Abstract
For the compact Drosophila genome, several factors mediating insulator function, such as su(Hw) and dCTCF, have been identified. Recent analyses showed that both these insulator-binding factors are functionally dependent on the same cofactor, CP190. Here we analysed genome-wide binding of CP190 and dCTCF. CP190 binding was detected at CTCF, su(Hw) and GAF sites and unexpectedly at the transcriptional start sites of actively transcribed genes. Both insulator and transcription start site CP190-binding elements are strictly marked by a depletion of histone H3 and, therefore, a loss of nucleosome occupancy. In addition, CP190/dCTCF double occupancy was seen at the borders of many H3K27me3 ‘islands'. As before, these sites were also depleted of H3. Loss of either dCTCF or CP190 causes an increase of H3 and H3K27 trimethylation at these sites. Thus, for both types of cis-regulatory elements, domain borders and promoters, the chromatin structure is dependent on CP190.
Keywords: chromatin domains, chromatin insulation, CP190, CTCF, transcription
Introduction
Promoter elements, enhancers, silencers and insulators control gene activity through their concerted action. Promoter sequences contain the information for the transcription start site (TSS) (Sandelin et al, 2007), they may harbour regulatory sequences and they are targets for action of and interaction with activating enhancers and repressing silencers. Genome-wide analyses of the sites occupied by DNA-binding factors and of specific histone modifications revealed that active promoters are characterised by a nucleosome-free region, which is flanked by well-positioned nucleosomes (Lee et al, 2004). These are marked by the H2A.Z/Htz1 histone variant (Zhang et al, 2005; Hawkins and Ren, 2006), the H3.3 variant (Mito et al, 2005) and by H3ac and H3K4me3 modification (Barski et al, 2007; Barrera et al, 2008). In addition to RNA polymerase II (Pol II) and components of the preinitiation complex (Kim et al, 2005), negative cofactor 2 also marks active promoters (Albert et al, 2007).
Enhancer elements activate promoters, whereas silencer elements repress transcription. Repressive elements bound by Polycomb group (PcG) proteins were first identified in Drosophila, which prevent inappropriate expression of homeotic (Hox) genes (Lewis, 1978). Genome-wide analysis of PcG-binding sites revealed an association with large genomic regions of up to 150 kb. PcG-bound chromatin is trimethylated at histone H3 Lys27 and is generally transcriptionally silent. Many of the genes bound by PcG proteins encode transcriptional regulators or components of signal transduction pathways and are developmentally regulated (Nègre et al, 2006; Schwartz et al, 2006; Tolhuis et al, 2006; Beisel et al, 2007; Schwartz and Pirrotta, 2007). Functionally, it is postulated that PcG-bound chromatin represses gene promoters or even larger domains by inducing H3K27 trimethylation and that the spreading of this histone modification is blocked by insulators, such as the gypsy transposon (Kahn et al, 2006).
The gypsy transposon is the most extensively studied Drosophila insulator that contains repeated binding sequences for the factor ‘suppressor of Hairy wing' [su(Hw) (Spana et al, 1988)]. The transposon or an isolated fragment with su(Hw)-binding sites was shown to mediate enhancer blocking within the yellow locus. Enhancer-blocking function by su(Hw) is mediated or affected by additional factors, such as Mod(mdg4) (modifier of mdg4), CP190 (centrosomal protein 190), the ubiquitin ligase dTopors and a putative RNA helicase Rm62 (Gerasimova et al, 1995; Pai et al, 2004; Capelson and Corces, 2005; Lei and Corces, 2006). Mapping of su(Hw)-binding sites in selected regions of the genome revealed hundreds of sites, for some of which an enhancer-blocking function was suggested (Parnell et al, 2006; Ramos et al, 2006; Adryan et al, 2007).
Further examples of insulator function can be found in the bithorax complex (BX-C), which encompasses three homeotic genes and nine distinct regulatory domains (Lewis, 1978; Sanchez-Herrero et al, 1985; Maeda and Karch, 2006). The highly conserved insulator factor CTCF is associated with six of the eight boundaries separating the regulatory domains (Holohan et al, 2007). dCTCF is the only one of the known Drosophila insulator factors with a conserved counterpart in vertebrates. This factor is structurally related to su(Hw) and both factors colocalise to nuclear speckles (Gerasimova et al, 2007). However, both factors have distinct binding targets (Mohan et al, 2007). In contrast to this difference in DNA binding, the su(Hw) interacting factor CP190 overlaps with dCTCF target sites (CTS) and interacts with dCTCF. In several cases, binding of dCTCF to targets requires CP190 (Mohan et al, 2007). dCTCF null mutations affect expression of Abdominal-B, cause pharate lethality and a homeotic phenotype. dCTCF as well as CP190 binding are required for the function of the Fab-8 insulator element (Moon et al, 2005; Mohan et al, 2007).
Here we found that CP190, besides showing overlapping binding to su(Hw) and to dCTCF, marks active promoters. Furthermore, dCTCF and CP190 mark and control nucleosome occupancy at the borders of histone H3K27 trimethylation domains often in the vicinity of PcG-bound sites. This points to a unique feature of CP190; the ability to regulate chromatin structure at both promoters and insulators.
Results
Genome-wide distribution of dCTCF- and CP190-binding sites
Previously identified binding sites for dCTCF have been selected based on their function as border or insulator elements (Moon et al, 2005; Mohan et al, 2007). Here we wanted to know in an unbiased, genome-wide manner whether dCTCF marks potential boundaries and whether the insulator cofactor CP190 is targeted in a similar fashion. We carried out chromatin immunoprecipitation with subsequent hybridisation to tiling arrays covering the complete non-repetitive genome of Drosophila melanogaster (Nimblegen). We used specific antibodies raised against dCTCF or CP190 for chromatin immunoprecipitation in comparison to DNA purified from input chromatin and analysed two biological replicates. Specificity of the antibodies used was verified by western blot and RNAi (Supplementary Figure 1A–C), lack of polytene chromosome staining in the null mutants p30.6 and CP1901 (Mohan et al, 2007; Supplementary Figure 1D–G and not shown) and by the sensitivity of ChIP results in the presence of RNAi against dCTCF or CP190 (see below). Biological replicates resulted in almost identical profiles (Supplementary Figure 1H).
We detected 3102 regions bound by CTCF at a false discovery rate of 5%. To validate the precipitation and array hybridisation, we analysed 10 very weak sites with chromatin precipitation and subsequent qPCR. We found that these sites were significantly precipitated as compared with control. Furthermore, S2 cells treated with dsRNA directed against dCTCF resulted in reduced signals when compared with chromatin from cells treated with RNAi against luciferase (Supplementary Figure 2B and D). On the basis of these findings, we conclude that the majority of identified CTCF-bound regions are genuine.
To analyse the genomic distribution of CTS, we determined the position of all dCTCF-bound tiles relative to gene proximal (500 bp upstream or downstream of transcription units) and intergenic positions and to gene coding, non-coding and intron positions. This distribution shows a bias towards gene proximal and non-coding regions as compared with the distribution of all sequences of the genome (Figure 1A). Nevertheless, CTS could be detected in all of these sequence classes. This distribution throughout the genome was confirmed by comparison to gene density. There is no correlation of dCTCF sites with transcriptional starts and gene density (Supplementary Figure 3), again suggesting that dCTCF sites can be found at many different locations.
Figure 1.
dCTCF- and CP190-binding sites in the genome. (A) Distribution of significantly bound tiles in comparison to genomic sequence classes. Gene proximal and non-coding tiles are significantly overrepresented for CTCF, CP190 and CTCF/CP190 (Fisher exact test: in all cases P<10−16). (B) Weblogo of the Drosophila CTCF-binding site consensus as determined by MEME motif search with the top 500 ChIP-chip regions in comparison to the human CTCF consensus (Kim et al, 2007). (C) Venn diagram of 3102 CTCF sites and 8862 CP190 sites (number of bound regions at 5% FDR). (D) Two thirds of the dCTCF sites (CTS) are bound by CP190 (heat map). Binding is shown within a 5-kb window sorted by binding strength (red) from top to bottom in each of the panels. Peak maxima of CTCF regions (left panel) and CP190 profiles sampled over the CTCF peak maxima (right panel).
To analyse whether regions bound by dCTCF are characterised by a specific sequence composition, we used the MEME algorithm with the strongest 500 enriched regions as input. The top identified motif (Figure 1B) is similar to the published binding consensus that was based on 33 sites (Holohan et al, 2007). This new optimised consensus shows a good similarity to the human CTCF consensus (Kim et al, 2007), but with a clear difference at nucleotide positions 3 and 13. Nevertheless, the core nucleotides from position 4 to 12 are identical. Such a minor difference between Drosophila and human CTCF-binding sites was expected, as 9 of 12 sequences tested for in vitro binding to human CTCF could also bind dCTCF (Moon et al, 2005). Next, we tested all of the dCTCF-enriched regions for the occurrence of this motif and found that 38% contained the consensus. Generally, we found that regions encompassing the binding consensus are bound significantly stronger and are, therefore, high affinity binding sites (Supplementary Figure 2A). Of the 1327 high affinity sites (average log2 enrichment >1.2) 70% show the consensus. Nevertheless, we identified many sites with lower log2R values lacking the consensus but with robust binding in individual ChIP/qPCR (Supplementary Figure 2D and data not shown).
A similar investigation of the genome-wide distribution of CP190 was also carried out. Analysis of the overall distribution revealed 8862 sites at a 5% false discovery rate and an enrichment for gene regions including coding and non-coding sequences (Figure 1A). In fact, we found a strong correlation between CP190-binding sites and transcriptional start sites (see below). To validate the precipitation and array hybridisation, we compared two biological replicates and found very similar profiles and analysed individual sites using two different CP190 antibodies and subsequent qPCR. We tested 20 CP190-binding sites and found those with strong log2 ratios to be both reproducibly precipitated with the two antibodies and sensitive to CP190 RNAi (Supplementary Figure 4). In contrast to CTCF, searching for a CP190-binding site consensus by the MEME algorithm did not reveal a robust consensus. To test the overlap between dCTCF and CP190, we sampled the CP190 profiles over the dCTCF peak maxima. From the 3102 CTS tested, about two thirds show CP190 binding. In addition to CP190 bound CTS, there are about 6800 CP190 sites without CTCF (Figure 1C). The colocalisation is indicated by the ‘heat map' (Figure 1D), showing a colocalisation with the peak centre of CP190 (right panel) for two thirds of the CTS. Relative genome-wide distribution of double-binding sites, CTCF plus CP190, shows an enrichment for gene proximal and non-coding sequences (Figure 1A).
CP190 colocalises with the insulator factors dCTCF, su(Hw), GAF and with cohesin and E(Z), whereas dCTCF does not overlap with su(Hw), GAF or cohesin
With the knowledge of all CP190- and dCTCF-binding sites, we wanted to know whether previously identified binding sites for other chromatin or DNA-binding factors might colocalise with CP190 or dCTCF. To address this issue, we compared the CP190 and dCTCF sites on the genome level with available data. The genome binding of su(Hw) has been determined for the three megabase Adh region (Adryan et al, 2007). When compared with dCTCF for most of the binding sites no overlap could be detected. In all, 45 dCTCF and 63 su(Hw) sites are located within the Adh region (Figure 2A and B), but only six sites overlapped. Furthermore, the dCTCF consensus (see above) is clearly different from the su(Hw) consensus (Adryan et al, 2007), suggesting that even the six overlapping sites harbour su(Hw) and dCTCF binding at different sequence elements. Thus, these functionally related factors bind to different genomic sites. In contrast, as predicted from the functional interaction of dCTCF or su(Hw) with CP190 (Pai et al, 2004; Mohan et al, 2007), CP190 largely overlaps with both dCTCF and su(Hw) (Figure 2A and B). The overlap between CP190 and su(Hw) was further stressed by the finding that approximately 50% of the top 200 su(Hw) consensus sequences on chromosome 2L showed binding of CP190, thereby validating our CP190 binding data (Supplementary Figure 5A).
Figure 2.
CP190 colocalises with dCTCF, su(Hw), GAF, E(Z) and cohesin. (A) Genome browser view of the 14.2–15.2 Mb region of chromosome 2L. Log2 ratios of dCTCF and CP190 precipitation are compared with su(Hw) (Adryan et al, 2007), the cohesin component Stromalin (Misulovin et al, 2008) and GAF (Lee et al, 2008). (B) Venn diagram of all 63 su(Hw) sites within the 3-Mb Adh region (Adryan et al, 2007) and 45 dCTCF and 130 CP190 sites in this area. (C) Venn diagram of all 3102 dCTCF sites with CP190 and GAF (Lee et al, 2008). (D) Venn diagram of all 308 E(Z)-binding sites with all dCTCF and CP190 sites (Schwartz et al, 2006).
A number of recently published reports showed a link between human CTCF and the sister chromatid cohesion complex cohesin. In all of these studies, cohesin binding was shown to coincide with CTCF binding (Parelho et al, 2008; Rubio et al, 2008; Stedman et al, 2008; Wendt et al, 2008). In contrast to vertebrates, Drosophila cohesin was shown to bind primarily to promoters and active genes. Therefore, we did not expect to find a strong overlap of dCTCF with cohesin throughout the Drosophila genome. To test this, we compared the dCTCF-binding profile with published profiles of the cohesin subunit Stromalin (Misulovin et al, 2008). Indeed, we found 75% of the dCTCF sites without cohesin binding (Figure 2A; Supplementary Figure 5B). When sampling all Stromalin peaks over dCTCF peaks in the overlapping cases, a clear separation between the two was detectable (not shown). This is clearly different from the vertebrate situation where cohesin and CTCF sites are identical, resulting in the same consensus (Parelho et al, 2008; Wendt et al, 2008). In contrast, 80% of the Stromalin sites overlapped with CP190 (Supplementary Figure 5B).
As a substantial number of sites bound by the insulator factors dCTCF or su(Hw) colocalise with CP190, we tested whether CP190 may bind to sites marked by other boundary factors. GAGA factor (GAF) has been implicated in insulator function (Ohtsuki and Levine, 1998; Belozerov et al, 2003; Melnikova et al, 2004). We compared the genome-wide distribution of GAF (Lee et al, 2008) with dCTCF and CP190 and found no striking overlap with CTCF, in contrast to CP190, where 80% of the GAF sites coincided with CP190 (Figure 2A and C; Supplementary Figure 5C).
Another possible association with dCTCF has been indicated by the previous finding that four of the dCTCF-binding sites in the bithorax locus are located next to binding sites for PcG proteins (Holohan et al, 2007). Therefore, we compared the dCTCF profile with the Enhancer of zeste (E(Z)) profile (Schwartz et al, 2006). We found that indeed about one third of the E(Z)-binding sites overlap with dCTCF binding. Again, most of the colocalised dCTCF/E(Z)-binding sites are marked by CP190 as well (Figure 2A and D).
Thus, one third of the genomic sites bound by the polycomb factor E(Z) are bound by dCTCF, whereas neither su(Hw) or GAGA insulator factors nor cohesin colocalise with dCTCF to a significant extent. In contrast, the unifying factor is CP190, which marks most of the E(Z) and cohesin sites and most of the insulator (dCTCF, su(Hw), GAF) sites.
CP190 and dCTCF mediate both gene activation and repression
Previously, it has been shown that dCTCF and CP190 are required for fly development (Butcher et al, 2004; Mohan et al, 2007). Here we wanted to identify non-developmental genes that respond to CP190 and dCTCF. To do so, we used RNAi to deplete Drosophila Schneider S2 cells of CP190 or dCTCF. With this treatment, we routinely achieved a reduction of both factors to about <10% of the basal level as measured by immunoblots (Supplementary Figure 1B and C). S2 cells treated in this way are not impaired in proliferation (Butcher et al, 2004; Mohan et al, 2007), a fact which is helpful in avoiding indirect effects caused by different proliferation rates when compared with cells targeted by control RNAi. With this material, we monitored changes in gene expression with the Fl002 (INDAC) 14 000 oligonucleotide containing array. After dCTCF depletion, we identified expression changes up to approximately four-fold. Ninety-seven genes were either activated or repressed (52 upregulated and 45 downregulated; P<0.05) by the loss of dCTCF (Figure 3A; Supplementary Table I; Supplementary Figure 6D). Reduction in dCTCF expression itself was seen, and served as a control. In all, 1237 genes are marked by CTCF binding. Of those that changed expression after CTCF depletion (97), 32 were bound by CTCF (transcribed region plus 1 kb flanking sequences; Figure 3C). The significance of this overrepresentation of CTCF-binding sites at deregulated genes was calculated to be P=2.5 × 10−11. Within the 18 genes showing the strongest effects (expression change >1.3-fold), we found 11 genes to be activated, whereas 7 genes were repressed by dCTCF reduction. As a hallmark of vertebrate CTCF is its multifunctional nature, we wondered whether these dCTCF-dependent genes show a unifying position of the CTS relative to the gene. Indeed when we combined the ChIP-chip data with the gene expression data, we found that most of the genes affected strongly by the loss of dCTCF show a CTS within the gene (Figure 3A). Either the CTS was located at the transcriptional start site or at the 3′end or at both positions. Only in two cases was no CTS found within the gene, rather the next CTS was located at +2.7 kb upstream (CG6126) or 60 kb downstream (Tango 7). This indicates that 60% of the genes affected strongly by the loss of dCTCF have a high affinity CTS within the TSS region in strong contrast to the genome-wide distribution of CTS which are in the vicinity off TSS in <5% of all genes (Supplementary Figure 6A–C and E). In addition, we analysed those genes found to be significantly deregulated (P<0.05) after RNAi but with weaker transcriptional effects (1.1-fold and higher). Correlation analysis revealed that within the total of 97 deregulated genes, in addition to the 32 genes with CTCF binding within the gene region (Figure 3C; Supplementary Figure 6E), most of the others are significantly enriched for CTCF binding within 10 kb around the respective genes in comparison to control genes (Supplementary Figure 6G). This finding suggests that an additional gene regulatory function of CTCF depends on remote CTCF sites located more distantly to the respective genes.
Figure 3.
dCTCF and CP190 binding in dCTCF/CP190-dependent genes. Heat map presentation of CTCF (A) or CP190 (B) binding (red) within a 2-kb window of the 3′end (CTCF) and the TSS (both factors) of the top 19 dCTCF-dependent genes and the top 40 CP190-dependent genes as identified in microarray analysis of S2 cells after specific RNAi treatment. Horizontal rows represent individual genes with the fold expression change (fc) after specific knock-down indicated. (C) Most of the genes responding to CP190 or CTCF knock-down harbour binding sites for either factors within the gene region (transcribed region plus 1 kb flanking sequences; for details see Supplementary Tables I and II).
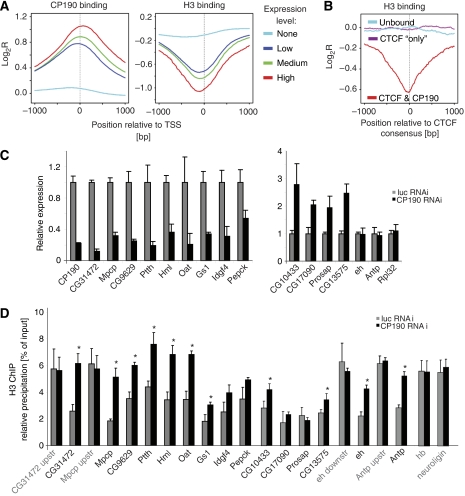
For CP190 knock-down, we also found both repressive and activating effects on gene activity. A total of 512 genes were affected with 235 genes being upregulated and 277 genes being downregulated (P<0.05; Figure 3B and C; Supplementary Figure 6F; Supplementary Table II). In total, 7380 genes are marked by CP190 binding. Of those that changed expression after CP190 depletion (512), 438 were bound by CP190. The significance of this overrepresentation of CP190-binding sites at deregulated genes was calculated to be P=3.4 × 10−37. Within the 38 genes showing the strongest expression changes (>1.4-fold), we found 19 activated and 18 (in addition to CP190) repressed genes after CP190 reduction (Figure 3B). Within both groups, 33 genes are marked by CP190 binding within a ±1 kb window around the TSS. Only for four genes (CG30269, CG30273, CG14946 and ninaD) was no CP190 binding detected within 2 kb of the TSS. When comparing both sets of genes controlled by dCTCF or by CP190, we found 25 genes to be regulated by both factors (Supplementary Tables I and II).
In summary, both CP190 and CTCF mediate gene activation and repression, a feature observed for almost all transcription factors tested in knock-down experiments. As CP190 is bound to three times as many sites as compared with dCTCF (see above), the number of genes affected by knock-down of CP190 is much larger as with dCTCF. The most striking difference between the factors is their binding location in target genes, with dCTCF found either upstream or downstream of the gene, but CP190 showing a strong preference for binding at the TSS.
CP190 marks active transcriptional start sites and shows a global anti-correlation with nucleosome occupancy
Intrigued by the finding that CP190 is bound at the TSS of genes (see above), we wanted to know whether this binding is correlated with gene activity. When clustering the genes for different expression levels in Schneider cells (Muse et al, 2007), we found a strong correlation with CP190 binding to the TSS of active genes (Figure 4A; Supplementary Figure 7). As a prominent mark for active genes is a lack of a nucleosome at the TSS and, therefore, a lack of H3 (Barski et al, 2007; Fu et al, 2008), we also sampled a published H3 data set (Larschan et al, 2007) over the same groups of genes with different expression levels (Figure 4A). Indeed, active genes with CP190 binding show a lack of H3 at the TSS in contrast to the genes with no expression (Figure 4A; Supplementary Figure 7). To confirm that H3 loss is an indication of nucleosome loss, we carried out Micrococcal nuclease digestion and tested several CP190-binding sites for nucleosome-mediated protection from nuclease digestion. In all cases tested, the H3 depletion indeed indicated nucleosome loss (Supplementary Figure 8A). Furthermore, we compared CP190 binding with the published regions of the BX-C locus devoid of nucleosomes (Mito et al, 2007) and again found CP190 to be inversely correlated with nucleosome occupancy (Supplementary Figure 8B). As we can distinguish between CP190 sites located at promoters (TSS) from sites bound by CTCF (CTS), we wondered whether the lack of H3 seen at CP190-bound promoters is also seen at CTS bound by CP190 or even at CTS in the absence of CP190. To do so, we used dCTCF consensus-containing sequences to generate three data sets: consensus bound by dCTCF and CP190, consensus bound by dCTCF only (‘CTCF only' sites) and control sites lacking CTCF and CP190. When sampling the H3 profiles over these three groups of sites, the dCTCF plus CP190-bound sites are regions lacking H3, whereas unbound sites or ‘CTCF only' sites did not reveal any differences in H3 occupancy (Figure 4B; Supplementary Figure 8D). This was confirmed by precipitation of CP190 sites with an antibody specific for histone H3 (Figure 4D).
Figure 4.
CP190 is associated with depletion of histone H3. (A) Genes were grouped by expression levels (Muse et al, 2007) into highly (red, 256 genes), medium (green, 1640 genes), low (dark blue, 1389 genes) and not expressed genes (light blue, 3673 genes) and cumulative binding profiles for CP190 and H3 (Larschan et al, 2007) relative to a 2-kb window at the TSS are shown as averaged mean enrichment ratios. (B) CP190 binds to CTS with a lack of H3. H3 profiles were sampled over a 2-kb window at the CTCF consensus for three data sets (200 sites each): CTCF plus CP190 binding (red), CTS devoid of CP190 (purple) and control CTCF consensus sites lacking both, CTCF and CP190 (blue). (C) Expression changes of individual genes after CP190-specific RNAi knock-down in Drosophila S2 cells result in upregulation (right panel) as well as downregulation (left panel). Firefly luciferase RNAi and genes Antp, eh and Rpl32 that showed no change in expression after loss of CP190 were used as controls. ChIP with anti-histone H3 antibody was analysed for promoters of downregulated genes (see above and Supplementary Table II) or for upregulated genes and for not affected genes (D). The genes hb and neuroligin are not bound by CP190 nor is their expression changed after RNAi. Error bars indicate the standard error of the mean of four independent experiments (*P<0.05 as calculated from a two-tailed t-test).
If CP190 contributes to the lack of H3 at the TSS, we would expect that after CP190 depletion, occupancy by H3 should be increased. Indeed in most cases, CP190 knock-down resulted in an increased binding of H3 (Figure 4D). This was observed for genes repressed by CP190 knock-down, but also to a lesser extent for activated genes (Figure 4C and D) and also for binding sites near genes not resulting in transcriptional change by CP190 knock-down (eh and Antp). This indicates that loss of CP190 binding leads to increased H3 binding. The resulting transcriptional changes cannot simply be attributed to H3 binding only, which should result in gene repression. Rather, the functional consequences may be combined effects involving other CP190-mediated effects resulting in gene activation upon CP190 depletion. Sites negative for CP190 binding, like the upstream or downstream controls of several genes, and the negative hb and neuroligin genes (Figure 4D) do not experience changes in H3 occupancy.
In summary, we can conclude that CP190 binds to active promoters and to most of the dCTCF target sites. In both cases, CP190 is associated with a lack of H3, whereas ‘CTCF only' sites exhibit a normal H3 occupancy.
dCTCF and CP190 mark borders of H3K27 trimethylation
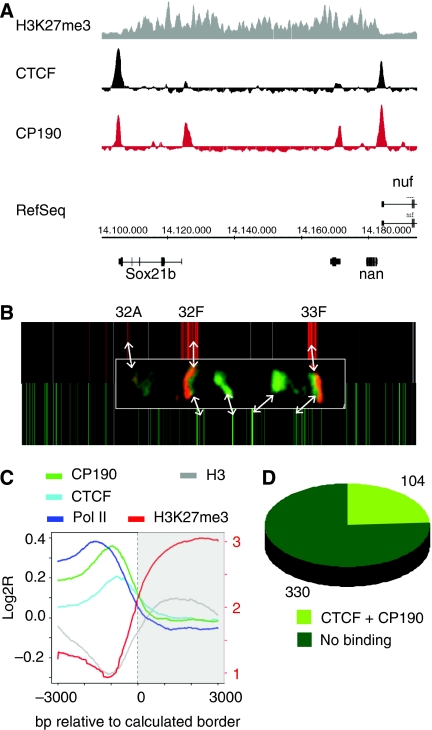
When we compared the location of dCTCF/CP190 genomic-binding sites with the distribution of known chromatin modifications, we found a striking correlation with borders of H3K27me3 domains. About 200 of these domains with the repressive chromatin mark have been identified in the Drosophila genome (Schwartz et al, 2006). One such domain is shown in Figure 5A. The borders of this H3K27me3 island are marked by both dCTCF and by CP190 binding. Previously, on the level of polytene chromosome binding, we have identified the distribution of CP190-bound sites (Mohan et al, 2007). Similarly, H3K27me3 islands have been shown on polytene chromosomes (Ringrose et al, 2004). We carried out antibody staining of polytene chromosomes and found adjacent staining for H3K27me3 and CP190 that was in agreement with our CP190 and published H3K27me3 (Schwartz et al, 2006) ChIP-chip data (Figure 5B; Supplementary Figure 10). Such an arrangement of H3K27me3 adjacent to CP190 suggests a possible border function of dCTCF/CP190. To test this hypothesis, we first searched genome wide for a similar arrangement at the molecular level. We compiled 6 kb of DNA flanking both borders of all 217 domains. Border positions were determined using the ChIPOTle algorithm and defined as position 0, from which nucleotides further outside the domain are counted with a negative sign and inside with a positive sign (Figure 5C). dCTCF and CP190 localise just outside the border at position −1000. This position is not only marked by a loss of H3K27me3 modification, but also by a dip in H3 occupancy. Within the 434 borders, 24% (104) are bound by both CP190 and dCTCF (Figure 5D). Further outside of the repressive domain, enrichment for RNA pol II binding can be observed. In total, 154 borders are marked by divergently transcribed genes pointing away from the H3K27me3 domains, of those 57 are marked by CTCF/CP190. Those divergently transcribed genes are generally active as compared with genes within the domains or those genes being transcribed towards the methylated domains. Within the top 11 CTCF RNAi downregulated genes, we find four genes (CG3358, Trx-2, CG10359 and CG13689) that are located just at the border position of H3K27me3 domains and are marked by strong CTCF binding. The H3 depletion at these sites reminded us of the situation of CP190 binding at TSS (see above). We, therefore, wanted to know whether the absence of H3 and H3K27me3 at repressive domain borders depends on dCTCF and CP190. To do so, we used homozygous third instar larvae of the dCTCF deficiency mutant p30.6 (Mohan et al, 2007) and of the CP190 loss of function mutant Cp1901 (Butcher et al, 2004). We tested four of the identified borders, which are bound by dCTCF as well as by CP190 for any change in factor occupancy and for the presence of H3 and H3K27me3. These domain borders are located close to the genes CG4389, CG1354, CG13689 and sbr (Figure 6). In all four cases, dCTCF and CP190 are bound to the wild-type chromatin at the CTS. The dCTCF deficiency (p30.6) shows no CTCF binding and no or reduced binding of CP190 and the strong hypomorph Cp1901 (Butcher et al, 2004) shows a strong reduction in CP190 binding and a weak reduction in dCTCF (Figure 6, CG4389 and sbr). Such interdependency between these two factors has already been seen at the level of polytene chromosome staining (Mohan et al, 2007). In all cases, the inside and outside sites flanking the border at +3 kb and −3 kb are negative for both factors. When precipitating H3 or the modified histone H3K27me3, the wild-type larvae show the expected pattern of a reduced amount of H3 at the CTS and, therefore, a reduced amount of H3K27me3. H3 reduction is not seen inside or outside, in contrast to H3K27me3 that is found to be enriched inside of the domain. The dCTCF mutant shows a two- to three-fold increase of H3 and of H3K27me3 at the CTS when compared with wild type, whereas all of the CTS flanking regions are not significantly changed in H3 precipitation. Similarly, the Cp1901 strain displayed an H3 and an H3K27me3 increase at most of the CTS (Figure 6). In three cases (CG4389, CG1354 and CG13689), the dCTCF mutant showed an increase of H3K27me3 outside of the domain, suggesting that the wild-type CTS prevents spreading of the H3K27me3 mark.
Figure 5.
dCTCF and CP190 mark borders of H3K27 trimethylation. (A) Genome browser view of ChIP-chip results for H3K27me3 (Schwartz et al, 2006) compared with dCTCF and CP190 precipitation in the 14.1–14.2 Mb region of chromosome 3L. (B) Alignment of CP190 (green) and H3K27me3 (red) ChIP-chip data (heat map) with polytene antibody staining (insert) for CP190 (green) and H3K27me3 (red) within the cytological region 32A to 33F of chromosome 2L. (C) H3 is underrepresented at CP190- and dCTCF-binding sites at domain borders. Compiled log2 ratios of ChIP results at the 434 borders of H3K27me3-enriched domains (217 in total) positioned to the right (grey). Graphs of the ChIP results for CP190, dCTCF (this study), H3 (Larschan et al, 2007), Pol II (Misulovin et al, 2008) and H3K27me3 (Schwartz et al, 2006). (D) Diagram of all borders of H3K27me3 domains, indicating that 24% are double occupied by both CP190 and dCTCF.
Figure 6.
CTCF and CP190 control chromatin structure at borders of H3K27me3 domains. Genome browser view (top) of H3K27me3 domain borders (Schwartz et al, 2006) in the vicinity of the genes CG4389, CG1354 CG13689 and sbr with the amplicons (filled boxes) ‘inside' (within shaded domain), at the CTS and ‘outside' of the H3K27me3 domain. These amplicons were used to detect binding of dCTCF, CP190, H3 and H3K27me3 in wild type (w−), dCTCF deficient (p30.6) and CP190 loss of function mutant third instar larvae (Cp1901). Error bars indicate the standard error of the mean of at least three independent experiments (a statistical confidence of P<0.05 for changes in H3 and H3K27me3 precipitation is indicated by asterisks (calculated from a two-tailed t-test)). To compare H3 and H3K27me3 between the different strains, relative precipitation was calculated relative to four invariant sites >10 kb separated from any CTCF- or CP190-binding site.
To distinguish between dCTCF and CP190 effects, which on combined binding sites influence each other (Figure 6), we also analysed dCTCF sites not bound by CP190. These sites do not show a depletion of H3, and the H3 content is not influenced by either mutant (Supplementary Figure 9).
Therefore, we conclude that dCTCF/CP190 double sites mark many borders of H3K27me3 domains and that CP190 causes the loss of H3 and, therefore, the absence of H3K27me3 at these particular sites.
Discussion
Genome-wide distribution of dCTCF- and CP190-binding sites
Using the ChIP-chip technique, we identified 3102 dCTCF-binding sites (CTS). As compared with human CTCF with about 27 000 sites (Jothi et al, 2008) and a 10 times larger genome, this number is in accordance with the genome size of Drosophila. Previous analysis of dCTCF binding to polytene chromosomes revealed about 300–400 bound CTS (Mohan et al, 2007). This difference very likely does not reflect a tissue specificity, because as tested for the bithorax locus (Holohan et al, 2007; Mohan et al, 2007) and other individual CTS (not shown), the overall dCTCF binding in different tissues is very similar or even identical. It is likely that the sensitivity of the cytological method is not comparable to the ChIP-chip technique. A similar difference with these two techniques has also been observed for su(Hw) (Adryan et al, 2007). In fact, when comparing polytene chromosomes with the ChIP-chip data side by side, as exemplified for CP190 in Figure 5B and Supplementary Figure 10, the difference in resolution becomes evident. The genome-wide distribution of the CTS reveals an approximately two-fold preference for promoter proximal sites in addition to many intergenic, coding, non-coding and intronic binding positions. This may reflect a multifunctional property of dCTCF (see below), which has also been found for vertebrate CTCF (Ohlsson et al, 2001). A search for a dCTCF consensus sequence revealed a nucleotide sequence similar to the vertebrate consensus. The only difference is seen at positions 3 and 13 resulting in two CpG sites in the Drosophila sequence. For mammalian CTCF, a subset of binding sites harbour an important CpG sequence at position 3, which upon methylation interferes with CTCF binding (Ohlsson et al, 2001). As the majority of the Drosophila CpG sequences are not methylated, there is no evolutionary pressure to deplete the Drosophila CTCF consensus from CpGs.
Genome-wide binding analysis of vertebrate cohesin resulted in the identification of a DNA consensus, which turned out to be identical to vertebrate CTCF (Parelho et al, 2008; Rubio et al, 2008; Stedman et al, 2008; Wendt et al, 2008). Because of the preferential detection of Drosophila cohesin at promoters of active genes (Misulovin et al, 2008), we were not surprised to see that dCTCF only partially overlaps with cohesin-bound genomic sites (Supplementary Figure 5B). Close inspection of the overlapping sites revealed that dCTCF and cohesin bind to separate DNA sequences. In vertebrates, cohesin apparently mediates the sister chromatid cohesion function independently of CTCF, whereas the insulator function of CTCF requires cohesin. It is possible that in Drosophila only the CTCF-independent sister chromatid cohesion function is used. A non-overlap is also found for two other insulator factors, su(Hw) and GAF, which only partially overlap with dCTCF without exact colocalisation (Figure 2B and C).
Faced with the lack of a substantial overlap of different insulator factors, we wondered whether the cofactor for dCTCF and su(Hw), CP190, would bind to other genomic sites besides insulators. The surprising result was that CP190 binds to most active promoters, in addition to most of the dCTCF, su(Hw) and GAF sites (Figures 2B, C and 4A; Supplementary Figure 5B). Such a sharing of factors between insulator factors and promoter elements has not been seen before. RNA pol II has been found at a subset of vertebrate CTCF sites (Chernukhin et al, 2007), but genome-wide mapping of Pol II did not reveal a colocalisation with CTCF or with insulators (Barski et al, 2007). Similarly, we could not detect a Pol II colocalisation with insulators in Drosophila (not shown).
dCTCF and CP190 mediate both gene activation and repression
To test the consequences of reduction of dCTCF or of CP190 levels for general cell function, we used a genome-wide microarray analysis after knock-down in Schneider cells. About 30% of the dCTCF-dependent genes harbour a CTS within the gene, in most cases in the promoter region (Figure 3A and C). This is similar to the insulator factor su(Hw), for which 20% of the genes affected by su(Hw) reduction are associated with a su(Hw)-binding site within 1 kb of the transcription unit (Adryan et al, 2007). This may suggest that in addition to the insulator function of CTCF, a promoter function controls gene repression as well as activation. Such an ambivalent effect on gene expression has been reported for su(Hw) (Adryan et al, 2007), and a similar situation is seen here with CP190. It is not surprising that the number of genes affected by CP190 knock-down is much higher as compared with the effect after depletion of dCTCF, as the cofactor function for both insulator proteins, CTCF and su(Hw), is impaired in addition to the promoter binding sites of CP190 identified here.
Although vertebrate CTCF localisation to intergenic regions suggests primarily an insulator function, both repressive and activating effects on gene activity have been reported as well (Wallace and Felsenfeld, 2007). Notably, 20% of all human CTCF sites were found to map to promoters (Kim et al, 2007).
CP190 is associated with H3 depletion and together with dCTCF marks boundaries of H3K27 trimethylation
On the basis of our finding that CP190 binds to active promoters, which are characterised by a lack of a nucleosome at the TSS, we have analysed both classes of CP190-binding sites (TSS and CTS) for depletion of H3. For both classes, H3 depletion was shown to be dependent on the presence of CP190. In contrast, CTS lacking CP190 binding were not depleted of H3 (Supplementary Figure 9). This suggests that CP190 can at least contribute to this H3 deficiency. Indeed, for both TSS and CTS, we could show that CP190 knock-down in Schneider cells, and in the case of CP190 deficient third instar larvae as well, an increase of H3 precipitation is detectable (Figures 4D and 6). CP190 is certainly not the only cause for the lack of H3 at these sites, as a more dramatic effect on gene expression by CP190 depletion would have been expected. Several mechanisms for H3 and, therefore, nucleosome depletion could be envisaged. These might include direct competitive effects of CP190 binding and nucleosome formation or indirect effects of CP190 recruiting nucleosome remodelling complexes or histone modification enzymes, which in turn may initiate remodelling.
Both CP190 and dCTCF are detected at borders of H3K27 trimethylation regions (Figure 5). This repressive chromatin mark has been found in outspread genomic domains in flies and vertebrates (Schwartz et al, 2006; Regha et al, 2007). In vertebrates, these are flanked by CTCF in several cases (Barski et al, 2007). In Drosophila, functional tests of binding sites for the su(Hw) insulator (Kahn et al, 2006) factor suggested that insulators interfere with spreading of the repressive H3K27me3 mark. This is supported by our finding that the CTS at boundaries of H3K27me3 domains bound by CP190 are depleted of H3 and that loss of CP190 or dCTCF causes an increased detection of histone H3 and H3K27me3 at these sites (Figure 6). Sites of nucleosome depletion at boundaries of cis-regulatory domains have been documented for the BX-C (Mito et al, 2007). These may now be explained by CP190, when considering our previous results that six of these boundaries are bound by dCTCF and by CP190 (Mohan et al, 2007). Indeed, we found that CP190 sites in the BX-C are marked by reduced nucleosome occupancy (Supplementary Figure 8B). Similarly, vertebrate CTCF binding was shown to coincide with nucleosome depletion in the context of TSS (Boyle et al, 2008). Here we show that CTCF sites, independent of their genomic context, are marked by depletion of H3 as long as CP190 is present (Figure 6; Supplementary Figure 9). Therefore, our data suggest that CP190 determines a nucleosome depleted chromatin conformation rather than CTCF. In addition to the binding of CTCF/CP190 and depletion of H3, another feature of the H3K27me3 borders is the presence of PolII binding (Figure 5C). In fact, about two thirds of the borders are marked by actively transcribed genes oriented away from the methylated domain. Only for some of the borders the simultaneous presence of CTCF and an active promoter can be found. This is similar for the borders of lamin-associated domains in vertebrates, which are characterised by gene repression and H3K27me3. For these borders, it has been shown that, besides the presence of CpG islands, either CTCF binding or actively transcribed genes are significant features (Guelen et al, 2008).
Similarity of insulators and promoters
Why should insulators and promoters share similar features? Two roles and mechanisms can be envisaged. One has been proposed previously (Geyer, 1997; Ohlsson et al, 2001), arguing that insulators function as promoters, which decoy enhancers and thereby interfere with the regular enhancer/promoter interaction. In fact, the promoters of tRNA genes (Noma et al, 2006) and SINE repeats (Lunyak et al, 2007) function as insulators.
Here we suggest another role and function for the similarity of promoters and insulators. In recruiting a similar subset of proteins, both types of sequence elements are poised for interaction. One could imagine that such an interaction is required for promoters controlled by insulators and enhancer blockers. This view is supported by the finding that CP190 is found in insulator bodies (Pai et al, 2004), suggesting a close distance for all elements bound by CP190. Furthermore, insulator–promoter interaction has been documented for the H19 ICR insulator (Yoon et al, 2007). Future experiments focusing on CP190 mediated long-range chromatin interaction will help to determine the role of CP190 at insulators and promoters.
Materials and methods
Fly strains
Fly crosses were maintained at 25°C on standard medium. For preparation of chromatin extracts, we used either w− animals as control, a CTCF null allele dCTCFp30.6 (Mohan et al, 2007) and a Cp1901 loss of function allele provided by WGF Whitfield and JW Raff (Butcher et al, 2004). Stocks were kept balanced over a TM6, Tb chromosome. Third instar larvae lacking the Tb marker were selected for preparation of chromatin extracts.
Antibodies
For CTCF-ChIP technique, we used the published antiserum directed against the N-terminal region of CTCF (Mohan et al, 2007). For CP190 detection and ChIP, we used either a monoclonal antibody (bx63) (Frasch et al, 1986), which was used for ChIP-chip analysis or a polyclonal serum as control (rb188), kindly provided by John Crang and David Glover (Cambridge).
Histone H3 and specific histone modification H3K27me3 were precipitated with 1 μg of Abcam antibodies Ab-1791 and Ab-6002, respectively. For control of H3K27me3 precipitation, we used additionally a polyclonal serum kindly provided by Thomas Jenuwein but found no differences.
ChIP-on-chip
Drosophila S2 cells were fixed with 1% formaldehyde and extracts were precipitated with antibodies specific to CTCF and CP190 essentially as described (Mohan et al, 2007). Chromatin shearing was adjusted to result in fragments with an average size ranging from 300–1000 bp. Immunoprecipitated DNA and input-DNA where amplified through whole genome amplification kit (Sigma GenomePlex Kit WGA2, SIGMA) following the protocol provided by the Farnham laboratory (http://www.genomecenter.ucdavis.edu/farnham/protocol.html). Two biological replicates per IP/input pair were prepared and labelled with Cy3/Cy5 or with swapped dyes and hybridised to 3 NimbleGen arrays representing the whole non-repetitive genome of Drosophila melanogaster (labelling, hybridisation and scanning performed through IMAGENES NimbleGen service). Data processing was essentially done as described (Straub et al, 2008). Data were normalised, and significantly enriched regions and respective false discovery rate were calculated following the HMM algorithm of the TileMap procedure (Ji and Wong, 2005). Probes were considered to be bound significantly if the posterior probability of the HMM was greater than 0.5. Subsequently, all coordinates were re-calculated according to the dm3 release of Drosophila melanogaster genome using the liftOver tool from the UCSC homepage (http://genome.ucsc.edu/). Profile smoothing was performed with the pseudomedian (Royce et al, 2007) script with a 375-bp sliding window. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE12765.
RNAi, reverse transcription and transcriptome analysis
S2 cell culture, dsRNA production and transfection were essentially done as described previously (Mohan et al, 2007). S2 cells were transfected with 2 μg dsRNA per ml of medium (Gibco Drosophila Schneider medium supplemented with 10% FCS) corresponding to the mRNA sequence of CTCF, CP190 or the firefly luciferase gene. After 6 days of transfection, cells were extracted with TRIZOL (Invitrogen) or processed for protein extraction as described previously (Mohan et al, 2007). Experiments were conducted with four replicates for both experimental conditions (luciferase RNAi versus specific RNAi). RNA was processed for hybridisation to FL002 oligonucleotide arrays (GEO: GPL5016) at FlyChip (http://www.flychip.org.uk). Information on spot finding, normalisation and averaging of biological replicates can be found at the FlyChip homepage (http://www.flychip.org.uk/services/analysis/) and in the corresponding GEO entry GSE12765. To control for off-target effects of the respective dsRNAs, we checked the sequences on the dsCheck (http://dscheck.rnai.jp/) homepage but did not find genes that we found deregulated in the results of the trancriptome analysis. For individual tests in RT–qPCR, we used alternative dsRNAs corresponding to non-overlapping sequences.
To test individual transcripts, extracted RNA samples were treated with 1 unit DNase I (Invitrogen) for 2 h at 37°C. Reverse transcription was done with 1 μg of purified RNA (GeneAmp reverse transcription kit, Roche). Typically 1/50 of the reaction was used for qPCR analysis. All analyses were carried out with a minimum of three independent experiments.
Supplementary Material
Supplementary Figures 1–10
Supplementary Materials and Methods
Supplementary Table I
Supplementary Table II
Acknowledgments
We thank Drs John Crang, David Glover and Thomas Jenuwein for antibodies and Helmut Dotzlaw for careful reading of the manuscript. Additionally, we thank Leni Schäfer-Pfeiffer for technical assistance. This work was supported by grants from the DFG (FOR 531 to RR) and by the Leibniz Programme of the DFG (PBB).
References
- Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R (2007) Genomic mapping of suppressor of Hairy-wing binding sites in Drosophila. Genome Biol 8: R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TK, Grote K, Boeing S, Stelzer G, Schepers A, Meisterernst M (2007) Global distribution of negative cofactor 2 subunit-alpha on human promoters. Proc Natl Acad Sci USA 104: 10000–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera L, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green R, Ren B (2008) Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res 18: 46–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R (2007) Comparing active and repressed expression states of genes controlled by the polycomb/trithorax group proteins. Proc Natl Acad Sci USA 104: 16615–16620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P, Cai HN (2003) A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22: 3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RD, Chodagam S, Basto R, Wakefield JG, Henderson DS, Raff JW, Whitfield WG (2004) The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J Cell Sci 117: 1191–1199 [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20: 105–116 [DOI] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E (2007) CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27: 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Glover DM, Saumweber H (1986) Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J Cell Sci 82: 155–172 [DOI] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z (2008) The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet 4: e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T, Lei E, Bushey A, Corces V (2007) Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Geyer PK (1997) The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev 7: 242–248 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Hawkins R, Ren B (2006) Genome-wide location analysis: insights on transcriptional regulation. Hum Mol Genet 15 (Spec No 1): R1–R7 [DOI] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R (2007) CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wong WH (2005) TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics 21: 3629–3636 [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K (2008) Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 36: 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V (2006) Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem 281: 29064–29075 [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching K, Loukinov DI, Green R, Zhang MQ, Lobanenkov VV, Ren B (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera L, Zheng M, Qu C, Singer M, Richmond T, Wu Y, Green R, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI (2007) MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28: 121–133 [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS (2008) NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lei EP, Corces VG (2006) RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941 [DOI] [PubMed] [Google Scholar]
- Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, Yung Y, Montoliu L, Glass CK, Rosenfeld MG (2007) Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317: 248–251 [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F (2006) The ABC of the BX-C: the bithorax complex explained. Development 133: 1413–1422 [DOI] [PubMed] [Google Scholar]
- Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P (2004) Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci USA 101: 14806–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D (2008) Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097 [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2007) Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411 [DOI] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, Renkawitz R (2007) The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26: 4203–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V (2005) CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K (2007) RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G (2006) Chromosomal distribution of PcG proteins during Drosophila development. Plos Biol 4: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI (2006) A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872 [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M (1998) GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev 12: 3325–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG (2004) The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson H, Jarmuz A, Canzonetta C, Webster Z, Nesterova T (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK (2006) Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol 26: 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E, Ghosh D, Baxter E, Corces VG (2006) Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172: 2337–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, Jenuwein T, Barlow DP (2007) Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell 27: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R (2004) Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell 16: 641–653 [DOI] [PubMed] [Google Scholar]
- Royce TE, Carriero NJ, Gerstein MB (2007) An efficient pseudomedian filter for tiling microrrays. BMC Bioinformatics 8: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche C, Filippova G, Baliga NS, Aebersold R, Ranish JA, Krumm A (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G (1985) Genetic organization of Drosophila bithorax complex. Nature 313: 108–113 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA (2007) Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 8: 424–436 [DOI] [PubMed] [Google Scholar]
- Schwartz Y, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Spana C, Harrison DA, Corces VG (1988) The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev 2: 1414–1423 [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil J, Bartolomei M, Lieberman P (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB (2008) The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4: e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M (2006) Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G (2007) We gather together: insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters J (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K (2007) Analysis of the H19ICR insulator. Mol Cell Biol 27: 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–10
Supplementary Materials and Methods
Supplementary Table I
Supplementary Table II