Abstract
Mutations in human homologues of the bacterial RecQ helicase cause diseases leading to cancer predisposition and/or shortened lifespan (Werner, Bloom, and Rothmund–Thomson syndromes). The budding yeast Saccharomyces cerevisiae has one RecQ helicase, Sgs1, which functions with Top3 and Rmi1 in DNA repair. Here, we report separation-of-function alleles of SGS1 that suppress the slow growth of top3Δ and rmi1Δ cells similar to an SGS1 deletion, but are resistant to DNA damage similar to wild-type SGS1. In one allele, the second acidic region is deleted, and in the other, only a single aspartic acid residue 664 is deleted. sgs1-D664Δ, unlike sgs1Δ, neither disrupts DNA recombination nor has synthetic growth defects when combined with DNA repair mutants. However, during S phase, it accumulates replication-associated X-shaped structures at damaged replication forks. Furthermore, fluorescent microscopy reveals that the sgs1-D664Δ allele exhibits increased spontaneous RPA foci, suggesting that the persistent X-structures may contain single-stranded DNA. Taken together, these results suggest that the Sgs1 function in repair of DNA replication intermediates can be uncoupled from its role in homologous recombinational repair.
Keywords: DNA repair, DNA replication, homologous recombination, Sgs1, yeast
Introduction
DNA repair and maintenance of genome stability are some of the most fundamental of all cellular processes. Critical to ensuring appropriate repair of DNA damage is the RecQ family of DNA helicases, which is conserved from bacteria to humans. In humans, mutations in members of this family lead to separate genetic diseases, Bloom, Werner, and Rothmund–Thomson syndromes. All of these syndromes have distinct features but are ultimately characterized by an increased susceptibility to cancer caused by chromosomal rearrangements. In the yeast Saccharomyces cerevisiae, there is only one homologue of the RecQ family of DNA helicases, Sgs1. Mutations in the SGS1 gene lead to defects similar to those seen in human cells from the RecQ family disorders, such as increases in the frequency of recombination and chromosome loss (Gangloff et al, 1994; Watt et al, 1995, 1996; Frei and Gasser, 2000b). Sgs1 functions in a conserved protein complex with Rmi1 and topoisomerase III, Top3 (Chang et al, 2005; Mullen et al, 2005; Yin et al, 2005; Raynard et al, 2006; Chen and Brill, 2007). When DNA is damaged by a double-strand break (DSB) or if replication forks are stalled, Sgs1 participates in repairing the damage and, in concert with Top3 and Rmi1, in resolving the DNA intermediates (such as double Holliday junctions) that arise during repair using the homologous recombination pathway (Cobb et al, 2002; Mullen et al, 2005; Mankouri and Hickson, 2007).
Sgs1 is a 1447-amino-acid-long protein that contains helicase motifs conserved with other RNA/DNA helicases (Figure 1A). The N-terminal half of Sgs1 includes a region that mediates a physical interaction with Top3 (Gangloff et al, 1994; Bennett et al, 2000; Fricke et al, 2001), and contains two regions with a high proportion of acidic amino acids (acidic regions or ARs) of unknown function. The Sgs1 helicase domain occupies a central tract of 300 amino acids. Two-hybrid interactions between Sgs1 and other proteins, such as topoisomerase II and the NER protein Rad16, are mediated by sequences that overlap with both the ARs and the helicase domain (Watt et al, 1995; Saffi et al, 2001). The sequences C-terminal to the helicase motifs are conserved among all RecQ homologues and are referred to as the RecQ conserved (RQC) domain. The RQC and HRDC domains (helicase and RNaseD C-terminal) have a function in nucleic acid binding. The HRDC domain partly overlaps the region mediating a two-hybrid interaction with the HR protein Rad51 (Wu et al, 2001).
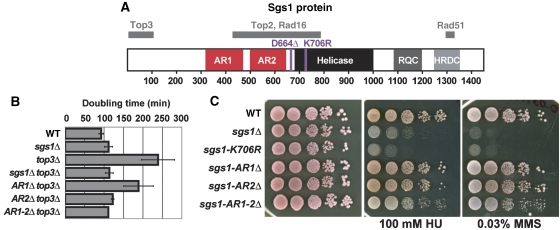
Figure 1.
Unlike sgs1Δ yeast, sgs1-AR2Δ yeast is not sensitive to DNA damage but can suppress top3Δ slow growth similar to sgs1Δ. Yeast with the acidic regions (AR1 or AR2) of Sgs1 deleted either separately or together (sgs1-AR1–2Δ) were analysed for SGS1 function. (A) Schematic representation of the functional motifs and domains of Sgs1. The grey bars above the protein represent the Sgs1 segments that mediate two-hybrid interactions with the indicated proteins. (B) sgs1-AR2Δ and sgs1-AR1–2Δ suppress top3Δ slow growth similarly to sgs1Δ, whereas sgs1-AR1Δ only slightly improves the growth rate of the top3Δ strain. (C) Unlike sgs1Δ or sgs1-AR1–2Δ, yeast with sgs1-AR1Δ or sgs1-AR2Δ is not sensitive to HU or MMS. Yeast were grown to log phase and plated in 10-fold serial dilutions onto YPD or YPD with 100 mM HU or 0.03% MMS.
As deletion of SGS1 leads to diverse phenotypes including sensitivity to genotoxic agents, hyper-recombination, chromosome missegregation, and meiotic defects, several mutational studies of SGS1 aimed to delineate the functions of the different parts of Sgs1. For example, as helicase action is dependent on ATP hydrolysis, it is possible to produce an intact yet catalytically inactive protein by mutating an invariant lysine residue within the ATP-binding pocket of the helicase domain, thereby creating sgs1-hd (helicase defective) mutants. In vivo studies of sgs1-hd mutants show that Sgs1 helicase activity is necessary for many of its cellular functions. For instance, sgs1-hd mutants are sensitive to DNA-damaging agents (Miyajima et al, 2000a; Mullen et al, 2000; Saffi et al, 2001; Weinstein and Rothstein, 2008), exhibit mitotic hyper-recombination (Mullen et al, 2000; Miyajima et al, 2000a), and suppress top3Δ slow growth (Mullen et al, 2000; Weinstein and Rothstein, 2008). On the other hand, sgs1-hd does not have the severe meiotic defects of the sgs1Δ allele (Miyajima et al, 2000a, 2000b). Furthermore, the helicase region is important for suppressing chromosome loss and missegregation independently of Sgs1 function in crossover and tract length control (Lo et al, 2006).
Another class of sgs1 alleles specifically interferes with the ability of Sgs1 to physically interact with Top3. This effect can result either from deleting 100–200 amino acids from the Sgs1 N terminus (sgs1-ΔN) or by creating point mutations within the N terminus (Mullen et al, 2000; Ui et al, 2001; Weinstein and Rothstein, 2008). These mutants exhibit DNA damage sensitivity, hyper-recombination, and sporulation defects similar or even greater than those of the sgs1Δ allele, suggesting that sgs1-ΔN alleles encode hypermorphic proteins (Mullen et al, 2000; Ui et al, 2001; Weinstein and Rothstein, 2008). Several studies have shown that the DNA damage sensitivity of the sgs1-ΔN alleles is suppressed either by overexpressing Top3 or by replacing the Top3 interaction domain with the entire Top3 protein covalently joined to the remaining Sgs1 (Bennett and Wang, 2001; Fricke et al, 2001). These results indicate that the ability to interact with Top3 is essential for normal Sgs1 function. Interestingly, in a top3 mutant background, removal of the Sgs1 N terminus has an effect opposite to that of sgs1Δ: instead of suppressing top3 defects, it exacerbates them (Mullen et al, 2000). This detrimental effect is helicase dependent, as concomitant mutation of the catalytic lysine 706 in the Sgs1-ΔN protein abolishes its toxicity in the top3 mutant (Mullen et al, 2000). This result suggests that deletion of the N terminus creates a ‘hyper-active' Sgs1 helicase, one that causes greater damage in the absence of Top3 than the wild-type Sgs1 protein. Overall, these results support a model, first proposed by Gangloff et al (1994), where a functional Top3 is needed to resolve a toxic DNA structure created by the Sgs1 helicase.
Here, we describe the phenotypes associated with deleting the two ARs found in the Sgs1 N-terminal region. We find that a separation-of-function phenotype results from deletion of AR2: sgs1-AR2Δ suppresses top3Δ slow growth, similarly to sgs1Δ, whereas in an otherwise wild-type background sgs1-AR2Δ is fully functional in its resistance to DNA damage. This finding prompted us to conduct a genetic screen for other sgs1 mutations that would confer the same phenotype. We found that deletion of a single amino acid, aspartic acid residue 664, confers the same separation of phenotype observed in sgs1-AR2Δ cells. The sgs1-D664Δ mutation does not alter DNA recombination or DNA damage resistance but can suppress top3Δ and rmi1Δ slow growth phenotype similar to sgs1Δ. We show that Sgs1-D664Δ has reduced protein levels. In addition, this sgs1 allele also causes a mis-localization of Rmi1 but not a change in abundance of its interacting partners Top3 and Rmi1. Finally, we show by two-dimensional (2D) gel analysis that sgs1-D664Δ causes an accumulation of replication-associated X-shaped structures at damaged replication forks, which are indicative of DNA replication defects. Our results suggest that Sgs1 activity in DNA repair can be uncoupled from its role in the repair of DNA replication intermediates (RIs).
Results
Characterization of the sgs1-ARΔ mutants
Sgs1 is a helicase that contains many conserved domains. Here, we analysed two of these domains, AR1 and AR2, located N-terminal to the helicase domains (Figure 1A). Although the ARs were previously identified (Kitao et al, 1998; Miyajima et al, 2000b), they have not been thoroughly described. In Miyajima et al, the two ARs were defined as AR1 spanning amino acids 400–474 (36% acidic residues) and AR2 spanning amino acids 510–596 (49% acidic residues). Upon protein sequence analysis, we believe that these domains are actually much larger where AR1 is 153 amino acids long (amino acids 321–474), 49 of which are aspartic or glutamic acid residues (32%), whereas AR2 is 146 amino acids long (amino acids 502–648), 44 of which are acidic (30%). By comparison, less than 15% of residues of the entire Sgs1 protein amino acids are acidic. These regions were not identified using standard bioinformatics tools such as PFAM, SMART, PROSITE, and Interpro search engines despite their enrichment for acidic amino acid residues.
Although these domains are thought to mediate protein interactions with Sgs1, these regions remain largely uncharacterized. To determine whether the ARs are important for Sgs1 function, we removed either AR separately (sgs1-AR1Δ or sgs1-AR2Δ) or both together including the intervening region (sgs1-AR1–2Δ; amino acids 321–648) at the SGS1 native chromosomal locus. As disruption of SGS1 suppresses top3Δ slow growth, we asked whether the AR deletions in SGS1 could similarly suppress top3Δ slow growth. By calculating the doubling times of exponentially growing yeast strains, we found that disruption of either sgs1-AR2Δ or sgs1-AR1–2Δ can suppress the slow growth of top3Δ-null yeast to the same extent as sgs1Δ (Figure 1B). In contrast, sgs1-AR1Δ only modestly suppresses the slow growth of top3Δ yeast (Figure 1B).
Next, these yeast strains were further analysed for cell growth in the presence of hydroxyurea (HU) and methylmethane sulphonate (MMS) (Figure 1C). HU causes stalled replication forks by depleting the dNTP pools, whereas MMS leads to DNA alkylation damage. We found that sgs1-AR1Δ and sgs1-AR2Δ yeast are not sensitive to HU and MMS, and sgs1-AR1–2Δ is only slightly sensitive to both these types of damage (Figure 1C). As expected, sgs1Δ and the catalytically inactive sgs1-K706R allele are sensitive to HU and MMS, and this sensitivity is much more severe than in the sgs1-ARΔ mutants (Figure 1C). Therefore, deletion of AR2 confers a separation-of-function phenotype: it resembles an sgs1 null for the suppression of top3Δ slow growth (Figure 1B), but behaves similar to wild-type SGS1 with respect to its sensitivity to DNA damage (Figure 1C).
Genetic screen for sgs1 mutations that confer an sgs1-AR2Δ-like phenotype
To pinpoint the Sgs1 function(s) that are dispensable in a wild-type background but become important in the absence of TOP3, we searched for alleles of SGS1 that mimic the sgs1-AR2Δ phenotype. Briefly, an SGS1 plasmid was introduced into an sgs1Δ top3Δ strain and 75 plasmid-borne sgs1 alleles were isolated that suppressed top3Δ slow growth (see Materials and methods). Of the 75 alleles, we identified one that fully complemented the HU and MMS sensitivity of an sgs1Δ strain. Sequencing of this sgs1 allele revealed that the plasmid-borne sgs1 ORF contained a deletion of three nucleotides, resulting in the loss of aspartic acid residue 664 (sgs1-D664Δ). As complementation by an SGS1 allele on a plasmid can differ from that of a genomically encoded allele (Weinstein and Rothstein, 2008), we introduced sgs1-D664Δ at the SGS1 chromosomal locus and subsequent analyses were performed with this allele.
As shown in Figure 2A, sgs1-D664Δ cells are not sensitive to DNA damage caused by HU or MMS. We next generated the sgs1-D664Δ top3Δ double mutants and measured their growth rate in rich medium, as well as their sensitivity to HU and MMS. The results show that sgs1-D664Δ indeed suppresses top3Δ slow growth similarly to sgs1Δ (Figure 2B). We then asked whether sgs1-D664Δ could also suppress top3Δ sensitivity to HU and MMS. As shown in Figure 2C, sgs1-D664Δ largely suppresses top3Δ HU and MMS sensitivity. Note that the concentration of HU and MMS in Figure 2C is much lower than in Figure 2A due to top3Δ hypersensitivity to DNA-damaging agents. Thus, the chromosomally encoded sgs1-D664Δ allele recapitulates the separation-of-function phenotype of sgs1-AR2Δ.
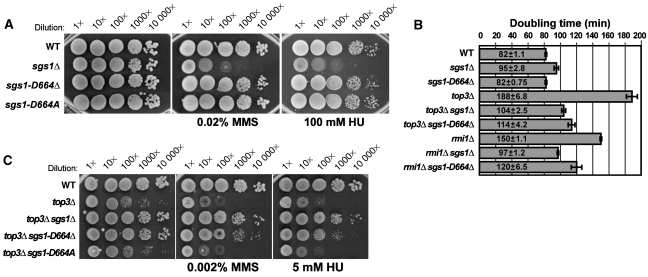
Figure 2.
Deletion of aspartic acid residue 664 in SGS1 causes a separation of function in SGS1. (A) Ten-fold serial dilutions were plated onto YPD or YPD with 0.02% MMS or 100 mM HU. Unlike sgs1Δ yeast, sgs1-D664Δ and sgs1-D664A yeast are not sensitive to HU or MMS. (B) sgs1-D664Δ suppresses the slow doubling times of top3Δ or rmi1Δ yeast. (C) sgs1-D664Δ suppresses the HU and MMS sensitivity of top3Δ. Ten-fold serial dilutions were plated onto YPD, YPD with 0.002% MMS, or YPD with 5 mM HU. Note, lower drug concentrations than in (A) were used due to the hypersensitivity of top3Δ.
To determine whether a substitution of aspartic acid residue 664 would also confer the same separation-of-function phenotype as the single amino-acid deletion, we created an SGS1 allele where aspartic acid residue 664 was changed to alanine (sgs1-D664A) and determined its phenotype. Similar to sgs1-D664Δ, the sgs1-D664A allele does not cause HU or MMS sensitivity (Figure 2A). However, in contrast to sgs1-D664Δ, sgs1-D664A suppresses neither top3Δ slow growth nor its HU or MMS sensitivity (Figure 2C; Supplementary Figure 1). These results show that sgs1-D664A behaves similar to a wild-type allele and the separation-of-function phenotype is specific to sgs1-D664Δ.
As Sgs1 functions in a complex with Top3 and Rmi1 (Chang et al, 2005; Mullen et al, 2005; Yin et al, 2005; Raynard et al, 2006; Chen and Brill, 2007), we also examined sgs1-D664Δ for the suppression of rmi1Δ slow growth. Figure 2B and Supplementary Figure 1 show that, consistent with previous results (Chang et al, 2005), sgs1Δ suppresses the slow growth of the rmi1Δ strain, whereas sgs1-D664Δ partially suppresses rmi1Δ slow growth.
sgs1-D664Δ does not affect DNA repair or recombination
As Sgs1 has an important function in DNA repair, its disruption in combination with loss of function of many DNA repair genes leads to a synthetic sick/lethal phenotype (Tong et al, 2004; Tong and Boone, 2005; Pan et al, 2006). Therefore, we examined the synthetic sick/lethal phenotype of sgs1-D664Δ allele with other DNA repair mutants by tetrad analysis. Supplementary Figure 2 shows that in contrast to sgs1Δ, sgs1-D664Δ is not synthetic sick/lethal when combined with disruptions in TOP1, SRS2, RRM3, or MUS81, four genes known to be important for DNA repair that also have a synthetic growth defect with sgs1Δ (Bennett et al, 2000; Mullen et al, 2000; Schmidt and Kolodner, 2004; Tong et al, 2004; Torres et al, 2004; Tong and Boone, 2005).
Deletion of SGS1 also leads to increased recombination rates. Therefore, we compared the hyper-recombination phenotype of sgs1Δ to that of sgs1-D664Δ and sgs1-AR2Δ. Recombination rates were measured at two loci: the SUP4 and rDNA. The SUP4-o gene, a tyrosine tRNA ochre suppressor, is located between five δ sequences, which are derived from long terminal repeats of the yeast Ty transposon and are oriented in direct and inverted orientation with respect to one another (Rothstein et al, 1987; Shor et al, 2002). Recombination through unequal crossing over, single strand annealing, or gene conversion between these approximately 300 bp repeats can lead to deletion of the intervening sequences (Figure 3A). The recombination rate of the parental strain (WT), sgs1Δ, sgs1-D664Δ, and sgs1-AR2Δ strains were calculated by measuring the loss of the URA3 marker. Figure 3A shows that sgs1-D664Δ and sgs1-AR2Δ have recombination rates similar to that of wild type. t-test analysis revealed that sgs1Δ has a significantly higher recombination rate compared with WT, sgs1-D664Δ, or sgs1-AR2Δ, which were not significantly different from each other.
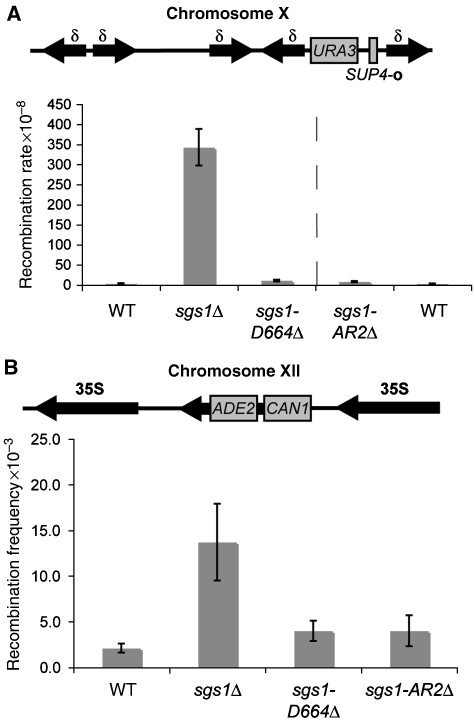
Figure 3.
Unlike sgs1Δ yeast, sgs1-D664Δ yeast do not have recombination defects. (A) A schematic representation of the SUP4-o locus with a URA3 insertion is shown, not to scale (Wallis et al, 1989). The rate of recombination events generating ura3 colonies was assayed and plotted for WT, sgs1Δ, sgs1-D664Δ, and sgs1-AR2Δ yeast strains. The recombination rate of sgs1-AR2Δ and WT were determined separately as indicated by the dotted line. Standard deviations are graphed. t-test analysis revealed that sgs1Δ has a significantly different recombination rate (P⩽0.001) when compared with WT, sgs1-D664Δ, or sgs1-AR2Δ, which were not significantly different from each other (P⩾0.05). (B) A schematic representation of the rDNA (35S repeated sequence) with ADE2 and CAN1 gene insertions is shown. Frequency of the generation of CANR, ade2 recombinants was measured in WT, sgs1Δ, sgs1-D664Δ, and sgs1-AR2Δ yeast strains. Standard deviations are graphed.
Next, we measured marker loss in the rDNA array, which is comprised of 100–200 repeated units that are 9.1 kb in length. To monitor the frequency of rDNA recombination in this array, an assay was used that disrupts one of those repeats with the ADE2 and CAN1 genes (Figure 3B). The parental strain (WT), sgs1Δ, sgs1-D664Δ, and sgs1-AR2Δ yeast were analysed for frequency of recombination by determining the number of yeast colonies that had simultaneously lost the ADE2 and CAN1 marker genes. The results show that sgs1-AR2Δ, sgs1-D664Δ, and the wild-type parental strain have similar recombination frequencies (Figure 3B). In contrast, sgs1Δ yeast exhibits an elevated recombination frequency at the rDNA locus, consistent with previous reports (Gangloff et al, 1994). Collectively, these results show that, unlike an sgs1 null, the sgs1-D664Δ allele does not alter mitotic recombination rate.
Sgs1-D664Δ exhibits altered protein levels and affects Rmi1 nucleolar localization
As deletion of aspartic acid residue 664 alters Sgs1 function, we measured whether its protein levels would be attenuated. To examine protein levels, we performed protein blot analysis on extracts from triple HA-tagged Sgs1 strains (Sgs1-3 × HA and Sgs1-D664Δ-3 × HA) and the untagged parent strain (WT). Figure 4 shows that Sgs1-D664Δ-3 × HA protein levels are reduced compared with Sgs1-3 × HA. To determine whether the separation-of-function phenotype of Sgs1-D664Δ was due to decreased protein levels, yeast strains where the Sgs1 AR-encoding regions are deleted (Sgs1-AR1Δ-3 × HA and Sgs1-AR2Δ-3 × HA) were analysed by protein blot. Yeast lacking Sgs1 AR2 confers a similar separation-of-function phenotype to Sgs1-D664Δ, whereas removal of Sgs1 AR1 does not disrupt its functions (Figure 1B and C). We found that both Sgs1-AR1Δ-3 × HA and Sgs1-AR2Δ-3 × HA protein levels are significantly reduced (Figure 4). Intriguingly, Sgs1-AR1Δ-3 × HA protein levels are undetectable at the exposure shown but are visible upon longer exposure times (Figure 4; data not shown). As sgs1-AR1Δ strains are not sensitive to HU and MMS, collectively these results suggest that low amounts of Sgs1 can still efficiently mediate DNA repair. Therefore, these results indicate that decrease in Sgs1 protein levels alone cannot account for the separation-of-function phenotype that we observe in the sgs1-D664Δ and sgs1-AR2Δ yeast strains. However, we cannot rule out the possibility that sgs1-AR1Δ has heightened activity despite its decreased protein levels.
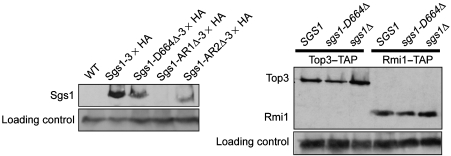
Figure 4.
Sgs1-D664Δ protein levels are reduced but the protein levels of Top3 and Rmi1 are unchanged. The protein levels were calculated using the ImageJ gel analysis tool by comparing the loading control (an unrelated abundant protein Adh1) to the protein of interest. Protein was extracted from equal cell numbers of the untagged parent strain (WT) and strains expressing Sgs1-3 × HA, Sgs1-D664Δ-3 × HA, Sgs1-AR1Δ-3 × HA, and Sgs1-AR2Δ-3 × HA, were analysed by protein blots using anti-HA antibodies. The expression of the mutant sgs1 alleles were 0.49 for Sgs1-D664Δ, 0.03 for Sgs1-AR1Δ, and 0.29 for Sgs1-AR2Δ to that of wild-type Sgs1. Top3–TAP and Rmi1–TAP protein levels were analysed by protein blot in SGS1, sgs1-D664Δ, and sgs1Δ strain backgrounds. Protein extracts were made from equal cell numbers and analysed for Top3 and Rmi1 levels by protein blot using anti-protein A antibodies and protein levels were quantitated from three experiments. For Top3–TAP strains, sgs1-D664Δ was 0.90±0.35 and sgs1Δ was 1.19±0.56 and for Rmi1–TAP strains, sgs1-D664Δ was 0.70±0.28 and sgs1Δ was 0.92±0.30 to that of SGS1. t-test analysis did not detect any significant difference between Top3 or Rmi1 protein levels in SGS1, sgs1Δ, or sgs1-D664Δ strains.
To ascertain the protein levels of both Top3 and Rmi1, protein extracts were made from Top3–TAP or Rmi1–TAP strains in SGS1, sgs1-D664Δ, or sgs1Δ genetic backgrounds and analysed by protein blot (Figure 4). Figure 4 shows that Top3 and Rmi1 protein levels are unaffected in the sgs1-D664Δ and sgs1Δ yeast strains when compared with wild type as determined by quantitation of Top3 and Rmi1 protein levels from three individual experiments and t-test analysis (data not shown).
As Sgs1-D664Δ has altered protein levels and functions in a complex with Rmi1 and Top3, we analysed the localization of Rmi1–YFP or Top3–YFP in the sgs1-D664Δ mutant. Both Rmi1–YFP and Top3–YFP constructs were fully functional for HU and MMS sensitivity (data not shown). As expected, Top3–YFP colocalizes with Nop1–CFP in the nucleolus (Supplementary Figure 3). When SGS1 is disrupted or D664 is deleted, Top3–YFP still colocalizes with Nop1–CFP. We next analysed the localization of Rmi1–YFP in sgs1Δ, sgs1-D664Δ, and sgs1-AR2Δ (Supplementary Figure 3). In sgs1Δ, sgs1-D664Δ, and sgs1-AR2Δ, the nucleolar localization of Rmi1–YFP is impaired: in these mutants, Rmi1–YFP shows a more diffuse nuclear localization compared with a wild-type SGS1 strain. These results show that disruption of SGS1 does not alter the localization of Top3 but does alter the localization of Rmi1.
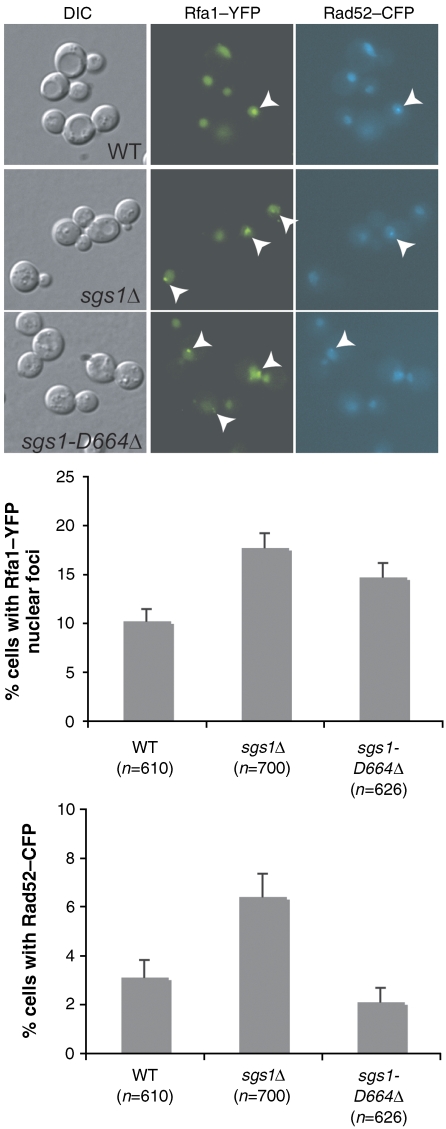
sgs1-D664Δ strains exhibit increased Rfa1 foci
Increasing evidence points to a role for Sgs1 in DNA replication. In conditions that induce replication fork stalling, Sgs1 interacts with the single-stranded DNA-binding protein, RPA and also promotes stabilization of DNA polymerases at the replication fork (Cobb et al, 2003). As sgs1-D664Δ clearly alters the activities of the Sgs1-D664Δ protein but has no detectable recombination phenotype, we analysed sgs1-D664Δ cells for the appearance of ssDNA as revealed by monitoring a YFP fusion protein to the large subunit of RPA (Rfa1–YFP). Figure 5 shows that spontaneous Rfa1–YFP nuclear foci in both sgs1Δ and sgs1-D664Δ are increased compared with wild type. In contrast, when the same cells are analysed for Rad52–CFP nuclear foci, only the sgs1Δ cells show increased Rad52-foci (Figure 5). These results suggest that the increased ssDNA observed in sgs1-D664Δ is not due to recombination defects as it is not correlated with an increased number or persistence of Rad52-foci.
Figure 5.
The number of cells with RPA foci, but not Rad52 foci, increases in sgs1-D664Δ. Spontaneous formation of Rfa1–YFP and Rad52–CFP nuclear foci was analysed in SGS1, sgs1Δ, and sgs1-D664Δ backgrounds. Images of Rfa1 and Rad52 are shown with white arrowheads indicating a focus. The average of three independent experiments is plotted with standard error bars.
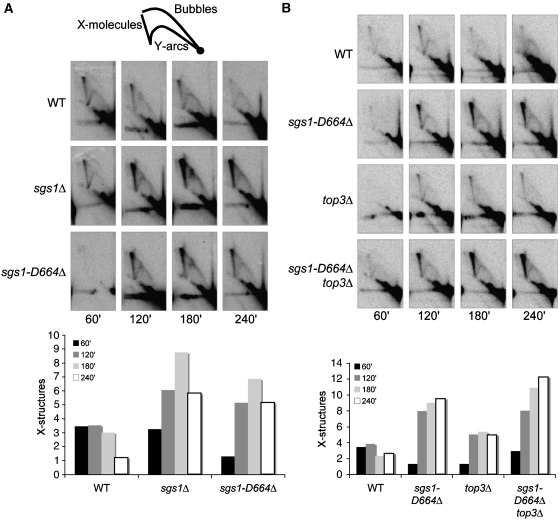
Persistent X-structures are observed in sgs1-D664Δ strains during replication of damaged templates
As sgs1-D664Δ has more ssDNA (Figure 5) and at the same time does not exhibit any measurable defects in damage sensitivity or mitotic recombination (Figures 2A and 3), we monitored these cells for defects in DNA replication. Previously it was found by 2D gel electrophoresis that sgs1Δ accumulates replication-associated and Rad51/Rad52-dependent X-shaped molecules specifically at damaged replication forks and not at forks stalled by HU treatment (Liberi et al, 2005). We analysed the pattern of RIs in sgs1Δ and sgs1-D664Δ during replication of damaged templates at ARS305, an early, efficient origin of replication on chromosome III as well as at its adjacent regions (Liberi et al, 2005; Branzei et al, 2006). Cells were first arrested in G2/M with nocodazole, a microtubule-destabilizing agent, and then released into YPD medium containing sublethal doses of MMS (0.033%). Subsequently, at 60, 120, 180, and 240 min, cells were collected and the RIs were extracted and analysed by 2D gel electrophoresis. Cell cycle progression into S phase was simultaneously measured by flow cytometry (Supplementary Figure 4A and B). Consistent with previous reports (Liberi et al, 2005; Branzei et al, 2006), we observe that in these conditions wild-type cells form X-structures but then resolve them (Figure 6A). In contrast, when sgs1Δ cells are compared with sgs1-D664Δ, both accumulate X-structures to a similar extent (Figure 6A; data not shown). Additionally, we found that the helicase dead sgs1-K706A allele similarly accumulated X-structures, which is not surprising as it largely resembles a null allele (Supplementary Figure 5). Furthermore, the replication-associated X-structures observed in sgs1-D664Δ cells likely reflect unresolved intermediates, indicating that these cells have certain replication defects that are similar to those observed in sgs1Δ. Similar to sgs1Δ top3Δ mutants (Liberi et al, 2005), sgs1-D664Δ top3Δ cells accumulate X-structures to the same extent as sgs1-D664Δ cells (Figure 6B). These results suggest that the X-structures that accumulate in sgs1-D664Δ form independently of TOP3.
Figure 6.
Replication-associated X-structures accumulate in sgs1-D664Δ that are independent of top3Δ. (A) Two-dimensional (2D) gel electrophoresis was conducted to visualize replication fork progression and processing at ARS305. Cells were first arrested in G2/M with nocodazole at 25°C and then released into YPD medium with 0.033% MMS at 30°C. ARS305 was analysed for replication intermediates after 60, 120, 180, and 240 min after release from nocodazole by Southern blotting. A cartoon of the replication intermediates observed is shown. X-structure accumulation was quantitated and graphed. Cells with sgs1-D664Δ accumulate X-structures. (B) Replication intermediates formed at ARS305 were analysed by 2D gel electrophoresis in WT, sgs1-D664Δ, top3Δ, and sgs1-D664Δ top3Δ strains as described for (A). Replication-associated X-structures accumulate in sgs1-D664Δ top3Δ to the same extent as in sgs1-D664Δ.
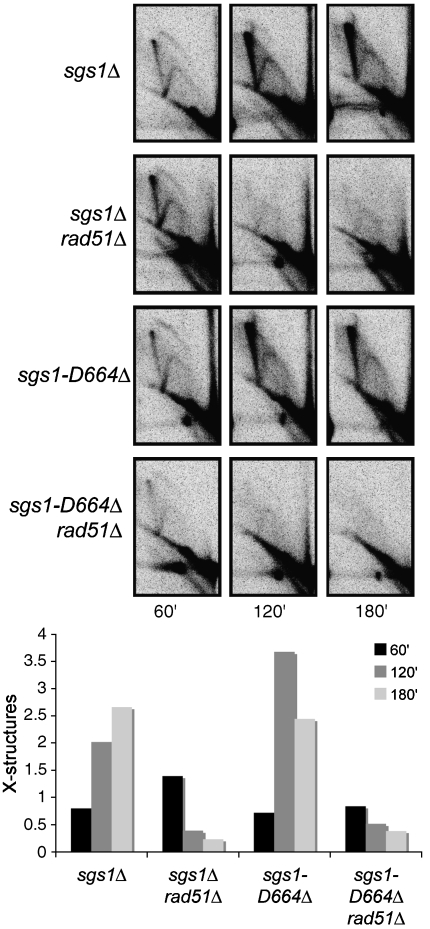
Previously, it was shown that the accumulation of replication-associated X-structures in sgs1Δ cells is Rad51 dependent (Liberi et al, 2005). To determine whether Rad51 is important for the accumulation of X-structures observed in sgs1-D664Δ strains, we performed 2D gel electrophoresis comparing X-structure formation of sgs1Δ and sgs1Δ rad51Δ cells to sgs1-D664Δ and sgs1-D664Δ rad51Δ cells (Figure 7). Cell cycle progression into S phase was measured by flow cytometry (Supplementary Figure 4C). Similar to the results observed with sgs1Δ strains, we found that accumulation of replication-associated X-structures in sgs1-D664Δ was dependent upon Rad51. These results suggest that Rad51 is important for the accumulation of the observed RIs.
Figure 7.
Replication-associated X-structure accumulation in sgs1Δ and sgs1-D664Δ cells is Rad51 dependent. Replication fork progression and processing at ARS305 was visualized by 2D gel electrophoresis and X-structure accumulation was quantitated.
Discussion
We have identified a region of the RecQ helicase, Sgs1, that uncouples its function with Top3/Rmi1 in DNA repair from its role in maintaining replication fork integrity. Removal of AR two (sgs1-AR2Δ) behaves similar to an sgs1-null allele in a top3Δ background, suppressing the slow growth of top3Δ. However, in a TOP3 wild-type background, the sgs1-AR2Δ mutant resembles wild-type SGS1 in that it confers HU and MMS resistance. As AR2Δ removes a large number of Sgs1 residues, this mutation may affect the global conformation of Sgs1 or interfere with several Sgs1 functions, unlike a more conservative mutation. Thus, we performed a genetic screen and found that deletion of a single codon, encoding aspartic acid residue 664, can mimic the phenotype of AR2Δ. Thus, sgs1-D664Δ confers a separation-of-function phenotype that resembles sgs1Δ in top3Δ and rmi1Δ backgrounds, but is similar to wild-type SGS1 for DNA recombination/repair-mediated functions. Sgs1-D664Δ protein levels are reduced, whereas the protein levels of its interacting partners, Top3 and Rmi1, are largely unaffected, despite altered Rmi1 localization. Finally, we show that sgs1-D664Δ cells exhibit an accumulation of replication-associated ssDNA and Rad51-dependent X-shaped structures at damaged replication forks independently of TOP3. Taken together, our results suggest that Sgs1 functions with Top3 and Rmi1 during DNA repair, but that Sgs1 function can be uncoupled from Top3 during repair of DNA RIs.
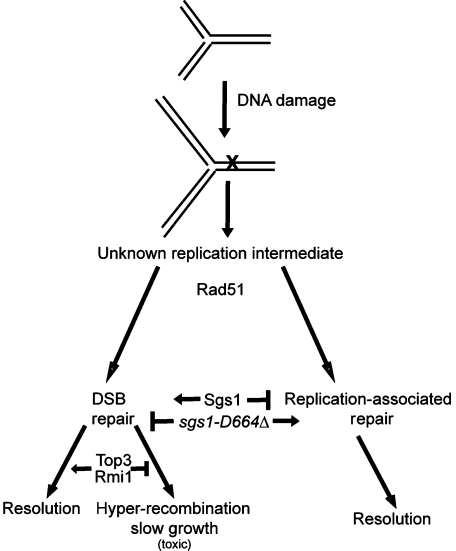
During replication, DNA damage can result from many sources such as radiation, oxidizing damage, or genotoxic chemicals, and cells must repair this damage before new DNA synthesis can progress. Depending on the type of damage and the cell cycle stage, cells process the DNA lesions differently (Barlow et al, 2008). Spontaneous DNA damage can also create DNA intermediates that utilize the Sgs1 protein for repair (Figure 8). Normally, Sgs1 will channel the processing of DNA lesions into a Top3-dependent DSB repair pathway. Thus, resolution of these intermediates requires Top3 and an accessory protein, Rmi1 (Figure 8). When TOP3 is disrupted, the DNA intermediates processed by Sgs1 remain unresolved and are ‘toxic' to the cell (Gangloff et al, 1994), leading to the slow growth, hyper-recombination, and DNA damage sensitivity characteristic of top3Δ cells. In the absence of TOP3, SGS1 gene deletion has a beneficial effect: it suppresses the slow growth, DNA damage sensitivity, and hyper-recombination phenotype of the top3Δ (Gangloff et al, 1994; Shor et al, 2002). It has been hypothesized that one of the toxic intermediates accumulating in these cells may be hemicatenane-like structures resulting from damage-bypass processes during replication or from conversion of recombination intermediates such as double Holliday junctions (Branzei and Foiani, 2007).
Figure 8.
Model of sgs1-D664Δ function in DNA repair and DNA replication. During DNA replication, DNA damage can occur that produces a DNA intermediate, the exact structure of which is unknown, but can be resolved in an Sgs1-dependent manner. In wild-type cells, the DNA intermediate is acted on by Top3, ultimately leading to resolution of the DNA damage (left arrows). When TOP3 is disrupted, Sgs1 creates a ‘toxic intermediate' that leads to hyper-recombination and slow growth. In the sgs1-D664Δ background, the replication lesions are repaired through a replication-associated repair pathway that produces replication-associated X-shaped structures as DNA intermediates but which can ultimately be resolved without the need for Top3 activity (right arrows). This model is in agreement with the findings that sgs1-D664Δ yeast is not sensitive to DNA-damaging agents but can suppress top3Δ slow growth as sgs1-D664Δ avoids shunting the DNA intermediate through the Top3-mediated DSB repair pathway.
Cells are thought to bypass the DNA damage at replication forks and to repair gaps and DSBs formed during replication, mainly by using homologous recombination, but alternative replication-associated repair mechanisms likely exist (Figure 8). In sgs1Δ and sgs1-D664Δ cells, we observed an accumulation of replication-associated X-shaped structures at damaged replication forks. X-structures are also visualized in wild-type cells but are ultimately resolved, suggesting that the accumulation of these X-shaped structures in sgs1Δ is likely due to their impaired resolution. However, it is possible that a fraction of the structures forming in sgs1 mutants may have different genetic dependencies and molecular or topological characteristics from the ones transiently forming in wild-type cells. These replication-associated X-structures may contain large stretches of ssDNA as we observe an increase in RPA foci in both sgs1Δ and sgs1-D664Δ mutants. Consistent with this conclusion, it has been shown that X-structures are sensitive to Mung Bean and S1 nucleases, which specifically degrade ssDNA (Liberi et al, 2005). The accumulation or stability of X-structures found in sgs1Δ and sgs1-D664Δ is dependent upon Rad51 (Liberi et al, 2005). These results suggest that homologous recombination proteins are likely shared by different pathways promoting damage-bypass during replication.
We find that sgs1-D664Δ alleviates the slow growth and DNA damage sensitivity of top3Δ. These results could reflect Sgs1-D664Δ shunting the DNA intermediate away from the DSB repair pathway and into a repair pathway that still leads to the formation of X-shaped intermediates during replication-associated repair (Figure 8). In this scenario, the ‘toxic' intermediate normally created by Sgs1, and subsequently needs Top3 for resolution, is not created. In support of this model, we find that the sgs1-D664Δ allele, similar to sgs1Δ, accumulates damaged RIs (X-structures) that are independent of TOP3. Sgs1 is important for DNA polymerase interaction with replication forks and is localized at S-phase-specific foci with other DNA replication proteins (Frei and Gasser, 2000a, b; Cobb et al, 2003). Although the Sgs1 protein has important functions in both DSB and replication-associated repair, sgs1-D664Δ is an SGS1 allele that has wild-type function during DSB repair while complexed with Top3/Rmi1, but is altered for other SGS1 cellular roles such as DNA replication-associated repair (Figure 8). The damaged DNA replication templates could be repaired by a post-replicative pathway utilizing single strand gap repair or template switching. Interestingly, both sgs1Δ and sgs1-D664Δ alter the localization but not abundance of Rmi1, whereas Top3 localization and abundance are unaffected (Figure 4; Supplementary Figure 3).
Similar to sgs1-AR2Δ or sgs1-D664Δ, C-terminal truncation of SGS1 also confers a separation-of-function phenotype (Lu et al, 1996; Miyajima et al, 2000b; Mullen et al, 2000). Truncation of 202 residues from the Sgs1 C terminus produces a similar separation-of-function phenotype, whereas the in vitro helicase activity of Sgs1 is unaffected (Lu et al, 1996). Another truncation allele, sgs1-ΔC200, suppresses top3Δ slow growth, but confers normal HU and MMS resistance and direct repeat recombination levels (Mullen et al, 2000). In another report, the C-terminal 254 amino acids of Sgs1 were found to be dispensable for HU and MMS resistance, as well as for normal levels of mitotic and meiotic recombination between homologous chromosomes (Miyajima et al, 2000b). Thus, the Sgs1 C terminus, as well as the AR2 domain, is dispensable for several functions of the protein in the wild type, but has an important function in the absence of TOP3.
Although aspartic acid residue 664 is located approximately 30 amino acids N-terminal to the first helicase motif of Sgs1, it is possible that the separation-of-function we observe is the result of a decrease in Sgs1 catalytic activity. However, our results show that in contrast to the helicase activity of SGS1 being essential for resistance to HU and MMS (Figure 1C), presence of aspartic acid residue 664 in SGS1 is not required for DNA damage resistance (Figure 2A). Interestingly, the helicase activity of a distinct separation-of-function mutant, Sgs1-ΔC202, has been measured and found indistinguishable from that of the wild-type protein (Lu et al, 1996). These results are consistent with a model where the sgs1-D664Δ mutation may attenuate the Sgs1 helicase activity in a lesion-specific manner.
A key aspect of the two sgs1 separation-of-function mutations described here is that they behave similar to the null allele in the absence of TOP3 but similar to the wild-type SGS1 allele in the presence of TOP3. As highly acidic stretches are known to mediate protein–protein interactions, the location of these mutations suggests that they may disrupt physical interactions between Sgs1 and in vivo binding partners. For instance, some transcription factors contain acidic clusters important for the recruitment of RNA polymerase complexes to gene promoters (Struhl, 1995). Thus, it is possible that deletion of aspartic acid residue 664 or of AR2 as a whole interferes with the ability of Sgs1 to interact with other proteins. Similarly, another separation-of-function allele, sgs1-ΔC202, may encode a protein incapable of interacting with Rad51. Thus, the ability of Sgs1 to physically interact with other proteins involved in DNA metabolism acquires additional importance for Sgs1 function in the top3Δ. In the future, it will be interesting to investigate whether other proteins help mediate the different roles of Sgs1 in DNA repair and DNA replication.
Materials and methods
Strains, plasmids, and media
The strains used were derived from the W303 background (Table I). Deletion of SGS1 ARs and D664Δ/D664A were created using a cloning-free allele replacement method (Erdeniz et al, 1997; Erdeniz and Rothstein, 2000). Standard procedures were used for tetrad dissection and LiOAc transformation (Sherman et al, 1986). The media were prepared as described, except that twice the amount of leucine was used (Sherman et al, 1986).
Table 1.
Strains
| Name | Description |
|---|---|
| W1588-4C | MATa |
| W1588-4A | MATα |
| W1958a | MATa top3∷TRP1 + |
| MATα + sgs1∷HIS3 | |
| J836b | MATa sgs1-AR1Δ (amino acids 321–474) |
| J752b | MATa sgs1-AR2Δ (amino acids 502–648) |
| J755b | MATa sgs1-AR1-2Δ (amino acids 321–648) |
| W1854-2C | MATα sgs1∷HIS3 |
| W1991-1Bc | MATα sgs1-K706R |
| J730c | MATa sgs1-K706A |
| W2886b | MATa top3∷LEU2 + |
| MATα + sgs1-AR1Δ | |
| W2284b | MATa top3∷HIS3 + |
| MATα +sgs1-AR2Δ | |
| W2287b | MATa top3∷LEU2 + |
| MATα +sgs1-AR1-2Δ | |
| W5927-20Ab | MATα sgs1-D664Δ |
| W5928-16Cb | MATα sgs1-D664A |
| W5904b | MATa top3∷TRP1 + |
| MATα + sgs1-D664Δ | |
| W6700b | MATa rmi1∷kanMX+ |
| MATα + sgs1-D664Δ | |
| W6701b | MATa rmi1∷kanMX + |
| MATα + sgs1∷HIS3 | |
| W6268b | MATa top1∷HIS3 + |
| MATα + sgs1-D664Δ | |
| W1956c | MATa top1∷HIS3 + |
| MATα + sgs1∷URA3 | |
| W8292b | MATa srs2∷LEU2 + |
| MATα + sgs1∷HIS3 | |
| W6669b | MATa srs2∷LEU2 + |
| MATα + sgs1-D664Δ | |
| W6672b | MATa rrm3∷URA3 + |
| MATα + sgs1∷HIS3 | |
| W6670b | MATa rrm3∷URA3 + |
| MATα + sgs1-D664Δ | |
| W6664b | MATa mus81∷kanMX + |
| MATα + sgs1∷HIS3 | |
| W6671b | MATa mus81∷kanMX + |
| MATα + sgs1-D664Δ | |
| W1594-2A | MATa sgs1∷HIS3 SUP4-o∷URA3 |
| W1868-8Ba | MATa SUP4-o∷URA3 |
| W6667-1Ab | MATa sgs1-D664Δ SUP4-o∷URA3 |
| W8607-12Ab | MATα sgs1-AR2Δ SUP4-o∷URA3 |
| W4314-2C | MATα rDNA∷ADE2-CAN1 |
| W6666-6Db | MATa sgs1∷HIS3 rDNA∷ADE2-CAN1 |
| W6668-10Cb | MATa sgs1-D664Δ rDNA∷ADE2-CAN1 |
| W8610-9Cb | MATα sgs1-AR2Δ rDNA∷ADE2-CAN1 |
| W8609-18Bb | MATa sgs1-AR2Δ RMI1-YFP RAD52-RFP pNOP1-CFP (pWJ1327) |
| W8370-1Ab | MATa sgs1∷HIS3 TOP3-YFP RAD52-RFP pNOP1-CFP (pWJ1327) |
| W8371-3Bb | MATα TOP3-YFP RAD52-RFP pNOP1-CFP (pWJ1327) |
| W8371-21Db | MATa sgs1-D664Δ TOP3-YFP RAD52-RFP pNOP1-CFP (pWJ1327) |
| W8372-8Db | MATa sgs1∷HIS3 RMI1-YFP RAD52-RFP pNOP1-CFP (pWJ1327) |
| W8372-1Ab | MATa RMI1-YFP RAD52-RFPpNOP1-CFP (pWJ1327) |
| W8373-7Bb | MATa sgs1-D664Δ RMI1-YFP RAD52-RFP pNOP1-CFP (pWJ1327) |
| W7588-8Cd | MATa SGS1-3 × HA-LEU2 |
| U2869b | MATa SGS1-3 × HA-LEU2 RMI1-TAP-TRP1 |
| W7740-1Ab | MATa SGS1-D664Δ-3 × HA-LEU2 |
| U2871b | MATα SGS1-D664Δ-3 × HA-LEU2 RMI1-TAP-TRP1 |
| U2870b | MATa SGS1-3 × HA-LEU2 TOP3-TAP-TRP1 |
| U2872b | MATa SGS1-D664Δ-3 × HA-LEU2 TOP3-TAP-TRP1 |
| W8111-9Db | MATα sgs1∷HIS3 TOP3-TAP-TRP1 |
| W8110-19Ab | MATα sgs1∷HIS3 RMI1-TAP-TRP1 |
| W8499-7Db | MATα sgs1-D664Δ rad51∷URA3 |
| All strains are RAD5 derivatives of the W303 (MATa BAR1 ade2-1 can1-100 ura3-1 his3-11,15leu2-3,112 trp1-1 rad5-535)/W1588 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) backgrounds, except for the ‘c' strains, which are library BY4741 (Brackmann et al, 1998) and were backcrossed with the W303 background between seven and nine times. | |
| aWagner et al (2006). | |
| bThis study. | |
| cWeinstein and Rothstein (2008). | |
| dChang et al (2005). | |
DNA damage sensitivity assays
HU (5 or 100 mM) and methyl methane sulphonate (MMS 0.002, 0.02, or 0.03%) were added to YPD medium. Yeast were grown in YPD overnight at 30°C, sonicated, and equal numbers of cells were 10-fold serially diluted onto plates containing YPD or YPD with HU or MMS. The plates were photographed after 3 days of growth at 30°C.
Growth curves
Yeast were grown to early log phase and the OD600 of exponentially growing cultures was measured every hour for at least 6 h. The natural logarithms of the OD600 values were plotted as a function of time and the slope of the best-fit line was obtained using Microsoft Excel. The doubling time was calculated by dividing ln 2 by this slope.
Screening for spontaneous sgs1 separation-of-function mutations
A centromeric plasmid containing a copy of SGS1 (pCEN-SGS1) was transformed into an sgs1Δ top3Δ double mutant. Five independent, slow-growing transformants were inoculated into selective liquid medium to ensure plasmid maintenance. The cultures were grown overnight and plated onto selective medium. The resulting plates contained small and large colonies. The 75 large, fast-growing colonies (top3Δ sgs1Δ pCEN-sgs1*) were crossed with an sgs1Δ TOP3 strain to create a TOP3-containing diploid whose only source of Sgs1 protein was the plasmid-borne sgs1* allele. These diploids were patched and replica-plated to plates containing 100 mM HU or 0.03% MMS. Three candidates that conferred HU and MMS resistance comparable to the wild-type strain were picked. One allele (sgs1-D664Δ) thus isolated confers the same degree of HU and MMS resistance as wild-type SGS1. Subsequent plasmid rescue and re-transformation into sgs1Δ and sgs1Δ top3Δ strains confirmed that the plasmid-borne sgs1-D664Δ allele indeed confers the desired phenotype.
Recombination assays
The SUP4 recombination assay was performed by analysing yeast for loss of the URA marker as described previously (Shor et al, 2002). The rDNA recombination assay was performed by analysing yeast for the loss of the ADE/CAN markers as described previously (Burgess et al, 2007).
Microscopy
Cells were grown in 5 ml SC medium plus 100 mg/l adenine at 23°C overnight, and harvested for microscopy as described previously (Lisby et al, 2001), except that images were captured under a × 100 magnification oil immersion objective (1.46NA). on a Leica DM5500B upright microscope illuminated with a 100 W mercury arc lamp and high-efficiency YFP and CFP filter cubes (Leica Microsystems). The images were captured with a Hamamatsu Orca AG cooled digital CCD camera, operated by Volocity software (Improvision). Stacks of 11 0.3-μm sections were captured using the following channels and exposure times: DIC (2 ms), YFP–Rmi1 (2000 ms), YFP–Top3 (2000 ms), Nop1–CFP (10 ms), Rfa1–YFP (500 ms), and Rad52–CFP (500 ms). Images were processed and enhanced identically using Volocity software.
Protein analysis
Here, 50 ml yeast cultures in YPD were grown to 0.5 OD600. Protein extracts were prepared by resuspending yeast in 200 μl of 20% TCA and 100 μl of glass beads and vortexing for 3 min. The supernatant was added to 600 μl of 5% TCA and pelleted. The pellet was resuspended in 100 μl of Laemmli buffer (150 mM Tris, 6% SDS, 30% glycerol, 0.6% bromophenol blue, and 15% beta-mercaptoethanol) and 50 μl 1 M Tris base was added. The protein was then pelleted and the supernatant was loaded onto a 10% SDS–PAGE gel. The gel was transferred to a membrane and protein blotted with anti-HA antibodies (1:1250; Covance, 16B12), anti-Adh1 antibodies (1:5000; Chemicon International, AB1202), or anti-protein A antibodies (1:5000; Sigma, P3775) to detect the TAP tag.
2D gel analysis
Cells were synchronized in G2/M by adding nocodazole to a final concentration of 10 μg/ml together with DMSO to a final concentration of 1% v/v, for about 2 h. The release from nocodazole arrest was performed as described previously, in YPD medium containing MMS at a final concentration of 0.033% v/v at 30°C (Branzei et al, 2006). Purification of DNA intermediates, FACS analysis, and 2D gel procedure were carried out as described (Branzei et al, 2004, 2006; Liberi et al, 2005). The DNA samples were digested with HindIII and EcoRV and analysed by 2D gel with a probe against ARS305. Quantitation of the X-structures was measured using IMAGEQUANT software. For each time point, the percentage of signal intensity was determined by selecting areas that correspond to the monomer spot, the spike signal (including additional RIs, bubbles, Ys, and double Ys) and also a region without any RIs for background reference. The background percentage, multiplied for the ratio between the dimension of the area for Xs (or the RI of interest) and for background, is then subtracted from the percentage of spike signal (or of the RI of interest). The resulting number is the absolute value to be plotted in the histogram.
Value for X=% (Xs)−[% (background) (area (Xs)/area (background))].
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure Legends
Acknowledgments
We thank Grant Brown for generously supplying yeast strains. We thank Peter Thorpe, Michael Chang, and David Alvaro for reading the article and helpful discussions. This study was supported by National Institute of Health grants GM50237 (RR), GM67055 (RR), GM078840 (KAB), and GM73567 (RCB), and by grants from the Associazione Italiana per la Ricerca sul Cancro and Association for International Cancer Research to MF and DB and by a National Science Foundation graduate fellowship to ES.
References
- Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC (2000) Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem 275: 26898–26905 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Wang JC (2001) Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins. Proc Natl Acad Sci USA 98: 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2007) RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev 21: 2019–3026 [DOI] [PubMed] [Google Scholar]
- Branzei D, Seki M, Enomoto T (2004) Rad18/Rad5/Mms2-mediated polyubiquitination of PCNA is implicated in replication completion during replication stress. Genes Cells 9: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M (2006) Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X (2007) The Slx5–Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol 27: 6153–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Rothstein R, Freyer GA, Boone C, Brown GW (2005) RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J 24: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ (2007) Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem 282: 28971–28979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Gasser SM (2002) RecQ helicases: at the heart of genetic stability. FEBS Lett 529: 43–48 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N, Mortensen UH, Rothstein R (1997) Cloning-free PCR-based allele replacement methods. Genome Res 7: 1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N, Rothstein R (2000) Rsp5, a ubiquitin-protein ligase, is involved in degradation of the single-stranded-DNA binding protein Rfa1 in Saccharomyces cerevisiae. Mol Cell Biol 20: 224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Gasser SM (2000a) RecQ-like helicases: the DNA replication checkpoint connection. J Cell Sci 113: 2641–2646 [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM (2000b) The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev 14: 81–96 [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Kaliraman V, Brill SJ (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem 276: 8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A (1998) Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54: 443–452 [DOI] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Paffet KS, Amit O, Clikeman JA, Sterk R, Brenneman MA, Nickoloff JA (2006) Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol 26: 4086–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff S, Romeo AM, Sternglanz R (1996) Human homologues of yeast helicase. Nature 383: 678–679 [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Hickson ID (2007) The RecQ helicase-topoisomerase III–Rmi1 complex: a DNA structure-specific ‘dissolvasome'? Trends Biochem Sci 32: 538–546 [DOI] [PubMed] [Google Scholar]
- Miyajima A, Seki M, Onoda F, Shiratori M, Odagiri N, Ohta K, Kikuchi Y, Ohno Y, Enomoto T (2000a) Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol Cell Biol 20: 6399–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima A, Seki M, Onoda F, Ui A, Satoh Y, Ohno Y, Enomoto T (2000b) Different domains of Sgs1 are required for mitotic and meiotic functions. Genes Genet Syst 75: 319–326 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Brill SJ (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ (2005) Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1–Top3 complex. Mol Cell Biol 25: 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD (2006) A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P (2006) A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem 281: 13861–13864 [DOI] [PubMed] [Google Scholar]
- Rothstein R, Helms C, Rosenberg N (1987) Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol Cell Biol 7: 1198–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffi J, Feldmann H, Winnacker EL, Henriques JA (2001) Interaction of the yeast Pso5/Rad16 and Sgs1 proteins: influences on DNA repair and aging. Mutat Res 486: 195–206 [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD (2004) Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol 24: 3213–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB (1986) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R (2002) Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K (1995) Yeast transcriptional regulatory mechanisms. Annu Rev Genet 29: 651–674 [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C (2005) Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol 313: 171–192 [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L et al. (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Torres J, Schnakenberg S, Zakian V (2004) Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and for restart activities. Mol Cell Biol 24: 3198–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui A, Satoh Y, Onoda F, Miyajima A, Seki M, Enomoto T (2001) The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol Genet Genomics 265: 837–850 [DOI] [PubMed] [Google Scholar]
- Wagner M, Price G, Rothstein R (2006) The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174: 555–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R (1989) A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58: 409–419 [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ (1996) SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Louis EJ, Borts RH, Hickson ID (1995) Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81: 253–260 [DOI] [PubMed] [Google Scholar]
- Weinstein J, Rothstein R (2008) The genetic consequences of ablating helicase activity and the Top3 interaction domain of Sgs1. DNA Repair (Amst) 7: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276: 19375–19381 [DOI] [PubMed] [Google Scholar]
- Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W (2005) BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J 24: 1465–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure Legends