Abstract
Lipocalin 24p3 is a secreted protein that can induce apoptosis in cells containing the 24p3 cell surface receptor, 24p3R. The oncoprotein BCR–ABL activates 24p3 and represses 24p3R expression. Thus, BCR–ABL+ cells synthesise and secrete 24p3, which induces apoptosis in normal 24p3R-containing cells but not in BCR–ABL+ cells. The cell signalling and transcription factor pathways by which BCR–ABL misregulates expression of 24p3 and 24p3R remain to be elucidated. Here we show that BCR–ABL upregulates 24p3 expression through activation of the JAK/STAT pathway, which culminates in binding of Stat5 to the 24p3 promoter. We find that 24p3R expression is regulated by Runx transcription factors, and that BCR–ABL induces a switch in binding from Runx3, an activator of 24p3R expression, to Runx1, a repressor of 24p3R expression, through a Ras signalling pathway. Finally, we show that repression of 24p3R by BCR–ABL is a critical feature of the mechanism by which imatinib kills BCR–ABL+ cells. Our results reveal diverse signalling/transcription pathways that regulate 24p3 and 24p3R expression in response to BCR–ABL and are directly relevant to the treatment of BCR–ABL+ disease.
Keywords: BCR–ABL, imatinib mesylate, Ras, 24p3, 24p3R
Introduction
Many hematopoietic cell types undergo programmed cell death (apoptosis) when deprived of specific trophic factors. For example, murine pro-B lymphocytic FL5.12 and myeloid 32D cells undergo apoptosis after withdrawal of the cytokine interleukin-3 (IL-3). We have previously identified a pro-apoptotic pathway that has an important function in hematopoietic cell apoptosis after cytokine withdrawal (Devireddy et al, 2001, 2005). The two major components of this pathway are the secreted protein lipocalin 24p3 (also called lipocalin 2 (Lcn2)) and its cell surface receptor, 24p3R (also called Slc22a17). Expression of 24p3 is normally repressed in the presence of IL-3 but is markedly induced after IL-3 withdrawal (Devireddy et al, 2001). Conditioned medium from IL-3-deprived FL5.12 and 32D cells contains 24p3 and induces apoptosis of naïve cells as well as a variety of hematopoietic cell types (Devireddy et al, 2001). On the basis of these results, we have proposed that IL-3 withdrawal results in 24p3 transcription, leading to synthesis and secretion of 24p3, which induces apoptosis through an autocrine/paracrine pathway.
Interestingly, expression of the oncoprotein BCR–ABL can confer resistance to apoptosis in cytokine-deprived, IL-3-dependent mouse cell lines (reviewed in Cross and Reiter, 2002; Skorski, 2002). BCR–ABL is a fusion protein created by a chromosomal translocation event that is frequently observed in a family of leukemias that includes chronic myelogenous leukemia (CML) (reviewed in Melo and Deininger, 2004). The translocation event creates a constitutively active ABL tyrosine kinase that stimulates several signalling pathways, the net effect of which is to promote cell survival and proliferation.
We have previously shown that BCR–ABL can counteract the 24p3/24p3R pro-apoptotic pathway, thus rendering BCR–ABL+ cells refractory to 24p3-mediated apoptosis (Devireddy et al, 2005). BCR–ABL inhibits the 24p3/24p3R pro-apoptotic pathway by profoundly misregulating the expression of both 24p3 and 24p3R (Devireddy et al, 2005). 24p3 is dramatically upregulated in BCR–ABL+ murine cell lines even in the presence of IL-3 as well as in BCR–ABL-transformed murine primary cells (Devireddy et al, 2005; Lin et al, 2005). Moreover, the human homolog of 24p3, LCN2 (also known as NGAL), is upregulated in BCR–ABL+ leukemic blasts isolated from CML patients (Kaneta et al, 2003; Devireddy et al, 2005). Conversely, 24p3R, which is normally expressed in the presence or absence of IL-3, is downregulated in BCR–ABL+ murine cell lines, and its human homolog, SLC22A17 (also called NGALR), is downregulated in CML leukemic blasts (Devireddy et al, 2005).
These results indicate that BCR–ABL+ cells synthesise and secrete 24p3, which induces apoptosis in normal 24p3R-containing cells but not in BCR–ABL+ cells. The constitutive expression and secretion of 24p3 by BCR–ABL+ cells is likely relevant to the pathophysiology of CML. Previous studies have suggested that BCR–ABL+ cells secrete a factor(s) that interferes with normal hematopoiesis (Lin et al, 2001). Experiments in mouse models of CML indicate that 24p3 has the expected properties of such a secreted factor (Lin et al, 2005; Leng et al, 2008).
BCR–ABL-induced misregulation of 24p3 and 24p3R can be reversed by addition of imatinib mesylate (also called Gleevec or STI571) (Devireddy et al, 2005), a specific small-molecule inhibitor of the ABL kinase (Buchdunger et al, 1996). Imatinib is used to treat CML and functions by inducing apoptosis selectively in BCR–ABL+ cells. Notably, ectopic expression of 24p3R can also induce apoptosis in BCR–ABL+ cell lines (Devireddy et al, 2005), suggesting that derepression of 24p3R after imatinib treatment of BCR–ABL+ cells may contribute to imatinib-induced apoptosis.
The cell signalling and transcription factor pathways by which BCR–ABL regulates expression of 24p3 and 24p3R remain to be determined. Here we study how BCR–ABL activates transcription of 24p3 while repressing transcription of 24p3R. Our results show that BCR–ABL regulates expression of 24p3 and 24p3R through distinct signalling and transcription factor pathways, and demonstrate an essential role for the 24p3/24p3R pathway in imatinib-induced apoptosis.
Results
Activation of 24p3 expression by BCR–ABL occurs through binding of activated Stat5 to the 24p3 promoter
We have previously shown that IL-3-dependent murine myeloid 32D cells contain a functional 24p3/24p3R pathway (Devireddy et al, 2001, 2005) that can be blocked by expression of the BCR–ABL oncoprotein (Devireddy et al, 2005). The experiments presented below were carried out in 32D cells stably transformed with BCR–ABL (32D/BCR–ABL cells) or, as a control, parental 32D cells.
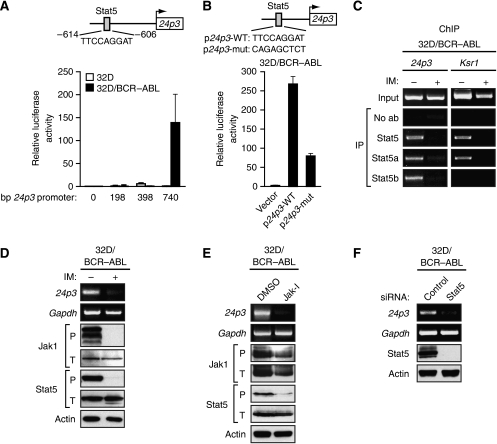
BCR–ABL is known to activate the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signalling pathway (reviewed in Danial and Rothman, 2000), and analysis of the 24p3 promoter revealed that it contains a consensus binding sequence for Stat5 at –606 to –614 bp upstream of the transcription start site (Figure 1A, top panel). To test the functional relevance of the putative Stat5 binding site, we first prepared a series of 24p3 promoter deletion derivatives and analysed their ability to drive a heterologous luciferase promoter after transient transfection in 32D and 32D/BCR–ABL cells. Figure 1A shows that a 24p3 promoter derivative containing 740 bp upstream of the transcription start site was active in 32D/BCR–ABL cells but not in 32D cells, consistent with the expression pattern of the endogenous 24p3 gene (Devireddy et al, 2005). Removal of the 24p3 promoter region harbouring the putative Stat5 binding site resulted in loss of transcriptional activity in 32D/BCR–ABL cells. Moreover, mutational analysis of the putative Stat5 binding site confirmed that it was required for high levels of 24p3 transcription in 32D/BCR–ABL cells (Figure 1B).
Figure 1.
Activation of 24p3 expression by BCR–ABL occurs through the JAK/STAT pathway. (A) (Top) Schematic diagram of the 24p3 promoter showing the location and sequence of the Stat5 binding site at −606 to −614 bp upstream of the transcription start site (indicated by the arrow). (Bottom) Promoter deletion analysis. 24p3 promoter fragments of the indicated size were tested for their ability to drive a heterologous firefly luciferase gene after transient transfection, together with an SV40-driven Renilla luciferase reporter, into 32D and 32D/BCR–ABL cells. Shown are firefly luciferase activities normalised to that of Renilla luciferase. Error bars indicate s.d. (B) A 740 bp 24p3 promoter fragment derivative in which the putative Stat5 binding site was mutated as shown (WT, wild type; mut, mutant) was tested for its ability to drive luciferase expression in 32D/BCR–ABL cells. (C) ChIP analysis monitoring binding of Stat5, Stat5a and Stat5b to the 24p3 promoter or, as a control, the Ksr1 promoter. 32D/BCR–ABL cells were treated in the presence or absence of imatinib (IM), as indicated. (D) 32D/BCR–ABL cells, treated in the presence or absence of imatinib, were monitored for expression of 24p3 by RT–PCR or for expression of phosphorylated (P) or total (T) Jak1 and Stat5 by immunoblot analysis. Gapdh and Actin were monitored as loading controls. (E) 32D/BCR–ABL cells were treated with Jak inhibitor I (Jak-I) or, as a control, DMSO, and monitored for expression of 24p3 by RT–PCR or Jak1 and Stat5 by immunoblot analysis. The concentration of Jak inhibitor I used (10 μM) was based on a titration experiment (Supplementary Figure S6A) and represents the minimal concentration required to obtain greater than 75% inhibition of Stat5 phosphorylation. (F) 32D/BCR–ABL cells were treated with a control (luciferase) or Stat5 siRNA and monitored for 24p3 expression by RT–PCR or Stat5 expression by immunoblot analysis.
To determine whether Stat5 bound to the 24p3 promoter in vivo, we carried out chromatin immunoprecipitation (ChIP) experiments. As a control, we analysed in parallel the promoter from a known Stat5 target gene, kinase suppressor of ras 1 (Ksr1) (Nelson et al, 2004). The results of Figure 1C show that Stat5 bound to the 24p3 promoter and that this interaction did not occur after imatinib treatment. Two Stat5 isoforms exist, Stat5a and Stat5b, which can play distinct roles on specific genes (e.g., Park et al, 1999). ChIP experiments revealed that both Stat5a and Stat5b were bound to the 24p3 promoter. By contrast, Stat5a but not Stat5b was bound to the control Ksr1 promoter.
We next tested whether BCR–ABL-mediated activation of the JAK/STAT pathway in 32D cells was required for Stat5 binding to the 24p3 promoter and 24p3 transcription. Figure 1D and Supplementary Figure S1 show that in 32D/BCR–ABL cells there were high levels of phosphorylated forms of Jak1 and Stat5 (phospho-Jak1 and phospho-Stat5, respectively), indicative of JAK/STAT pathway activation. Activated Jak2 and Jak3 were also detectable in 32D/BCR–ABL cells (Supplementary Figure S2). The levels of phospho-Jak1, -Jak2, -Jak3 and -Stat5 were substantially reduced after imatinib treatment (Figure 1D; Supplementary Figure S2), indicating that BCR–ABL was responsible for activation of the JAK/STAT pathway. As we reported previously (Devireddy et al, 2005), imatinib treatment of 32D/BCR–ABL cells also resulted in the loss of 24p3 expression (Figure 1D). In addition, after treatment of 32D/BCR–ABL cells with a JAK/STAT pathway inhibitor, Jak inhibitor I (Thompson et al, 2002), the levels of phospho-Jak1 and phospho-Stat5 were also decreased, accompanied by loss of 24p3 expression (Figure 1E). Finally, the RNA interference (RNAi) experiment of Figure 1F shows that siRNA-mediated depletion of Stat5 also resulted in loss of 24p3 expression. Collectively, these results indicate that BCR–ABL stimulates the JAK/STAT pathway, leading to activation of Stat5, which then binds to the 24p3 promoter resulting in transcription activation.
Repression of 24p3R expression by BCR–ABL occurs through a Runx protein binding switch
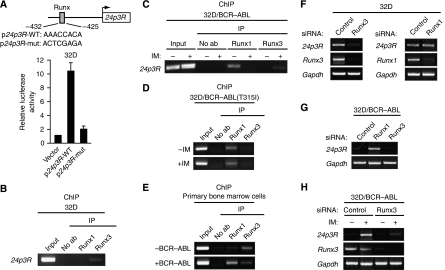
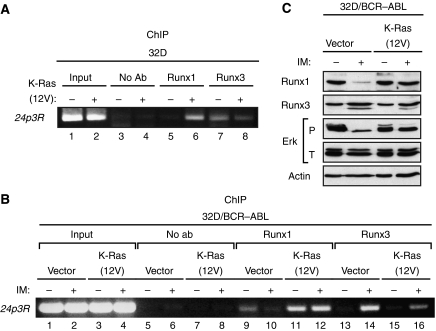
As described earlier, BCR–ABL represses 24p3R expression. Analysis of the 24p3R promoter revealed the presence of a putative Runx binding site at –425 to –432 bp upstream of the transcription start site (Figure 2A, top panel). Mutational analysis confirmed that the Runx binding site was required for 24p3R transcriptional activity in 32D cells (Figure 2A, bottom panel). Of the three Runx family members (Runx1, Runx2 and Runx3), only Runx1 and Runx3 are expressed in cells of hematopoietic origin (reviewed in Levanon and Groner, 2004). As an initial step to determine whether Runx proteins have a role in BCR–ABL-mediated regulation of 24p3R expression, we performed ChIP analysis for Runx1 and Runx3. The results of Figure 2B show that in 32D cells, in which 24p3R is transcriptionally active, binding of Runx3 but not Runx1 could be detected at the 24p3R promoter. By contrast, in 32D/BCR–ABL cells, in which 24p3R is transcriptionally inactive, binding of Runx1 but not Runx3 was detected at the 24p3R promoter (Figure 2C). Moreover, after treatment of 32D/BCR–ABL cells with imatinib, binding of Runx1 to the 24p3R promoter decreased, which was accompanied by increased Runx3 binding. To rule out the possibility that the effects we observed on imatinib treatment resulted from inhibition of protein kinases other than BCR–ABL, we monitored binding of Runx1 and Runx3 to the 24p3R promoter in 32D cells expressing the imatinib-resistant BCR–ABL(T315I) mutant. Figure 2D shows that imatinib failed to alter the binding pattern of Runx1 and Runx3 in 32D/BCR–ABL(T315I) cells. Finally, we found that the switch from Runx3 to Runx1 at the 24p3R promoter also occurred in BCR–ABL-transformed primary bone marrow cells (Figure 2E). Collectively, these results reveal that BCR–ABL induces a Runx3 to Runx1 binding switch on the 24p3R promoter.
Figure 2.
Repression of 24p3R expression by BCR–ABL occurs through a Runx protein binding switch. (A) (Top) Schematic diagram of the 24p3R promoter showing the location of the Runx binding site at −425 to −432 bp upstream of the transcription start site (indicated by the arrow). (Bottom) An ∼2 kb 24p3R promoter fragment in which the putative Runx binding site was mutated was tested for its ability to drive luciferase expression in 32D cells. (B) ChIP analysis monitoring binding of Runx1 and Runx3 to the 24p3R promoter in 32D cells. (C) ChIP analysis monitoring binding of Runx1 and Runx3 to the 24p3R promoter in 32D/BCR–ABL cells in the presence or absence of imatinib. (D) ChIP analysis monitoring binding of Runx1 and Runx3 to the 24p3R promoter in 32D/BCR–ABL(T315I) cells in the presence or absence of imatinib. (E) ChIP analysis monitoring binding of Runx1 and Runx3 to the 24p3R promoter in primary bone marrow cells transduced with BCR–ABL. (F) 32D cells were treated with a control, Runx3 or Runx1 siRNA and monitored for 24p3R, Runx3 and Runx1 expression by RT–PCR. (G) 32D/BCR–ABL cells were treated with a control, Runx3 or Runx1 siRNA and monitored for 24p3R expression by RT–PCR. (H) 32D/BCR–ABL cells were treated with a control or Runx3 siRNA in the presence or absence of imatinib and monitored for 24p3R and Runx3 expression by RT–PCR.
To determine whether the differential binding of Runx proteins contributed to 24p3R transcriptional activity, we carried out RNAi experiments using siRNAs that selectively knocked down expression of either Runx1 or Runx3. Figure 2F shows that treatment of 32D cells with a Runx3 siRNA resulted in loss of 24p3R expression, whereas a Runx1 siRNA had no effect. By contrast, treatment of 32D/BCR–ABL cells with a Runx1 siRNA activated expression of 24p3R, whereas a Runx3 siRNA had no significant effect (Figure 2G). After activation of 24p3R expression in 32D/BCR–ABL cells by imatinib treatment, knock-down of Runx3 substantially decreased 24p3R expression (Figure 2H). Collectively, these results show that Runx3 is an activator and Runx1 a repressor of 24p3R expression.
Runx1 represses 24p3R expression by recruiting the Sin3a–HDAC corepressor complex
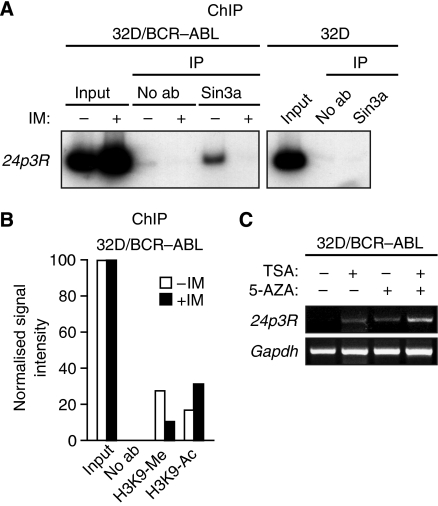
Previous studies have suggested that Runx1 is physically associated with Sin3a (Lutterbach et al, 2000), a component of a histone deacetylase (HDAC)-containing corepressor complex (reviewed in Ayer, 1999). We, therefore, considered the possibility that in 32D/BCR–ABL cells, Runx1 recruits the Sin3a–HDAC complex to the 24p3R promoter and the resultant histone deacetylation contributes to 24p3R repression. As a first test of this idea, we used a ChIP assay to analyse association of Sin3a with the 24p3R promoter. The results of Figure 3A show that Sin3a was bound to the 24p3R promoter in 32D/BCR–ABL but not in 32D cells. After treatment of 32D/BCR–ABL cells with imatinib, which results in the loss of Runx1 binding (see Figure 2C), association of Sin3a with the 24p3R promoter was also abrogated (Figure 3A). The decrease in Runx1 and Sin3a binding on imatinib treatment was accompanied by modifications of lysine 9 in histone H3 (H3K9) that are associated with transcriptional activation: increased H3K9 acetylation and decreased H3K9 methylation (Figure 3B). Finally, treatment of 32D/BCR–ABL cells with the HDAC inhibitor trichostatin A resulted in activation of 24p3R expression, which was further increased by treatment with the DNA methylation inhibitor 5-azacytidine (Figure 3C). Collectively, these results indicate that in 32D/BCR–ABL cells, binding of Runx1 to the 24p3R promoter recruits Sin3a, resulting in histone deacetylation and methylation, and transcriptional repression.
Figure 3.
Runx1 represses 24p3R transcription by recruiting the Sin3a–HDAC corepressor complex. (A) ChIP analysis monitoring binding of Sin3a to the 24p3R in 32D/BCR–ABL cells treated in the presence or absence of imatinib (left) or in 32D cells (right). (B) ChIP analysis monitoring the presence of H3K9 methylation and acetylation at the 24p3R in 32D/BCR–ABL cells treated in the presence or absence of imatinib. (C) RT–PCR analysis monitoring 24p3R expression in 32D/BCR–ABL cells treated with trichostatin A (TSA) and/or 5-azacytidine (5-AZA).
BCR–ABL signals through the Ras/MAPK pathway to repress 24p3R transcription
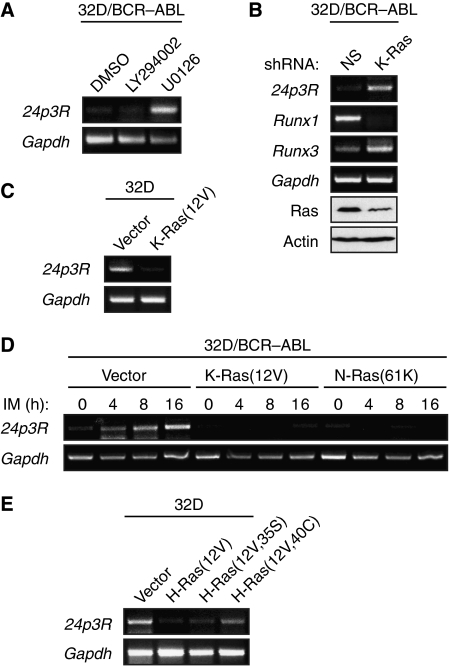
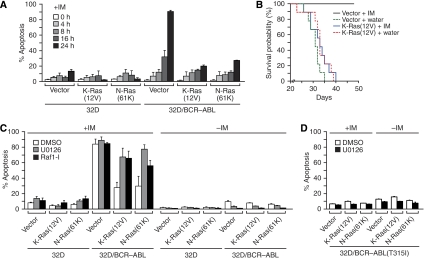
We next attempted to gain insight into how BCR–ABL controls the Runx protein binding switch. As discussed earlier, BCR–ABL is known to function through several signalling pathways, including the Ras/mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/Akt pathways (reviewed in Melo and Deininger, 2004). As an initial step to determine the specific pathway involved in the Runx protein binding switch, we treated 32D/BCR–ABL cells with a chemical inhibitor of either Ras/MAPK signalling (U0126, a selective inhibitor of MEK1 and MEK2; Favata et al, 1998) or PI3K/Akt signalling (LY294002, a selective PI3K inhibitor; Vlahos et al, 1994). The results of Figure 4A show that U0126 but not LY294002 activated 24p3R expression, implicating a critical role for the Ras/MAPK pathway in repression of 24p3R by BCR–ABL. To confirm this result, we carried out an RNAi experiment. Previous studies have shown that K-Ras is the major Ras isoform that mediates signalling through the MAPK pathway (Yan et al, 1998). Figure 4B shows that shRNA-mediated knock-down of K-Ras in 32D/BCR–ABL cells activated 24p3R expression. Significantly, after knock-down of K-Ras, Runx1 mRNA levels markedly decreased, which was accompanied by an increased level of Runx3 mRNA. Collectively, these results indicate that the Ras/MAPK pathway is necessary for repression of 24p3R expression by BCR–ABL.
Figure 4.
BCR–ABL signals through the Ras/MAPK pathway to repress 24p3R transcription. (A) RT–PCR analysis monitoring expression of 24p3R in 32D/BCR–ABL cells treated with a PI3K inhibitor (LY294002) or a MEK1/2 inhibitor (U0126). The concentrations of LY294002 (25 μM) and U0126 (10 μM) used were based on titration experiments (Supplementary Figures S6B and S6C, respectively) and represents the minimal concentrations required to obtain greater than 75% inhibition of Akt and Erk phosphorylation, respectively. (B) 32D/BCR–ABL cells were treated with a non-silencing (NS) or K-Ras shRNA, and expression of 24p3R, Runx1 and Runx3 were monitored by RT–PCR. Knock-down efficiency of Ras was monitored by immunoblot analysis. (C) 32D cell lines stably expressing a constitutively activated K-Ras allele (K-Ras(12V)) or, as a control, the empty expression vector were monitored for expression of 24p3R by RT–PCR. (D) 32D/BCR–ABL cells stably expressing a constitutively activated K-Ras(12V) or N-Ras(61K) mutant were monitored for expression of 24p3R by RT–PCR at various timepoints after treatment with imatinib, as indicated. (E) RT–PCR analysis monitoring 24p3R expression in 32D cells stably expressing the activated K-Ras(12V) mutant, or effector domain derivatives defective for signalling through either the PI3K/AKT pathway (H-Ras(12V,35S)) or MAPK pathway (H-Ras(12V,40C)).
We next carried out a series of experiments to determine whether activation of Ras signalling was sufficient for repression of 24p3R. We constructed 32D cell lines stably expressing a constitutively activated K-Ras allele (K-Ras(12V)) (Barbacid, 1987) or, as a control, the empty expression vector. Figure 4C shows that expression of K-Ras(12V) in 32D cells resulted in loss of 243pR expression. In a complementary set of experiments, we stably expressed K-Ras(12V) or another constitutively activated Ras allele, N-Ras(61K) (Taparowsky et al, 1983), in 32D/BCR–ABL cells. We then inactivated BCR–ABL by addition of imatinib, and monitored 24p3R expression. The results of Figure 4D show, as expected, that in control cells expressing vector alone, imatinib activated 24p3R expression. However, 24p3R expression was not activated in imatinib-treated 32D/BCR–ABL cells expressing K-Ras(12V) or N-Ras(61K), indicating that activation of Ras signalling is sufficient to repress 24p3R expression.
Activated Ras stimulates several downstream signalling pathways including the MAPK and PI3K/Akt pathways (reviewed in Steelman et al, 2004). To understand in greater detail the basis of Ras-mediated silencing of 24p3R, we analysed activating H-Ras effector domain mutants that are defective for signalling through either the MAPK pathway (H-Ras(12V,40C)) or the PI3K/Akt pathway (H-Ras(12V,35S)) (White et al, 1995; Rodriguez-Viciana et al, 1997). Figure 4E shows, as expected, that in 32D cells stably expressing H-Ras(12V), 24p3R was repressed. However, 24p3R was not efficiently repressed in 32D cells expressing the MAPK signalling mutant H-Ras(12V,40C), whereas the PI3K/Akt signalling mutant H-Ras(12V,35S) repressed 24p3R. Thus, consistent with the chemical inhibition experiments described earlier (Figure 4A), Ras represses 24p3R expression predominantly through the MAPK signalling pathway.
Activated Ras induces the Runx protein binding switch
We next asked whether, similarly to BCR–ABL, Ras-mediated 24p3R repression occurred through a Runx protein binding switch. As described earlier, expression of K-Ras(12V) in 32D cells is sufficient to repress 24p3R. The ChIP experiment of Figure 5A shows that in control 32D cells, the 24p3R promoter was preferentially bound by Runx3 (compare lanes 5 and 7), whereas in K-Ras(12V)-expressing 32D cells the 24p3R promoter was preferentially bound by Runx1 (compare lanes 6 and 8). Thus, Ras induces a Runx protein binding switch analogous to that observed with BCR–ABL.
Figure 5.
Activated Ras induces the Runx protein binding switch. (A) ChIP analysis monitoring binding of Runx1 and Runx3 to the 24p3R promoter in 32D cells in the presence or absence of the activated K-Ras(12V) allele. (B) ChIP analysis monitoring binding of Runx1 and Runx3 to the 24p3R promoter in 32D/BCR–ABL cells expressing either the activated K-Ras(12V) allele or, as a control, empty vector. Cells were treated in the presence or absence of imatinib, as indicated. (C) Immunoblot analysis monitoring levels of Runx1, Runx3, phosphorylated (P) Erk and total (T) Erk in 32D/BCR–ABL cells expressing either K-Ras(12V) or empty vector, treated in the presence or absence of imatinib.
We next carried out a complementary set of experiments in 32D/BCR–ABL cells. Consistent with the results described earlier, treatment of 32D/BCR–ABL cells with imatinib led to a loss of Runx1 association with the 24p3R promoter (Figure 5B, compare lanes 9 and 10), with a concomitant increase in Runx3 binding (compare lanes 13 and 14). However, after stable expression of K-Ras(12V), Runx1 remained associated with the 24p3R promoter after imatinib treatment (compare lanes 11 and 12).
To investigate the basis of the Runx protein binding switch, we analysed steady-state levels of Runx1 and Runx3 in 32D/BCR–ABL cells in the presence or absence of imatinib. The immunoblot experiment of Figure 5C shows that imatinib treatment markedly reduced Runx1 levels, whereas Runx3 levels were largely unaffected. By contrast, imatinib had no effect on Runx1 levels in 32D cells expressing the BCR–ABL(T315I) mutant (Supplementary Figure S3). Moreover, in 32D/BCR–ABL cells stably expressing activated K-Ras(12V), Runx1 levels did not decrease after imatinib treatment (Figure 5C). These results, in conjunction with those in Figure 4B, indicate BCR–ABL stimulation of the Ras pathway increases the steady-state levels of Runx1, which accounts, at least in part, for the Runx protein binding switch.
Constitutive downregulation of 24p3R results in imatinib resistance
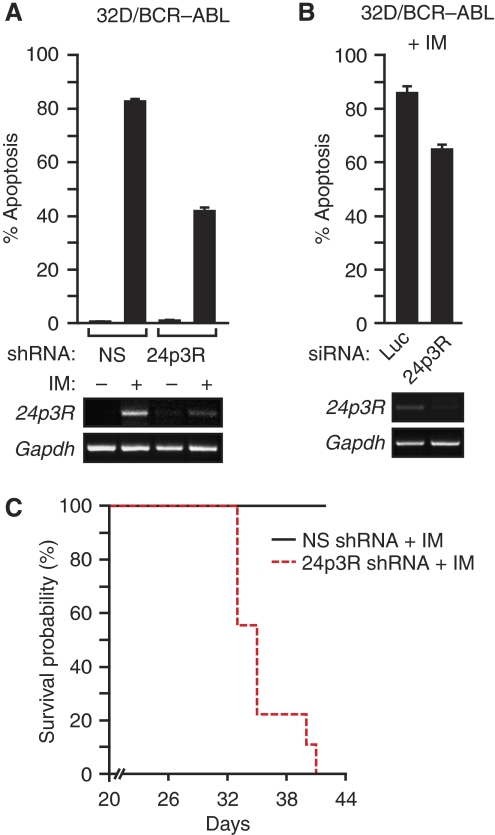
As described earlier, ectopic expression of 24p3R can induce apoptosis in BCR–ABL+ cell lines (Devireddy et al, 2005), suggesting that derepression of 24p3R after imatinib treatment of BCR–ABL+ cells may contribute to imatinib-induced apoptosis. To test this possibility, we asked whether blocking 24p3R derepression could affect the ability of imatinib to kill BCR–ABL+ cells. As a proxy for blocking 24p3R derepression, we used an shRNA to knock-down 24p3R in 32D/BCR–ABL cells, and then monitored imatinib-induced apoptosis. Figure 6A shows, as expected, that knock-down of 24p3R in 32D/BCR–ABL cells resulted in decreased 24p3R expression, which was evident after imatinib treatment. Significantly, the 24p3R knock-down 32D/BCR–ABL cells were markedly more imatinib resistant than 32D/BCR–ABL cells expressing a control non-silencing shRNA.
Figure 6.
Constitutive downregulation of 24p3R results in imatinib resistance. (A) 32D/BCR–ABL cells treated with either an NS or 24p3R shRNA in the presence or absence of imatinib were monitored for apoptosis by annexin V-PE staining (top) or 24p3R expression by RT–PCR (bottom). Error bars indicate s.d. (B) 32D/BCR–ABL cells treated with either a control luciferase (Luc) or 24p3R siRNA in the presence of imatinib were monitored for apoptosis (top) or 24p3R expression by RT–PCR (bottom). Error bars indicate s.d. The difference between the samples was analysed using the Student's t-test and gave a P-value of 0.003. (C) Kaplan–Meier survival analysis. 32D/BCR–ABL cells stably expressing a 24p3R or NS shRNA were injected into the tail vein of myeloablated mice. The mice were then treated with imatinib and monitored for survival.
To rule out possible off-target effects of the 24p3R shRNA, we carried out two experiments. First, knock-down of 24p3R by an siRNA, unrelated in sequence to the 24p3 shRNA used in Figure 6A, rendered 32D/BCR–ABL cells more resistant to imatinib (Figure 6B). Second, ectopic overexpression of a cDNA encoding 24p3R increased imatinib sensitivity of 32D/BCR–ABL cells after 24p3R knock-down (Supplementary Figure S4).
We next asked whether 24p3R knock-down could also confer imatinib resistance in mice. 32D/BCR–ABL cells stably expressing a 24p3R or non-silencing shRNA were injected into the tail vein of myeloablated mice, and starting 3 days later, the mice were treated daily with imatinib. The Kaplan–Meier analysis of Figure 6C shows, remarkably, that imatinib did not promote survival of mice bearing 32D/BCR–ABL cells in which 24p3R had been knocked down. Thus, derepression of 24p3R after imatinib treatment is an essential part of the mechanism by which imatinib kills BCR–ABL+ cells.
Activated Ras induces imatinib resistance
The fact that activated Ras represses 24p3R independently of BCR–ABL (Figures 4C–E), and that 24p3R knock-down results in imatinib resistance (Figure 6), raised the possibility that Ras-mediated repression of 24p3R might provide a mechanism for imatinib resistance. As a first test of this idea, we asked whether activated Ras would render 32D/BCR–ABL cells resistant to imatinib-induced apoptosis. Figure 7A shows, as expected, that 32D/BCR–ABL cells expressing vector alone efficiently underwent apoptosis after addition of imatinib. However, expression of a constitutively activated Ras allele (K-Ras(12V) or N-Ras(61K)) significantly reduced the level of apoptosis in imatinib-treated 32D/BCR–ABL cells, indicating that Ras activation was sufficient to confer imatinib resistance. The Kaplan–Meier analysis of Figure 7B confirmed that expression of activated Ras could also confer imatinib resistance in mice.
Figure 7.
Activated Ras induces imatinib resistance. (A) Apoptosis assays. 32D or 32D/BCR–ABL cells stably expressing an activated Ras allele, either K-Ras(12V) or N-Ras(61K), were treated with imatinib for various lengths of time, and analysed for apoptosis by annexin V-PE staining. Error bars indicate s.d. (B) Kaplan–Meier survival analysis. 32D/BCR–ABL cells expressing activated K-Ras(12V) or, as a control, empty vector were injected into the tail vein of myeloablated mice. The mice were then treated with imatinib or, as a control, water and monitored daily for survival. (C) 32D or 32D/BCR–ABL cells stably expressing K-Ras(12V) or N-Ras(61K) were treated with either DMSO, the MEK inhibitor U0126 or Raf1 kinase inhibitor I (Raf1-I) in the presence or absence of imatinib, as indicated. Cells were monitored for apoptosis 16 h later, as described in (A). Error bars indicate s.d. (D) 32D/BCR–ABL(T315I) cells expressing K-Ras(12V) or N-Ras(61K) were treated with either DMSO or U0126 and monitored for apoptosis, as described in (C). Error bars indicate s.d.
As described earlier, Ras-mediated repression of 24p3R occurs through the MAPK pathway. We, therefore, tested whether Ras induced imatinib resistance of BCR–ABL+ cells through the MAPK pathway. 32D/BCR–ABL cells expressing an activated Ras allele were treated with an inhibitor of the MAPK pathway, either a MEK inhibitor (U0126) or a Raf1 inhibitor (Raf1 kinase inhibitor I) (Lackey et al, 2000). Figure 7C shows that treatment with either U0126 or the Raf1 inhibitor increased sensitivity of activated Ras-expressing 32D/BCR–ABL cells to imatinib relative to the control (DMSO). Significantly, in the absence of imatinib treatment neither U0126 nor the Raf1 inhibitor induced apoptosis in 32D/BCR–ABL cells. As expected, activated Ras-expressing 32D/BCR–ABL(T315I) cells treated with U0126 failed to undergo apoptosis after imatinib treatment (Figure 7D). Thus, Ras induces imatinib resistance in cells expressing BCR–ABL through the MAPK pathway.
Discussion
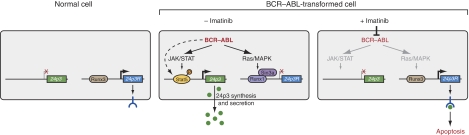
On the basis of the results presented in this study, we propose the following model for the reciprocal misregulation of 24p3 and 24p3R expression by BCR–ABL (summarised in Figure 8). BCR–ABL induces 24p3 expression by stimulating the JAK/STAT pathway, leading to phosphorylation and activation of Stat5, which then binds to the 24p3 promoter, resulting in transcription activation. We have found that in 32D/BCR–ABL cells, Jak1, Jak2 and Jak3 are all activated and that phospho-Jak1 levels are particularly high. It has been reported previously that Jak2 is the major effector of BCR–ABL-mediated signalling (Samanta et al, 2006), and thus may have an important function in 24p3 misregulation. Activation of Stat5 through Jak kinase-independent mechanisms (Ilaria and Van Etten, 1996; Klejman et al, 2002; Tao et al, 2008) may also contribute to induction of 24p3 transcription in 32D/BCR–ABL cells. With regard to 24p3R, BCR–ABL functions through the MAPK pathway to increase Runx1 levels, resulting in a switch from binding of Runx3 to Runx1 to the 24p3R promoter. Runx1 recruits the Sin3a–HDAC corepressor complex, resulting in 24p3R transcriptional repression. Thus, BCR–ABL regulates expression of 24p3 and 24p3R through distinct signalling and transcription factor pathways.
Figure 8.
A model for the reciprocal misregulation of 24p3 and 24p3R expression by BCR–ABL. (Left) In normal cells in the presence of IL-3, 24p3 is repressed and 24p3R is expressed. (Middle) BCR–ABL activates 24p3 expression by stimulating the JAK/STAT pathway, leading to phosphorylation and activation of Stat5, which binds to the 24p3 promoter and results in transcription activation. Activation of Stat5 through Jak kinase-independent mechanisms (dashed line) may also contribute to induction of 24p3 transcription in 32D/BCR–ABL cells. Concomitantly, BCR–ABL represses 24p3R expression by increasing the levels of Runx1, which recruits the Sin3a–HDAC corepressor complex, resulting in 24p3R transcriptional repression. In the absence of 24p3R, BCR–ABL+ cells are resistant to 24p3-mediated apoptosis. (Right) In the presence of imatinib, which inhibits BCR–ABL kinase activity, Stat5 is inactive and absent from the 24p3 promoter. Inhibition of BCR–ABL also results in decreased Ras/MAPK signalling, which results in a switch from binding of Runx1 to Runx3 to the 24p3R promoter, thereby derepressing 24p3R transcription. In the presence of 24p3R, the pool of pre-existing 24p3 triggers apoptosis in BCR–ABL+ cells.
Misregulation of 24p3 and 24p3R expression by BCR–ABL is directly relevant to the basis by which imatinib induces apoptosis of BCR–ABL+ cells. By inhibiting BCR–ABL, imatinib derepresses 24p3R expression allowing the pool of pre-existing 24p3 to trigger apoptosis in BCR–ABL+ cells. Accordingly, abrogation of 24p3R expression, either through RNAi-mediated knock-down of 24p3R or expression of an activated Ras allele, substantially reduces the ability of imatinib to kill BCR–ABL+ cells (Figures 6A and 7A, respectively). We suspect that a portion of the pre-existing pool of 24p3 that induces apoptosis may function without actually being released into the extracellular medium. For example, we have found that siRNA-mediated depletion of 24p3 inhibits apoptosis more efficiently than addition of an anti-24p3 antibody to the extracellular medium (compare Figure 7H in Devireddy et al, 2005 and Supplementary Figure S5 in this study).
Misregulation of 24p3 and 24p3R expression by BCR–ABL may also be relevant to mechanisms of imatinib resistance. Imatinib is a first-line treatment for patients with BCR–ABL+ CML (reviewed in Peggs and Mackinnon, 2003). The treatment is particularly successful in chronic phase CML, either newly diagnosed or previously treated, in which the majority of patients achieve a robust response (reviewed in Kantarjian et al, 2007). However, imatinib treatment is less effective in patients in the accelerated and blast-crisis phases of disease. This reduced efficacy is due to the acquisition of imatinib resistance, which occurs in approximately 70–90% of patients in the accelerated-blast phases, and 20% of patients with chronic phase CML. Acquired imatinib resistance is typically associated with reactivation of BCR–ABL due to gene amplification or, more commonly, to mutations in the kinase domain that prevent imatinib binding. However, it has been estimated that approximately 45% of imatinib-resistance cases are not the result of qualitative or quantitative alterations in BCR–ABL (Hochhaus et al, 2002).
Several mechanisms underlying BCR–ABL independent imatinib resistance have been proposed, including overexpression of other tyrosine kinases such as LYN (Donato et al, 2003, 2004) and autocrine/paracrine signalling by secreted granulocyte-macrophage colony-stimulating factor (Wang et al, 2007). The results presented here, in conjunction with several previous studies, reveal constitutive Ras signalling as an additional alternative mechanism for the development of imatinib-resistant CML. Previous studies have detected activating Ras mutations in leukemic blasts from CML patients including cases in which imatinib-resistance has developed (LeMaistre et al, 1989; Agarwal et al, 2008). In particular, a recent study described an imatinib-resistant CML patient lacking BCR–ABL mutations and harbouring an activating KRAS T58I allele. Moreover, expression of KRAS T58I in 32D/BCR–ABL cells was shown to result in imatinib resistance (Agarwal et al, 2008).
The major therapeutic strategy to overcome imatinib resistance has been the development of alternative BCR–ABL inhibitors, such as Nilotinib and Dasatinib (Kantarjian et al, 2007). Our findings suggest that inhibition of the Ras/MAPK pathway may be an alternative therapeutic strategy in a definable subset of imatinib-resistant patients. Consistent with this idea, farnesyl transferase inhibitors, which suppress Ras activity, have been reported to induce apoptosis in some imatinib-resistant BCR–ABL+ cells (Hoover et al, 2002; Tauchi and Ohyashiki, 2004; Copland et al, 2008).
Materials and methods
Cell culture and transfection
32D cells were obtained from ATCC and maintained in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) and 1 ng/ml murine IL-3 (PeproTech). 32D/BCR–ABL cells, kindly provided by T. Skorski (Temple University, PA), were maintained in RPMI1640 with 10% FBS. 32D/BCR–ABL(T315I) cells were generated by stable transfection of 32D cells with a plasmid expressing BCR–ABL(T315I), kindly provided by C Sawyers (Memorial Sloan-Kettering Cancer Center, NY). In experiments involving BCR–ABL inhibition, imatinib mesylate (Novartis) was added at a concentration of 5 μM for 16 h. For inhibiting other kinases, the following inhibitors were added to cells for 24 to 48 h: JAK inhibitor I (CalBiochem, 10 μM), Raf1 kinase inhibitor I (CalBiochem, 10 μM), LY294002 (CalBiochem, 25 μM) or U0126 (Cell Signaling Technology Inc., 10 μM). Trichostatin A (Sigma) was added at a concentration of 10 nM, and 5-azacytidine (Sigma) was added at a concentration of 1.25 μM for 10 days.
To construct cell lines stably expressing Ras mutants, HEK293T packaging cells were transfected with 2 μg pBABE-K-Ras(12V), pBABE-N-Ras(61K), pBABE-H-Ras(12V), pBABE-H-Ras(12V,35S) or pBABE-H-Ras(12V,40C) (all obtained from Addgene) using Effectene (QIAGEN) according to the manufacturer's instructions. Viral supernatants were collected 48 h later and used to infect 32D or 32D/BCR–ABL cells followed by selection using 2 μg/ml puromycin. Cell lines stably expressing Ras mutants were maintained in culture media containing 1 μg/ml of puromycin.
24p3/24p3R promoter activity assays
24p3 and 24p3R promoter fragments were PCR amplified from BAC clones (ATCC) using primers containing KpnI and SacI (for 24p3) or XhoI and BglII (for 24p3R) restriction enzyme sites. PCR fragments were then digested and cloned into the vector pGL4.14[luc2/Hygro] (Promega). 24p3 and 24p3R promoter mutations were generated using the QuikChange XL site-directed mutagenesis kit (Strategene) according to the manufacturer's instructions. Mutagenesis of the putative Stat5 binding site in the 24p3 promoter (740 bp fragment) and the Runx binding site in the 24p3R promoter (∼2 kb fragment) was carried out to create SacI and XhoI sites, respectively, and confirmed by restriction enzyme analysis and DNA sequencing. Plasmids containing 24p3 or 24p3R promoter fragments (2 μg) were transfected together with 50 ng of pGL4.73[hRluc/SV40] (Promega) into 32D or 32D/BCR–ABL cells by electroporation (Amaxa) according to the manufacturer's instructions. Luciferase activity was measured 24 h later using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocols. Firefly luciferase activity was normalised to that of Renilla luciferase. Experiments were carried out three times.
Chromatin immunoprecipitation
ChIP assays were carried out as described previously (Raha et al, 2005), with the following minor modifications. Briefly, 3 × 107 cells were incubated with 1% formaldehyde for 10 min at room temperature before crosslinking was quenched by addition of 0.125 M glycine. Cells were collected by centrifugation and lysed in lysis buffer containing 50 mM Tris–HCl pH 8.0, 10 mM EDTA, 0.5% SDS, proteinase inhibitors (Roche) and phosphatase inhibitors (Sigma). The cell suspension was sonicated seven times for 10 s each with 2-min intervals on ice using a Misonix Sonicator 3000 at output 8. Sonicated chromatin was then incubated at 4°C overnight with 5 μg of the appropriate antibody: α-Stat5 (Santa Cruz), α-Stat5a (Santa Cruz), α-Stat5b (Santa Cruz), α-Runx1 (Santa Cruz), α-Runx3 (Santa Cruz), α-Sin3a (Santa Cruz), α-H3K9-Me (Upstate) or α-H3K9-Ac (Upstate). Immunoprecipitated chromatin DNA was analysed by PCR. For some experiments, 32P-dATP was added and the radiolabelled PCR products were analysed using ImageJ software (NIH). Primers used in ChIP were as follows Stat5-chip-for (5′-GCAGCCACATCTAAGGACTAC-3′), Stat5-chip-rev (5′-ACCCTGTGCAGCTTCCTTGTC-3′), Runx-chip-for (5′-GCAGAGACTTGTTCCCCTGAA-3′) and Runx-chip-rev (5′-TGTATCCCAGGCAGTGTGCGA-3′). Primers used to detect Stat5 binding on the Ksr promoter were reported previously (Nelson et al, 2004).
RT–PCR
Total RNA was isolated using TriZol (Invitrogen) according to the manufacturer's instructions and cDNA synthesis was performed using SuperScript reverse transcriptase (Invitrogen). Primers to detect 24p3, 24p3R, and Gapdh expression were described previously (Devireddy et al, 2005). Other primers used were as follows Runx1-rt-for (5′-GAGCGGCTCAGTGAATTGGAG-3′), Runx1-rt-rev (5′-TAGATGGTAGGAGGGCGAGCC-3′) Runx3-rt-for (5′-CTACCC AAGTGGCTACCTACC-3′) and Runx3-rt-rev (5′-GTAGGTGTGGTGGAACGGCT-3′).
Immunoblot analysis
Cells were lysed in lysis buffer containing 20 mM HEPES pH 6.8, 140 mM NaCl, 2.5 mM MgCl2, 2.5 mM CaCl2, 1% NP40, 0.5% sodium deoxycholate, proteinase inhibitors (Roche), and phosphatase inhibitors (Sigma). Total protein (100 μg) was separated by SDS–PAGE and then transferred onto PVDF membrane (Millipore), which was incubated at 4°C overnight with one of the following antibodies: phospho-Jak1(Tyr1022/1023) (Cell Signaling Technology Inc., at 1:500 dilution), Jak1 (Cell Signaling Technology Inc., 1:1000), Phospho-Stat5(Tyr694) (Cell Signaling Technology Inc., 1:500), Stat5 (Zymed, 1:2000 or Cell Signaling Technology Inc., 1:1000), phospho-Erk1/2(Thr202/Tyr204) (Cell Signaling Technology Inc., 1:500), and Erk1/2 (Cell Signaling Technology Inc., 1:2000), Ras (Abcam, 1:1000), Runx1 (Abcam, 1:500), Runx3 (Abcam, 1:1000) or Actin (Sigma, 1:5000). Proteins were visualised using SuperSignal West Pico or Femto chemiluminescent substrate (Pierce).
RNA interference
32D or 32D/BCR–ABL cells (1 × 106) were transfected by electroporation (Amaxa) with 200 pmol of the following siRNAs: luciferase/control (5′-TGATCAAATACAAGGGATA-3′), Stat5 (5′-AGACCTGGCTGCAGCGAGA-3′), Runx1 (5′-AGAACCAGGTAGCGAGATT-3′), Runx3 (5′-GGGAAGAGTTTCACGCTCA-3′) or 24p3R (5′-GGAUCCUGGCUGAGCGAAA-3′) (all synthesised by CFAR, UMass Medical School). Cells were collected 24 to 48 h later and RT–PCR or immunoblotting was carried out as described earlier. To knock-down K-Ras, 5 μg of the K-Ras shRNA (catalog no. RHS1764-9685820, Open Biosystems) or non-silencing shRNA (catalog no. RHS1707, Open Biosystems) was used to transfect 1 × 106 32D/BCR–ABL cells as described earlier. For shRNA-mediated knock-down of 24p3R, retroviral supernatants prepared from HEK293T cells 48 h after transfection with 2 μg of either non-silencing or 24p3R shRNA (5′-AGCCCAGGAAGCTTTGCATGAA-3′; catalog no. RMM1766-97042505, Open Biosystems) were immediately used to infect 32D/BCR-ABL cells. Stable clones were selected using 2.5 μg/ml puromycin. Knock-down of 24p3R was examined using RT–PCR as described previously.
Bone marrow transduction and transplantation
Male donor 6- to 10-week-old 129SvEv mice (Jackson Laboratory) were treated with 150 mg/kg 5-fluorouracil for 8 days and then sacrificed by CO2 asphyxiation. Femur and tibia bones were collected, and bone marrow was harvested by flushing the bones with bone marrow growth medium [RPMI1640, 20% FBS, 1% penicillin/streptomycin, 6 ng/ml recombinant mouse IL-3 (PeproTech), 10 ng/ml recombinant human IL-6 (PeproTech), 5 ng/ml recombinant mouse SCF (PeproTech)]. Erythrocytes were removed using FCM Lysing Solution (Santa Cruz Biotechnology, Inc.) according to the manufacturer's instructions. After overnight incubation at 37°C with 5% CO2, bone marrow cells were infected with a retrovirus expressing BCR–ABL and green fluorescent protein (GFP) that was packaged in 293T cells using the plasmid pMSCV–BCR–ABL–IRES–GFP (provided by W. Pear, University of Pennsylvania, PA) or, as a negative control, with a retrovirus expressing GFP only (pMSCV–IRES–GFP). Bone marrow cell infection was carried out using two rounds of cosedimentation of cells and virus suspension, one round per day. For the GFP virus control, infected bone marrow cells were cultured in growth medium for two additional days, before carrying out the ChIP assay. Infected bone marrow cells were then transplanted into lethally irradiated (2 × 550 cGy) syngeneic female recipient mice (0.75 × 106 cells per animal) through tail vein injection. Thirty days later, mice were diagnosed to have CML-like leukemia based on hallmark features of the disease such as increased peripheral blood cells, splenomegaly, and extramedullary haematopoiesis in liver (Ren, 2005). Peripheral blood was collected and erythrocytes were removed as described earlier. More than 80% of the cells were BCR–ABL–GFP positive and were used for the subsequent ChIP assay.
Apoptosis assays
Cells (1 × 106) were incubated in the presence of 5 μM of Gleevec for 16 h unless otherwise noted. Cells were then washed with ice-cold PBS, stained with annexin-V-PE and 7-AAD (BD Pharmingen) according to the manufacturer's instructions, and evaluated using a Guava Personal Cell Analysis system (Guava Technologies) or by flow cytometry by the Flow Cytometry Core Facility at UMMS. Experiments were carried out three times.
Animal experiments
Six to eight-week-old SCID/Beige Mice (Taconic) were injected through the tail vein with 1 × 106 32D/BCR–ABL cells expressing either K-Ras(12V) or empty vector, or stably expressing either a 24p3R or non-silencing shRNA. Three days later, the mice injected with either K-Ras or vector-expressing cells or shRNA-treated cells were each divided into two groups (n=9 mice per group), with each group receiving either water or imatinib (50 mg/kg twice a day, dissolved in 100 μl water) orally by gavage. Mice were monitored daily for viability, and statistical analysis of survival curve data was carried out using Kaplan–Meier survival analysis. The experiment was discontinued after 48 days (K-Ras experiment) or 42 days (24p3R knock-down experiment), at which time any remaining mice were humanely killed. All mouse experiments were carried out in accordance with the Institutional Animal Care and Use Committee guidelines.
Supplementary Material
Supplementary Information
Acknowledgments
We thank T Skorski, C Sawyers and W Pear for providing reagents, the Center for AIDS Research (CFAR) Molecular Biology Core at UMass Medical School for siRNA synthesis, S Griggs for excellent technical assistance, members of the Green lab for critical discussion, M Deininger for communicating results before publication and S Evans for editorial assistance. This work was supported in part by a grant from the National Institutes of Health to MRG. MRG is an Investigator of the Howard Hughes Medical Institute.
References
- Agarwal A, Eide CA, Harlow A, Corbin AS, Mauro MJ, Druker BJ, Corless CL, Heinrich MC, Deininger MW (2008) An activating KRAS mutation in imatinib-resistant chronic myeloid leukemia. Leukemia 22: 2269–2272 [DOI] [PubMed] [Google Scholar]
- Ayer DE (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol 9: 193–198 [DOI] [PubMed] [Google Scholar]
- Barbacid M (1987) ras genes. Annu Rev Biochem 56: 779–827 [DOI] [PubMed] [Google Scholar]
- Buchdunger E, Zimmermann J, Mett H, Meyer T, Muller M, Druker BJ, Lydon NB (1996) Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res 56: 100–104 [PubMed] [Google Scholar]
- Copland M, Pellicano F, Richmond L, Allan EK, Hamilton A, Lee FY, Weinmann R, Holyoake TL (2008) BMS-214662 potently induces apoptosis of chronic myeloid leukemia stem and progenitor cells and synergizes with tyrosine kinase inhibitors. Blood 111: 2843–2853 [DOI] [PubMed] [Google Scholar]
- Cross NC, Reiter A (2002) Tyrosine kinase fusion genes in chronic myeloproliferative diseases. Leukemia 16: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Danial NN, Rothman P (2000) JAK-STAT signaling activated by Abl oncogenes. Oncogene 19: 2523–2531 [DOI] [PubMed] [Google Scholar]
- Devireddy LR, Gazin C, Zhu X, Green MR (2005) A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 123: 1293–1305 [DOI] [PubMed] [Google Scholar]
- Devireddy LR, Teodoro JG, Richard FA, Green MR (2001) Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science 293: 829–834 [DOI] [PubMed] [Google Scholar]
- Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, Talpaz M (2003) BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood 101: 690–698 [DOI] [PubMed] [Google Scholar]
- Donato NJ, Wu JY, Stapley J, Lin H, Arlinghaus R, Aggarwal BB, Shishodia S, Albitar M, Hayes K, Kantarjian H, Talpaz M (2004) Imatinib mesylate resistance through BCR-ABL independence in chronic myelogenous leukemia. Cancer Res 64: 672–677 [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM (1998) Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem 273: 18623–18632 [DOI] [PubMed] [Google Scholar]
- Hochhaus A, Kreil S, Corbin AS, La Rosee P, Muller MC, Lahaye T, Hanfstein B, Schoch C, Cross NC, Berger U, Gschaidmeier H, Druker BJ, Hehlmann R (2002) Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia 16: 2190–2196 [DOI] [PubMed] [Google Scholar]
- Hoover RR, Mahon FX, Melo JV, Daley GQ (2002) Overcoming STI571 resistance with the farnesyl transferase inhibitor SCH66336. Blood 100: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Ilaria RL Jr, Van Etten RA (1996) P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem 271: 31704–31710 [DOI] [PubMed] [Google Scholar]
- Kaneta Y, Kagami Y, Tsunoda T, Ohno R, Nakamura Y, Katagiri T (2003) Genome-wide analysis of gene-expression profiles in chronic myeloid leukemia cells using a cDNA microarray. Int J Oncol 23: 681–691 [PubMed] [Google Scholar]
- Kantarjian HM, Giles F, Quintas-Cardama A, Cortes J (2007) Important therapeutic targets in chronic myelogenous leukemia. Clin Cancer Res 13: 1089–1097 [DOI] [PubMed] [Google Scholar]
- Klejman A, Schreiner SJ, Nieborowska-Skorska M, Slupianek A, Wilson M, Smithgall TE, Skorski T (2002) The Src family kinase Hck couples BCR/ABL to STAT5 activation in myeloid leukemia cells. EMBO J 21: 5766–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey K, Cory M, Davis R, Frye SV, Harris PA, Hunter RN, Jung DK, McDonald OB, McNutt RW, Peel MR, Rutkowske RD, Veal JM, Wood ER (2000) The discovery of potent cRaf1 kinase inhibitors. Bioorg Med Chem Lett 10: 223–226 [DOI] [PubMed] [Google Scholar]
- LeMaistre A, Lee MS, Talpaz M, Kantarjian HM, Freireich EJ, Deisseroth AB, Trujillo JM, Stass SA (1989) Ras oncogene mutations are rare late stage events in chronic myelogenous leukemia. Blood 73: 889–891 [PubMed] [Google Scholar]
- Leng X, Lin H, Ding T, Wang Y, Wu Y, Klumpp S, Sun T, Zhou Y, Monaco P, Belmont J, Aderem A, Akira S, Strong R, Arlinghaus R (2008) Lipocalin 2 is required for BCR-ABL-induced tumorigenesis. Oncogene 27: 6110–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Groner Y (2004) Structure and regulated expression of mammalian RUNX genes. Oncogene 23: 4211–4219 [DOI] [PubMed] [Google Scholar]
- Lin F, Monaco G, Sun T, Liu J, Lin H, Stephens C, Belmont J, Arlinghaus RB (2001) BCR gene expression blocks Bcr-Abl induced pathogenicity in a mouse model. Oncogene 20: 1873–1881 [DOI] [PubMed] [Google Scholar]
- Lin H, Monaco G, Sun T, Ling X, Stephens C, Xie S, Belmont J, Arlinghaus R (2005) Bcr-Abl-mediated suppression of normal hematopoiesis in leukemia. Oncogene 24: 3246–3256 [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW (2000) A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol Chem 275: 651–656 [DOI] [PubMed] [Google Scholar]
- Melo JV, Deininger MW (2004) Biology of chronic myelogenous leukemia—signaling pathways of initiation and transformation. Hematol Oncol Clin North Am 18: 545–568, vii–viii [DOI] [PubMed] [Google Scholar]
- Nelson EA, Walker SR, Alvarez JV, Frank DA (2004) Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J Biol Chem 279: 54724–54730 [DOI] [PubMed] [Google Scholar]
- Park SH, Liu X, Hennighausen L, Davey HW, Waxman DJ (1999) Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J Biol Chem 274: 7421–7430 [DOI] [PubMed] [Google Scholar]
- Peggs K, Mackinnon S (2003) Imatinib mesylate—the new gold standard for treatment of chronic myeloid leukemia. N Engl J Med 348: 1048–1050 [DOI] [PubMed] [Google Scholar]
- Raha T, Cheng SW, Green MR (2005) HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol 3: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R (2005) Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 5: 172–183 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J (1997) Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89: 457–467 [DOI] [PubMed] [Google Scholar]
- Samanta AK, Lin H, Sun T, Kantarjian H, Arlinghaus RB (2006) Janus kinase 2: a critical target in chronic myelogenous leukemia. Cancer Res 66: 6468–6472 [DOI] [PubMed] [Google Scholar]
- Skorski T (2002) Oncogenic tyrosine kinases and the DNA-damage response. Nat Rev Cancer 2: 351–360 [DOI] [PubMed] [Google Scholar]
- Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA (2004) JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 18: 189–218 [DOI] [PubMed] [Google Scholar]
- Tao WJ, Lin H, Sun T, Samanta AK, Arlinghaus R (2008) BCR-ABL oncogenic transformation of NIH 3T3 fibroblasts requires the IL-3 receptor. Oncogene 27: 3194–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky E, Shimizu K, Goldfarb M, Wigler M (1983) Structure and activation of the human N-ras gene. Cell 34: 581–586 [DOI] [PubMed] [Google Scholar]
- Tauchi T, Ohyashiki K (2004) Imatinib mesylate in combination with other chemotherapeutic agents for chronic myelogenous leukemia. Int J Hematol 79: 434–440 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Cubbon RM, Cummings RT, Wicker LS, Frankshun R, Cunningham BR, Cameron PM, Meinke PT, Liverton N, Weng Y, DeMartino JA (2002) Photochemical preparation of a pyridone containing tetracycle: a Jak protein kinase inhibitor. Bioorg Med Chem Lett 12: 1219–1223 [DOI] [PubMed] [Google Scholar]
- Vlahos CJ, Matter WF, Hui KY, Brown RF (1994) A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 269: 5241–5248 [PubMed] [Google Scholar]
- Wang Y, Cai D, Brendel C, Barett C, Erben P, Manley PW, Hochhaus A, Neubauer A, Burchert A (2007) Adaptive secretion of granulocyte-macrophage colony-stimulating factor (GM-CSF) mediates imatinib and nilotinib resistance in BCR/ABL+ progenitors via JAK-2/STAT-5 pathway activation. Blood 109: 2147–2155 [DOI] [PubMed] [Google Scholar]
- White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH (1995) Multiple Ras functions can contribute to mammalian cell transformation. Cell 80: 533–541 [DOI] [PubMed] [Google Scholar]
- Yan J, Roy S, Apolloni A, Lane A, Hancock JF (1998) Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem 273: 24052–24056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information