombinant ROP2 and ROP8 as well as undergo autophosphorylation in vitro. Whether ROP18 also phosphorylates these ROP proteins in vivo is uncertain as its natural substrates remain to be identified.
Figure 2.
ROP18 phosphorylates other ROPs in vitro and is regulated by autophosphorylation. (A) Recombinant ROPs purified by nickel-affinity chromatography following expression in E. coli. Coomassie blue-stained gel. (B) Immunoprecipitation of wild-type ROP18 and inactive ROP18 (D/A) containing a mutation of D394A. Immunoprecipitated with mAb BB2 and immunoblotted with rabbit anti-ROP18. Anti-actin (ACT) is provided as a loading control. SUP, supernatant. (C) In vitro kinase reactions using immunoprecipitated ROP18. Addition of ROP18 immunoprecipitated from parasites expressing the wild-type (WT) enzyme leads to phosphorylation of recombinant ROP2, ROP8 and truncated ROP18 (* indicates phosphorylated protein). Labelling was not obtained using catalytically inactive ROP18 (D/A mutation of D394A). Additionally, recombinant ROP2, ROP8 and ROP18 were not active alone (− lanes). Control is immunoprecipitated ROP18 alone, which shows autophosphorylation (bracket). Phosphorimage analysis following resolution of samples by 12% SDS–PAGE.
Heterologous expression and structures of ROP8 and ROP2
Using an E. coli-based platform (Vedadi et al, 2007), heterologous expression of ROP2, ROP8 and ROP18 yielded only soluble protein from cloned constructs including an additional region in the N terminus preceding the predicted boundary of subdomain I of the kinase domain. Purified and unphosphorylated ROP2 and ROP8 protein samples crystallized under conditions summarized in Table I. The structural solution for ROP8 was refined to 1.8-Å resolution using single wavelength anomalous diffraction data from selenomethionine-derivatized protein and submitted to the Protein Data Bank (PDB accession 3BYV). Using ROP8 as a search model, the ROP2 structure was determined to 1.8-Å resolution by molecular replacement (PDB accession 3DZO). The crystallographic statistics are summarized in Table I. Previous attempts to model ROP kinases by homology modelling have been largely incomplete; hence, our crystallographic structures illustrate, for the first time, the true fold of a rhoptry kinase.
Table 1.
Crystallographic parameters for ROP8 and ROP2
| Crystallization | ROP8 Selmet | ROP8 Native | ROP2 Native |
|---|---|---|---|
| 20% PEG3350, 0.2 M Mg(NO3)2, 1 mM MgCl2, 0.1 M Tris–HCl, pH 8.5 | 24% PEG3350, 0.2 M Mg(NO3)2, 0.1 M Tris–HCl, pH 8.4 | 21–24% PEG4K, 0.2 M MgCl2, 0.1 M Tris–HCl, pH 8.2–8.6 | |
| Data collection | |||
| Space group | P212121 | P212121 | P21 |
| Cell dimensions | |||
| a (Å) | 59.65 | 59.07 | 41.26 |
| b (Å) | 70.83 | 69.82 | 44.29 |
| c (Å) | 85.89 | 85.42 | 105.30 |
| Alpha (deg) | 90.00 | 90.00 | 90.00 |
| Beta (deg) | 90.00 | 90.00 | 98.90 |
| Gamma (deg) | 90.00 | 90.00 | 90.00 |
| Wavelength (Å) | 0.9777 | 0.9179 | 0.96749 |
| Resolution (Å) | 40–2.3 | 40–1.8 | 40–1.8 |
| Measured reflections | 426 258 | 490 721 | 214 991 |
| Unique reflections | 16 727 | 63 327 | 34 415 |
| Rsym | 0.080 (0.315) | 0.069 (0.486) | 0.049 (0.200) |
| I/σI | 43.8 (8.11) | 28.5 (4.3) | 22.4 (4.2) |
| Completeness (%) | 99.2 (98.7) | 99.9 (100.0) | 97.6 (81.4) |
| Redundancy | 7.0 (7.4) | 7.3 (7.4) | 3.6 (2.6) |
| Refinement | |||
| Resolution (Å) | 40–1.8 | 35–1.8 | |
| Number of reflections | 31 731 | 32 620 | |
| Test set | 1692 | 1717 | |
| Rwork/Rfree | 0.233/0.202 | 0.269/0.246 | |
| Number of atoms | |||
| Protein | 2864 | 2845 | |
| Water | 171 | 107 | |
| Ligands | 13 (3 EDO, 1 Mg) | 1 Mg | |
| Mean B-factor | 13.03 (TLS) | 29.87 | |
| Ramachandran favoured | 97.1 | 95.9 | |
| Ramachandran disallowed | 0 | 0 | |
| RMS deviations | |||
| Bond lengths (Å) | 0.028 | 0.008 | |
| Bond angles (deg) | 2.367 | 1.069 |
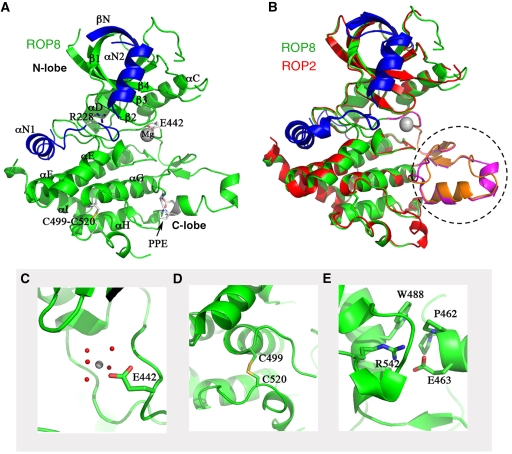
Despite extensive sequence variation from well-known S/T kinases, ROP2 and ROP8 contain the classic bilobed protein kinase fold (Figure 3A and B), consisting of an N-lobe, narrow hinge and large C-lobe (Figure 3A). The additional N-terminal region needed for the expression of soluble protein turns out to be a subdomain of two helices and a beta strand (αN1, αN2 and βN in Figure 3A). The structural backbones of ROP2 and ROP8 deviate by only 0.6 Å RMS, consistent with 92.5% sequence identity between them (sequence alignment shown in Supplementary Figure S1). Their major discrepancies are the somewhat unstructured activation loop and the substrate-binding loop highlighted in Figure 3B and discussed further below.
Figure 3.
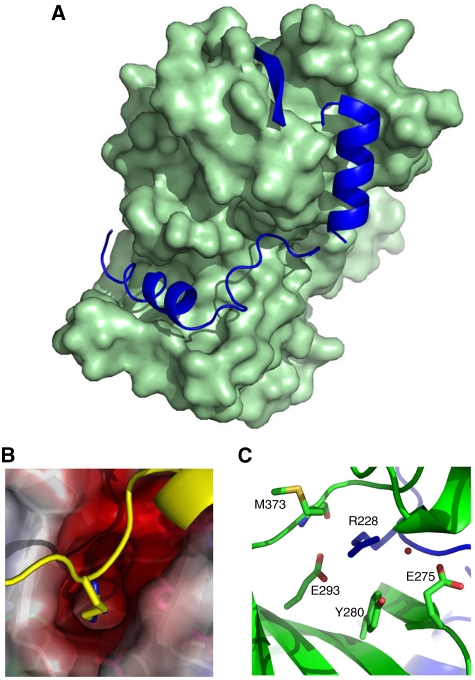
Models of ROP2 and ROP8 structures. (A) Ribbon model of ROP8 structure shows a typical kinase fold consisting of N- and C-lobes. N-terminal extension provides two additional alpha helices (αN1, αN2) and an extra beta sheet (βN) capping the N-lobe (blue). Major helices, including αC, are labelled, as are the magnesium ion, the inhibitory arginine (R228), the intrachain disulphide bond (C499–C520), Glu442 (E442) and the PPE motif at the end of the activation loop. (B) Superposition of ROP2 (red) on ROP8 (green) shows a tight alignment, including both the N- and C-lobes and the N-terminal regulatory domain. Major region of difference is in the activation loop and substrate-binding region (dotted circle, magenta=ROP8, orange=ROP2). (C) Enlarged view of Glu442 in ROP8 that coordinates the Mg2+ ion. (D) Intrachain disulphide bond in ROP8 stabilizes some of the C-lobe helices. (E) Enlarged view of the PPE motif stabilizing the substrate-binding region.
In ROP8 and ROP2 structures, the 12 subdomains of the protein kinase catalytic domain as defined previously (Hanks and Hunter, 1995) are clearly conserved (Supplementary Figure S1). However, the VAIK motif in subdomain II of the kinase domain, in which the lysine residue interacts with the α- and β-phosphates of ATP to anchor and orient the nucleotide, is replaced by the quartet of FEVH. Most significantly, the replacement of lysine by histidine makes these kinases unlikely to interact with ATP. Moreover, the DFG motif in subdomain VII, in which the aspartic acid binds Mg2+ ions and coordinates the β- and γ-phosphates of ATP, is replaced in both structures by an equivalent triad in reverse order: GFE. This results in Glu442 interacting with one magnesium ion and five water molecules similarly to aspartate of the DFG motif from the kinases, but from a different direction (Figure 3C). The commonly found HRD motif in subdomain VIb that interacts with the magnesium-binding loop is replaced by HTY in both ROP2 and ROP8. Collectively, these features are consistent with ROP2 and ROP8 being inactive and highly degenerate kinases.
Two highly conserved cysteine residues, from subdomains IX and X, respectively, form a disulphide bond (C489–C510 in ROP2, C499–C520 in ROP8; Figure 3D). MS analysis confirmed the presence of this bridge in purified samples of ROP2, ROP8 and ROP18. Its role appears to be stabilization of the last two helices of the kinase domain, which are connected by a partially disordered loop in both ROP2 and ROP8. As these cysteines are conserved throughout the ROP2 family (data not shown), we predict the same stabilizing disulphide bond in all the members.
Substrate-binding region
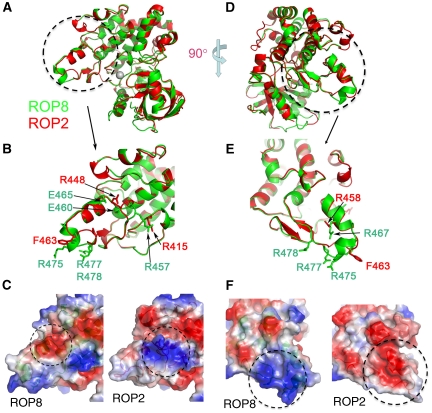
As noted above, the triad of APE at the end of a kinase substrate recognition loop in subdomain VIII is present but with alanine replaced by proline in ROP2, ROP4, ROP7, ROP8 and ROP18. In the structures of ROP2 and ROP8, this substitution does not alter the orientation of the loop in comparison to PKA (PDB accession 1ATP). The two prolines (Pro451 and Pro452 in ROP2, Pro461 and Pro462 in ROP8) interact hydrophobically with a conserved tryptophan (Trp478 in ROP2 and Trp488 in ROP8) from subdomain IX. Finally, the last residue of the PPE triad, namely Glu463 (in ROP8), forms an ion pair with an invariant Arg542 from subdomain XI, further stabilizing the conformation of the triad (Figure 3E). Overall, these conserved interactions indicate that subdomain VIII in ROP2 and ROP8 likely maintains the capability to bind substrates, although electrostatic differences in this region predicts that they will have different specificities. Where Glu460 and Glu465 produce a negative surface in ROP8, ROP2 features a positive charge primarily generated by Arg448 (Figure 4B and C). Similarly, arginines in positions 450, 467, 475, 477 and 478 form a highly positively charged surface in ROP8 (Figure 4D) that is replaced by a partly neutral and negative surface on ROP2 (Figure 4E and F). Hence, although not catalytically active, these pseudokinases may serve a scaffolding role or modulate kinase activation similarly to how some human pseudokinases function (Boudeau et al, 2006).
Figure 4.
Comparison of substrate-binding region between ROP2 and ROP8. (A) Rotated view of ROP2 (red) and ROP8 (green) superposed. (B) Enlarged view of substrate-binding region for ROP2 (red) and ROP8 (green) highlights the main region of deviation. (C) Surface map shows the substrate-binding region of ROP8 that is mostly negative due to Glu460 and Glu465, whereas ROP2 is mostly positive primarily due to Arg448 and Arg415. (D) The superposed structures of ROP2 (red) and ROP8 (green) rotated 90° to show another variable surface of the substrate-binding region. (E) Enlarged view of another divergent substrate-binding region between ROP2 (red) and ROP8 (green). (F) Surface map shows ROP8 is mostly positive due to Arg at positions 450, 467, 475, 477 and 478, whereas ROP2 is mostly neutral or negative attributable to Glu458.
Unique N-terminal subdomain
The crystallographic structures of the ROP2 and ROP8 pseudokinases revealed a unique N-terminal subdomain consisting of two helices (αN1: D212-R221 and αN2: E232-L246 in ROP8) and a beta sheet βN (G250-S257 in ROP8) buttressing the N-terminal beta lobe of the kinase domain (Figure 5A). Helix αN1 fills a hydrophobic cleft mainly formed by helices αE from subdomain V and αF from subdomain VIa. In ROP8, the side chain of Asp212 forms a salt bridge with side chain of Arg335, stabilizing the helix αN1. Most interestingly, the aliphatic side chain of Arg228 in ROP8 (Arg220 in ROP2) protrudes from between the two helices into the negatively charged nucleotide-binding pocket (Figure 5B). The conformation of this arginine is stabilized by two salt bridges with Glu293, one hydrogen bond with the backbone carbonyl oxygen from Met373 as well as water-mediated hydrogen bonds with Glu275 and Tyr280 (Figure 5C). Thus, positioned, this amino-acid side chain encroaches on the nucleotide-binding site.
Figure 5.
Model of the N-terminal extension in ROP8. (A) Surface representation of the main body of the kinase domain of ROP8 with helices αN1 and αN2 as well as βN from the N-terminal regulatory subdomain (all blue) tucked into hydrophobic clefts. (B) The side chain of Arg228, which lies between αN1 and αN2, protrudes into the negatively charged ATP-binding pocket and potentially hinders nucleotide binding. (C) The inhibitory Arg228 is stabilized by two salt-bridges with Glu293, one hydrogen bond with the backbone carbonyl oxygen from Met373, as well as water-mediated hydrogen bonds with Glu275 and Tyr280.
The N-terminal subdomain also interacts with the main kinase domain in other regions conserved not just in ROP2 and ROP8, but also ROP18 (see the sequence alignment in Supplementary Figure S2 and structural alignment in Supplementary Figure S3). Specifically, an aspartate located ahead of αN1 (Asp200 on ROP2, Asp209 on ROP8 and Asp180 on ROP18) interacts with a conserved triad on αF of the kinase domain—Gln392-Gln396-Arg399 on ROP2, Gln401-Gln405-Arg408 on ROP8 and Tyr373-Gln377-Lys380 on ROP18. Tucked between αN2 and βN is a tryptophan (Trp238 in ROP2, Trp247 in ROP8 and Trp218 in ROP18) that extends into a conserved hydrophobic pocket made of Leu272-Val285-Val287-Phe358 in ROP2's β-lobe, Leu281-Val294-Val296-Phe367 in ROP8 and Tyr252-Val267-Val269-Phe338 in ROP18. Finally, a conserved aspartate on αN2 (Asp230 on ROP2, Asp239 on ROP8 and Asp210 on ROP18) interacts with an arginine on the beta strand β1 (Arg259, Arg268 and Arg239, respectively, in ROP2, ROP8 and ROP18). Sequence alignment indicates that these interaction motifs are also found in ROP4, ROP5 and ROP7, but not ROP16 and ROP17 (data not shown), consistent with the latter two being more phylogenetically divergent as shown in Figure 1A. This suggests that the rhoptry kinase fold revealed by our ROP2 and ROP8 structures, including the N-terminal subdomain, is representative of a number of rhoptry kinases, but possibly not all.
Homology model of ROP18
Using ROP8 as a template, we generated a homology model of the kinase domain of ROP18 (Figure 6A). This model facilitated identification and location of important catalytic motifs and understanding the mechanism of autoactivation. As discussed above, the N-terminal extension of ROP18 forms a subdomain consisting of αN1 and αN2 regions and βN that caps the N-lobe (Figure 6A). ROP2 and ROP8 share ∼92% sequence identity with each other but only 27% with ROP18. Nevertheless, several lines of evidence suggest that the N-terminal subdomain found in the structures of the pseudokinases also forms an integral and essential part of ROP18. First, its inclusion in ROP18 was essential for the expression of soluble recombinant protein and detectable kinase activity. As discussed above and shown in Supplementary Figures S2 and S3, residues that impart structurally similar contacts between the kinase domain and the N-terminal extension are conserved between ROP18 and the two pseudokinases. Finally, as discussed below, a conserved threonine on beta strand β1 (Thr256, Thr265 and Thr236, respectively, in ROP2, ROP8 and ROP18) is a phosphorylation site with regulatory implications in ROP18.
Figure 6.
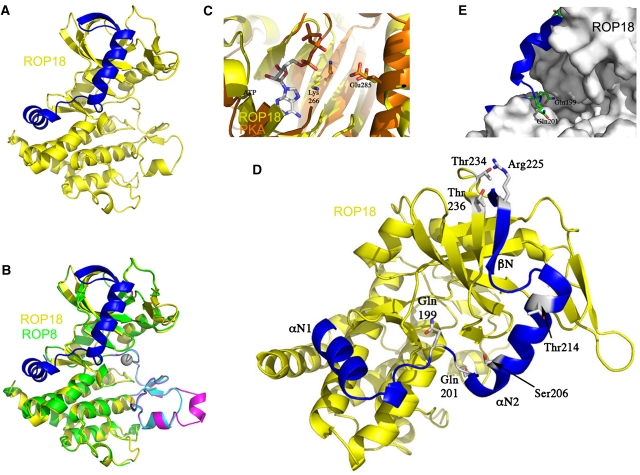
Homology model of ROP18 and regulatory phosphorylation. (A) ROP18 contains the conserved kinase fold as well as an N-terminal extension (blue). (B) Supposition of ROP18 model (yellow) on ROP8 structure (green) shows deviation in substrate-binding region (magenta in ROP8 and cyan in ROP18). (C) ROP18 model (yellow) superimposed on PKA (orange): Lys266 on β4 and Glu285 on αC are responsible for coordinating α- and β-phosphates on the ATP molecule. (D) The N-terminal regulatory domain of ROP18 (blue), with autophosphorylation sites shown: Ser206, Thr214, Thr 234 and Thr236. In the inactive form, Gln199 and Gln201 potentially interfere with ATP binding and inhibit activity. The N-terminal extension is also stabilized by interaction of Arg225 with Thr234 and Thr236. (E) Close-up view of active site of ROP18 showing Gln199 and Gln201 side chains that occlude the ATP-binding site.
ROP18 differs from ROP8 substantially in the nucleotide- and substrate-binding regions (Figure 6B). Unlike ROP2 and ROP8, ROP18 features a lysine on β4 in the N-terminal lobe, situated at the end of a Leu-Ala-Val-Lys (LAVK) motif to interact with the α- and β-phosphates of ATP. In a typical active kinase conformation, this lysine should be within hydrogen bonding distance of an essential glutamine on αC. To locate this lysine–glutamine pair, we superposed the ROP18 model (Figure 6C) on the structure of PKA with ATP bound (PDB accession 1ATP). In ROP18, Lys266 and Glu285 are slightly farther apart than the corresponding catalytic pair in PKA but are close enough to position the helix αC in its active conformation. Furthermore, the DFG motif at the start of the activation loop in ROP18 is positioned to interact with magnesium in the familiar way seen in most protein kinases (Johnson et al, 1996). These features further support ROP18 as an active S/T kinase with standard catalytic motifs.
Autophosphorylation of ROP18
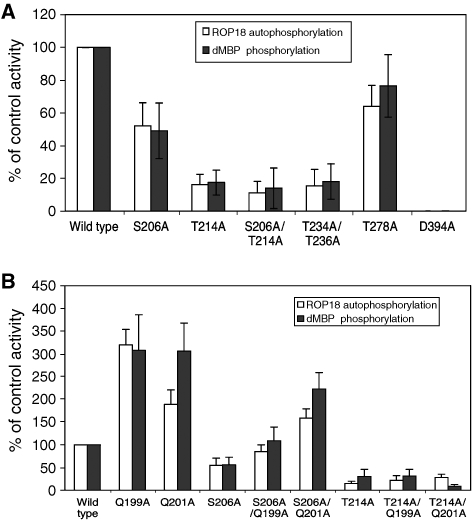
Protein kinases are often regulated by autophosphorylation. We performed an in vitro kinase reaction using purified recombinant ROP18 and analysed the resulting tryptic peptides by LC/MS/MS to locate the autophosphorylation sites at Ser206 and threonine at positions 214, 234, 236 and 278 (amino-acid numbering based on second ATG of CAJ27113). We generated point mutants in these residues using the active form of ROP18 produced in E. coli. The resulting wild-type and mutant enzymes were tested for their ability to undergo autophosphorylation and to phosphorylate the heterologous substrate dephosphorylated myelin basic protein (d-MBP). The wild-type enzyme was highly active in autophosphorylation and against d-MBP (Figure 7A). In contrast, both autophosphorylation and activity against dMBP were reduced by ∼80% when Thr214 was mutated to Ala (Figure 7A). A more modest decrease in activity was observed when Ser206 was mutated to Ala, whereas the combined mutant (S206A and T214A) was decreased by >90% (Figure 7A). When both Thr234 and Thr236 were mutated, the enzyme also showed decreased activity, whereas mutation of Thr278, which lies in the kinase domain, had no meaningful effect (Figure 7A). These results indicate that phosphorylation of certain N-terminal amino acids regulates ROP18 activity. The role that such autoactivation might have in vivo is uncertain, although concentration at the surface of the parasite-containing vacuole (Taylor et al, 2006; El Hajj et al, 2007), might locally induce activation by increasing the likelihood of trans-phosphorylation.
Figure 7.
Mutation analysis of autophosphorylation in the N-terminal extension of ROP18. (A) In vitro kinase reactions of ROP18 autophosphorylation versus transphosphorylation of dMBP by wild-type and mutant ROP18 kinases. Mutation of Ser206A partially blocked activity, whereas mutations of Thr214A or Thr234A/Thr236A lead to greater decreases in activity. Mutation of T278A, which lies in the kinase domain had no effect. For comparison, mutation of D394A, which is in the catalytic triad, abolished all activity. (B) Enhanced activity of ROP18 bearing mutations in Gln199 or Gln201 and influence on restoring activities of Ser206A and Thr214A mutations. Activities were determined by phosphorimager analysis of proteins resolved on SDS–PAGE gels following in vitro kinase reactions. Data are averages±s.d. for three or more similar experiments.
A model for regulation by N-terminal phosphorylation
In experiments described above, we have established that the integral N-terminal subdomain in rhoptry kinases has a key role in activating ROP18. Examination of the homology model provides a possible explanation for the activation mechanism. The residues Ser206 and Thr214 are located on αN2 and their phosphorylation would potentially disturb the position of this helix and consequently of the entire N-terminal regulatory domain (Figure 6D). Likewise, phosphorylating one or both of Thr234 and Thr236 may alter their interaction with Arg240, so as to further mobilize the regulatory domain (Figure 6D). In the homology model of ROP18 (Figure 6D and E), the side chain of Gln199 encroaches on the ATP pocket in a manner analogous to Arg228 in ROP8 (Figure 5). Two residues downstream, Gln201 further occludes the active site. In their positions shown in Figure 6D and E, these two glutamines may conspire to limit kinase activity by hindering ATP binding. To render ROP18 active, this subdomain must be sufficiently displaced as to shift the two glutamines—Gln199 and Gln201—out of their inhibitory positions and allow binding of ATP.
Our data suggest a model where the N-terminal extension functions as a flexible lever arm, clamping down on the kinase domain to inhibit activity when closed and swinging out to allow activity when phosphorylated. Such a model predicts that mutation of the side chains of Gln199 and Gln201 would relieve autoinhibition. To test this hypothesis, we systemically mutated Gln199 and Gln201 to Ala. These single-point mutations elevated kinase activity by 3- to 4-fold, supporting the observation from the homology model that the side chains of these residues disrupt ATP binding and limit activity of ROP18 (Figure 7B). Similarly, mutation of either Gln199A or Gln201A was able to partially overcome the inhibition of the Ser206A mutation, which otherwise impairs phosphorylation-mediated activation (Figure 7B). Similar mutations of the Gln residues did not overcome the inhibition of Thr214A (Figure 7B). This latter result might be expected if the N-terminal extension functions as a lever with the fulcrum at the top of the N-lobe, such that more distal residues such as Ser206 contribute less to the inhibition than those more proximal such as Thr214 (hence phosphorylation of Thr214 is more critical to activation).
Rhoptry kinases are unique to the apicomplexan parasite T. gondii. Our findings show that their structures and regulatory mechanism are also distinct from known S/T kinases. Most of the members of this family are predicted to be pseudokinases—that is, inactive, including ROP2 and ROP8. In contrast, ROP18 is an active kinase capable of phosphorylating itself as well as other ROP family members. Crystallographic structures of ROP2 and ROP8, along with the homology model of ROP18, reveal a rhoptry kinase fold with an N-terminal regulatory domain that distinguishes it from other kinases as well as previously predicted models. Hence, specific inhibitors to ROP18 might be developed either based on its unique ATP-binding pocket or by identifying non-competitive inhibitors that disrupt this activation mechanism.
Materials and methods
Phylogenetic analysis
Selected human kinases were retrieved from GenBank for comparison. The following ROP kinases corresponding to the genes from the type I RH strain were used for analysis: ROP2 (GenBank accession no. CAA85377), ROP4 (AAU87405), ROP5 (AAZ73240), ROP7 (CAJ20842), ROP8 (AAC47797), ROP16 (AAZ73239), ROP17 (CAJ27112) and ROP18 (CAJ27113). Several representative FIKK kinases (Schneider and Mercereau-Puijalon, 2005; Nunes et al, 2006) were obtained from the P. falciparum genome (http://PlasmoDB.org), including PF10_0160, PFL0040C, PF1165W and PFI0100C. Predicted protein sequences were aligned in Clustal using a gap penalty of 15 with an elongation penalty of 0.3 (Higgins et al, 1996). The resulting alignment was analysed in PAUP* 4.0 (Swofford, 2002) and the informative characters were used to generate phylogenetic trees using distance with neighbour-joining BioNJ. Trees were bootstrapped for 1000 replicates using a 50% majority rule.
Domain analysis
The coding regions of ROPs were analysed for conserved domains using InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) and for signal peptides using SignalP (http://www.cbs.dtu.dk/services/SignalP/). Comparison of S/T domains was conducted by Clustal alignment of ROP kinases with the representative human kinases above.
Production of recombinant proteins
For crystallography, ROP2, ROP8 and ROP18 proteins were expressed with an N-terminal His6 tag as described previously (Vedadi et al, 2007). Expression constructs were subcloned from full-length synthetic templates ordered from Codon Devices (Cambridge, MA, USA) and based on coding sequences from ToxoDB (www.toxodb.org) for the genes 33.m00005 (ROP8 starting at Pro210) and 33.m01389 (ROP2 starting at Phe192). cDNA from type I RH strain of T. gondii was used to clone ROP18 (starting from Ser181 based on the second ATG of GenBank protein CAJ27113). ROPs were expressed in BL21(DE3)-V2R-pACYC LamP, which includes a plasmid for co-expression of lambda phosphatase to suppress protein phosphorylation (provided by Opher Gileadi, SGC, University of Oxford).
For functional assays, ROP2 (beginning at Pro197) and ROP18 (beginning at Ser228) were amplified using primers to introduce an ATG with an NdeI restriction site at the N terminus and a C-terminal His6 tag. ROP genes were amplified from cDNA of the type I RH strain of T. gondii using PfuUltra high-fidelity DNA polymerase (Stratagene, La Jolla, CA). Following overnight culture in LB at 37 °C, cultures were diluted 100-fold in fresh LB, induced with 1 mM IPTG and grown for an additional 3 h at 37 °C. Induced soluble proteins were purified using ProBond Ni-affinity resin (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations.
Immunoprecipitation of ROP18
Transgenic parasite clones expressing the type I allele of ROP18 expressing a C-terminal TY epitope tagged (i.e. CTG-ROP18I clone V1), a well as the kinase dead version (D394A) (i.e. CTG-ROP18I clone L1) were described previously (Taylor et al, 2006). Parasites were propagated in human foreskin fibroblast monolayers and harvested following natural egress, as described previously (Taylor et al, 2006). ROP18 was immunoprecipitated using the mAb BB2 (Bastin et al, 1996) coupled to protein G as follows: protein G sepharose (Pierce Biotechnology Inc., Rockford, IL) (25 μl) was preincubated with mAb BB2 (5 μl) for 1 h at RT and washed with PBS. Precharged beads were then incubated with parasite proteins made by lysing ∼3 × 107 parasites in 1% NP-40, 50 mM Tris–HCl, 150 mM NaCl, pH 8.0, followed by centrifugation at 1000 g for 5 min. Following binding of the lysate supernatant at 4°C for 16 h, beads were washed with PBS three times.
Western blotting
Recombinant ROP18 was produced in E. coli and purified under denaturing conditions using ProBond resin, as described above. Recombinant ROP18 was used to produce a high-titre antiserum in a rabbit using a commercial vendor (Covance, Denver, PA). For western blot analysis, epitope-tagged ROP18 was immunoprecipitated as described above, resuspended in non-denaturing SDS sample buffer, resolved in 10% acrylamide gels, transferred to nitrocellulose and probed with rabbit anti-ROP18 or rabbit anti-Tg-actin at 1:10 000. Blots were washed, incubated with goat anti-rabbit IgG conjugated to HRP (1:10 000), detected using SuperSignal (Pierce Biotechnology Inc.), and exposed to BX autoradiography film (MidSci, St Louis, MO).
In vitro kinase reactions
Epitope-tagged ROP18 was immunoprecipitated from parasites expressing either wild-type kinase or D394A mutant kinase and added to in vitro kinase reactions containing 5–7 μg of purified ROP proteins in reaction buffer: 25 mM Tris–HCl pH 7.5, 15 mM MgCl2 and 2 mM MnCl2. Each reaction contained 10 μCi of 32P-ATP (specific activity: 3000 Ci/mmol) (Perkin Elmer, Waltham, MA) in addition to 33 μM unlabelled ATP. The reaction was allowed to proceed at 30°C for 60 min. Proteins were resolved on 12% SDS–PAGE gels, dried and imaged using a FLA5000 phosphorimager (Fuji Life Sciences, Stamford, CT).
Mutagenesis
The expression plasmid pROP18KD3 (beginning at Ser181 numbered from second ATG of CAJ27113) was used to purify wild-type ROP18 for kinase assays and as backbone for making point mutants. The ROP18 gene was inserted into vector pET22b (Novagen) NdeI site (5′ end) and HindIII (3′end) resulting in the addition of seven extra amino acids (KLAAALE) preceding the C-terminal His6 tag. Mutants were introduced into the pROP18KD3 plasmid by QuickChange site-directed mutagenesis (Stratagene) using oligonucleotides bearing nucleotide mismatches. Changes were verified by PCR-based cycle sequencing using BigDye (SeqWright DNA Technology Services, Houston, TX). Plasmids were transformed into BL21-CodonPlus(DE3)-RP cells (Stratagene) and expression was induced by culture in 1 mM IGTP for 3–4 h. Bacterial cell pellets were lysed in CelLytic B cell lysis reagent (Sigma-Aldrich, St Louis, MO). His-tagged proteins were purified using ProBond resin (Invitrogen) under native conditions, and proteins were dialysed against 250 mM NaCl, 10 mM MgCl2 and Tris–HCl pH 8.0. In vitro kinase reactions were performed as described above.
Identification of phosphorylation sites in ROP18
Recombinant ROP18 at about 10 mg/ml was incubated overnight at 4°C in a buffer of 10 mM HEPES pH 7.5, 500 mM NaCl, 2 mM MgCl2 and 2 mM ATP. Samples were analysed on an Agilent LC/MS mass spectrometer and found to identify autophosphorylated peaks based in deconvoluted ESI spectra. In-solution digestion was performed on 5 μg of sample protein using 0.02 μg of trypsin in 25 mM ammonium bicarbonate solution (a total volume of 10 μl), or with 0.05 μg of Asp-N proteinase in 10 mM Tris–HCl buffer (pH 7.6). Each enzymatic reaction was incubated at 37°C for 3 h. Proteolytic products were analysed by peptide mapping and MS/MS measurements on an Applied Biosystems/MDS Sciex QSTAR XL MALDI-QTOF mass spectrometer.
Crystallization, data collection and structure solution
Crystals for SeMet-derivatized ROP8 were obtained using the hanging-drop vapour diffusion method in 24% PEG3350, 0.2 M Mg(NO3)2, 0.1 M Tris–HCl and pH 8.4. They were harvested with a cryo-protectant buffer containing 15% glycerol and crystallization buffer solution mentioned above and flash frozen in liquid nitrogen. Diffraction data were collected at the A1 beamline at the Cornell High Energy Synchrotron Source (CHESS, Cornell University, Ithaca, USA), where an additional high-resolution data set was collected using native crystals at beamline F1. The SeMet data were integrated and scaled using the HKL2000 package (Minor et al, 2006) with weak initial phase information at 2.5 Å obtained from four selenomethionine atoms using the program BnP (Weeks et al, 2002). Phases were improved and initial models were built using the program RESOLVE (Terwilliger, 2001, 2003). Incorporating the high-resolution native data, extension and map improvement were performed with the program Arp/Warp (CCP4) (Murshudov et al, 1997, 1999; Pannu et al, 1998; Winn et al, 2001). Manually rebuilding using the program COOT (Emsley and Cowtan, 2004) and refining using REFMAC5 from CCP4, a final model ROP8 at 1.8 Å was obtained and deposited in the Protein Data Bank (PDB accession 3BYV).
Diffracting crystals of unlabelled ROP2 protein were also obtained using the hanging-drop vapour diffusion technique in 21–24% PEG4K, 0.2 M MgCl2, 0.1 M Tris–HCl and pH 8.2–8.6. A complete data set was collected at beamline 23ID at the Advance Photon Source (APS, Argonne National Laboratory), processed using HKL2000 (Minor et al, 2006) and phased by PHASER (McCoy, 2007; McCoy et al, 2007) using ROP8 as a search model. The ROP2 model was built and refined again using COOT and CCP4, with a final resolution of 1.8 Å (PDB accession 3DZO).
Both the ROP8 and ROP2 models have excellent stereochemistry with no residues in disallowed or unfavourable regions from Ramachandran plots as validated by the program MOLPROBITY (Davis et al, 2004).
Homology modelling
The ROP8 structure was used as a template to build by homology a predictive model of the kinase domain of ROP18 using the software package ICM (Molsoft, CA). The ROP18 sequence was threaded on the ROP8 structure based on sequence alignment and the energy of the system was minimized in the internal coordinates space by a biased probability Monte Carlo simulation, utilizing ECEPP/3 and solvation energy terms (Abagyan and Totrov, 1994; Cardozo et al, 1995). Three independent simulations run in parallel converged towards the same global energy minimum.
Accession numbers
Coordinates for ROP8 (3BYV) and ROP2 (3DZO) have been deposited in the Protein Data Bank.
Supplementary Material
Supplementary Information
Acknowledgments
We thank David Roos and Lucia Peixoto for helpful discussions, Julie Nawas for expert technical assistance, Leslie Hicks and Sophie Alvarez for assistance with the mass spectrometry. Work at Washington University School of Medicine was supported by NIH grant AI036629. The SGC is a registered charity (no. 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co. Inc., the Novartis Research Foundation, the Petrus and Augusta Hedlund's Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
References
- Abagyan R, Totrov M (1994) Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol 235: 983–1002 [DOI] [PubMed] [Google Scholar]
- Baas AF, Boudeau J, Sapkota GP, Smit L, Medema R, Morrice NA, Alessi DR, Clevers HC (2003) Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J 22: 3062–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh Z, Matthews KR, Gull K (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol 77: 235–239 [DOI] [PubMed] [Google Scholar]
- Beckers CJM, Dubremetz JF, Mercereau-Puijalon O, Joiner KA (1994) The Toxoplasma gondii rhoptry protein ROP2 is inserted into the parasitophorous vacuole membrane, surrounding the intracellular parasite, and is exposed to the host cell cytoplasm. J Cell Biol 127: 947–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd JC, Dubremetz JF (2008) Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat Rev Microbiol 6: 79–88 [DOI] [PubMed] [Google Scholar]
- Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR (2006) Emerging roles of pseudokinases. Trends Cell Biol 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Sibley LD (2007) Rhoptries: an arsenal of secreted virulence factors. Curr Opin Microbiol 10: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC (2005) Proteomic analysis of rhoptry organelles reveals many novel constituents for host–parasite interactions in T. gondii. J Biol Chem 280: 34245–34258 [DOI] [PubMed] [Google Scholar]
- Cardozo T, Totrov M, Abagyan R (1995) Homology modeling by the ICM method. Proteins 23: 403–414 [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD (1997) Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol 73: 114–123 [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj H, Lebrun M, Arold ST, Vial H, Labesse G, Dubremetz JF (2007) ROP18 is a rhoptry kinase controlling the intracellular proliferation of Toxoplasma gondii. PLoS Pathog 3: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj H, Lebrun M, Fourmaux MN, Vial H, Dubremetz JF (2006) Inverted topology of the Toxoplasma gondii ROP5 rhoptry protein provides new insights into the association with the parasitophorous vacuole membrane. Cell Microbiol 9: 54–64 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Håkansson S, Charron AJ, Sibley LD (2001) Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J 20: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 382–402 [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP (1995) Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature 376: 313–320 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Kannan N, Neuwald AF, Taylor SS (2008) Analogous regulatory sites within the alphaC–beta4 loop regions of ZAP-70 tyrosine kinase and AGC kinases. Biochim Biophys Acta 1784: 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Applied Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62: 859–866 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Südhof TC, Wahl MC (2008) CASK functions as a Mg2+-independent neurexin kinase. Cell 133: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dosdson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Nichols BA, O'Connor RG (1981) Penetration of mouse peritoneal macrophages by the protozoan Toxoplasma gondii. Lab Invest 44: 324–335 [PubMed] [Google Scholar]
- Nunes M, Goldring JPD, Doerig C, Scherf A (2006) A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol Microbiol 63: 391–403 [DOI] [PubMed] [Google Scholar]
- Pannu NS, Murshudov GN, Dodson EJ, Read RJ (1998) Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr D Biol Crystallogr 54: 1285–1294 [DOI] [PubMed] [Google Scholar]
- Saeij JPJ, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC (2006) Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314: 1780–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JPJ, Coller S, Boyle JP, Jerome ME, White ME, Boothroyd JC (2007) Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445: 324–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P, Silvennoinen O (2002) The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem 277: 47954–47963 [DOI] [PubMed] [Google Scholar]
- Schneider AG, Mercereau-Puijalon O (2005) A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD, Charron AJ, Hakansson S, Mordue DG (2007) Invasion and intracellular survival by Toxoplasma. In Protozoans in Macrophages, Denkers EY, Gazzibelli RT (eds) pp 46–24 Austin: Landes Bioscience [Google Scholar]
- Sinai AP, Joiner KA (2001) The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J Cell Biol 154: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suss-Toby E, Zimmerberg J, Ward GE (1996) Toxoplasma invasion: the parasitophorous vacuole is formed from host cell plasma membrane and pinches off via a fusion pore. Proc Natl Acad Sci USA 93: 8413–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (2002) Phylogenetic Analysis Using Parsimony (* and Other Methods). Sunderland: Sinauer Associates [Google Scholar]
- Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Haijj EL, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD (2006) A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314: 1776–1780 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2001) Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr 57: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374: 22–37 [DOI] [PubMed] [Google Scholar]
- Vedadi M, Lew J, Artz J, Amani M, Zhao Y, Dong A, Wasney GA, Gao M, Hills T, Brokx S, Qiu W, Sharma S, Diassiti A, Alam Z, Melone M, Mulichak A, Wernimont A, Bray J, Loppnau P, Plotnikova O et al. (2007) Genome-wide protein expression and structural biology of Plasmodium falciparum and related apicomplexan organisms. Mol Biochem Parasitol 151: 100–110 [DOI] [PubMed] [Google Scholar]
- Ward P, Equinet L, Packer J, Doerig C (2004) Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks CM, Blessing RH, Miller R, Mungee R, Potter SA, Rappleye J, Smith GD, Xu H, Furey W (2002) Towards automated protein structure determination: BnP the SnB–PHASES interface. Z Kristalogra 217: 686–693 [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information