Abstract
The contractile tail of bacteriophage T4 is a molecular machine that facilitates very high viral infection efficiency. Its major component is a tail sheath, which contracts during infection to less than half of its initial length. The sheath consists of 138 copies of the tail sheath protein, gene product (gp) 18, which surrounds the central non-contractile tail tube. The contraction of the sheath drives the tail tube through the outer membrane, creating a channel for the viral genome delivery. A crystal structure of about three quarters of gp18 has been determined and was fitted into cryo-electron microscopy reconstructions of the tail sheath before and after contraction. It was shown that during contraction, gp18 subunits slide over each other with no apparent change in their structure.
Keywords: bacteriophage T4, cryo-electron microscopy, crystallography, tail sheath contraction
Introduction
For many bacteriophages, including T4, one viral particle is sufficient for infecting a single bacterial cell (Goldberg et al, 1994). A major reason for such high infection efficiency is a specialized organelle called a tail. It is present in about 96% of all bacteriophages and is designed to attach to bacteria, to penetrate their cell walls and to deliver the viral genome into the host (Ackermann, 2006). There are three groups of tailed bacteriophages based on the tail morphology: Siphoviridae (long, non-contractile tails), Myoviridae (contractile tails) and Podoviridae (short, non-contractile tails). In all three groups of phages, the signal that initiates genome ejection is passed through the tail by virtue of conformational changes. In Siphoviridae, these changes occur in the tail tube (Plisson et al, 2007), whereas in Myoviridae, the signal is transmitted through the tail sheath. In Podoviridae, the inner proteins might be ejected from the head to generate a delivery channel (Kemp et al, 2005).
Bacteriophage T4 belongs to the Myoviridae family. Myoviridae phages have the most complex tail structures, generally consisting of a baseplate with tail fibres and a long, non-contractible tube surrounded by a contractile sheath. Bacteriophage T4 has a tail sheath that is composed of 138 copies of gene product (gp) 18 (Leiman et al, 2004). The tail tube inside the sheath is estimated to be assembled from as many gp19 subunits as there are gp18 subunits in the sheath (Moody and Makowski, 1981).
During infection, following the attachment of the tail fibres to the host cell, the baseplate changes its conformation from a hexagonal dome-shaped to a planar star-shaped structure, causing the sheath to contract from an initial length of 925 Å to a final length of 420 Å, whereas its diameter increases from 240 to 330 Å (Leiman et al, 2004; Kostyuchenko et al, 2005). The contraction of the sheath drives the central tube through the outer membrane, creating a channel for DNA ejection from the capsid into the host cell (Leiman et al, 2003).
In the absence of the baseplate or the tail tube, the tail sheath protein can self-assemble both in vivo and in vitro into tubular structures of variable lengths called polysheaths that have the same helical parameters as the contracted tail sheath (Moody, 1967). Furthermore, different gp18 mutants with deletions of about 250 C-terminal amino-acid residues can still assemble into tubular structures, although with different helical parameters (Poglazov et al, 1999).
Here, we present a crystal structure of about ¾ of the tail sheath protein of bacteriophage T4. The structure is composed of three domains, each having a novel protein fold. To our knowledge, this is the first known atomic structure of a tail sheath protein. Combining the new structural information with the earlier cryo-electron microscopy (cryo-EM) reconstructions and biochemical data, we propose a mechanism for sheath contraction.
Results and discussion
Crystal structure of tail sheath mutants
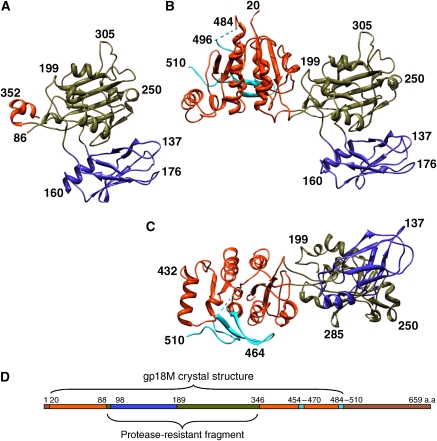
Wild-type gp18 consists of 659 residues and assembles into tubular polymers of variable lengths, which makes crystallization difficult. Several C-terminal deletion mutants that lack polymerization properties (Efimov et al, 2002) were used in extensive crystallization screens. Crystal structures of two gp18 fragments have been determined. One of these is the protease-resistant fragment (gp18PR) and consists of residues 83–365 (Figure 1A), whereas the other deletion mutant (gp18M) consists of residues 1–510 with the last C-terminal residue replaced by a proline (Figure 1B and C) (Efimov et al, 2002). The crystal structure of gp18PR fragment was determined with the multiple anomalous dispersion (MAD) method, refined to 1.8 Å resolution, and subsequently used as a molecular replacement (MR) model to solve the structure of the bigger gp18M fragment to 3.5 Å resolution.
Figure 1.
Structures of the gp18 deletion mutants. (A) A ribbon diagram of the protease resistant fragment (gp18PR). (B, C) A ribbon diagram of the gp18M mutant (¾ of the total protein length) in two orientations. The three domains are shown in blue (domain I), olive green (domain II) and orange red (domain III); the β-hairpin (residues 454–470) and the last 27 C-terminal residues of gp18M are shown in cyan. (D) Domain positions on the amino-acid sequence, using the same colour scheme as in (A), (B) and (C). Brown indicates the part of gp18 for which the atomic structure remains unknown.
The structure of gp18M includes that of gp18PR and consists of domains I, II and III (Figure 1). Domain I (residues 98–188) is a six-stranded β-barrel plus an α-helix. Domain II (residues 88–97 and 189–345) is a two-layer β-sandwich, flanked by four small α-helices. Taken together, domains I and II form the protease-resistant fragment (Figure 1A). Domain III (residues 20–87 and 346–510) consists of a β-sheet with five parallel and one anti-parallel β-strands plus six α-helices, which surround the β-sheet (Figure 1B and C). The 20 N-terminal amino-acid residues as well as residues 484–496 were not ordered in the gp18M crystal structure. The N- and C-termini of the structure are close in space, suggesting that the first 20 residues and residues 510–659 form an additional domain, domain IV, of the full-length protein. Individual domains were used to search for similar structures in the Protein Data Bank (PDB) using the DALI server (Holm and Sander, 1996), but no significant hits were found.
Fitting of the crystal structure into the cryo-EM map of the extended and contracted tail sheaths
The tail of bacteriophage T4 was one of the first objects for electron microscopy and image reconstruction, establishing that gp18 molecules (tail sheath subunits) are arranged into a six-start helix (De Rosier and Klug, 1968). The tail sheath structure can also be described as a stack of disks (or annuli), each with six subunits. Recently, the cryo-EM structures of the T4 tail sheaths in the extended and contracted conformation had been determined to 15 and 17 Å resolution, respectively (Leiman et al, 2004; Kostyuchenko et al, 2005).
The gp18 structure was computationally fitted into the cryo-EM densities reported previously. For both the contracted and the extended tail sheath reconstructions, the best fit was about 5 standard deviations better than any other fit (see Materials and methods). The crystal structure of gp18M fits into the cryo-EM densities of both the extended and the contracted tail sheath as a rigid body (Figure 2). Although the previous segmentation of the cryo-EM densities is consistent with the fitting of the crystal structure, the resolution was insufficient to conclude that there was no conformational change in the gp18 structure during contraction (Leiman et al, 2004). Nevertheless, at the present resolution, small changes in domain–domain positions cannot be excluded.
Figure 2.
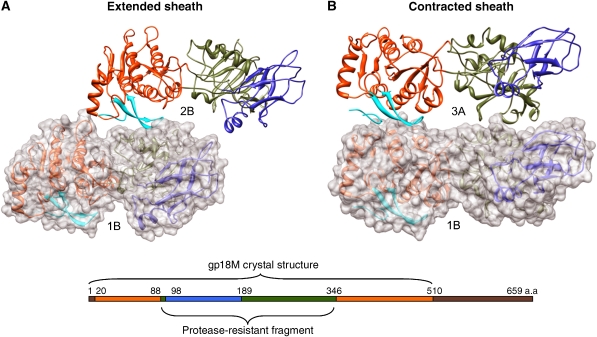
Fit of the gp18M structure into (A) the extended (left) and (C) the contracted (right) tail cryo-EM densities. Shown for each conformation is a one-disk-thick slab (top left and right), a two-disk-thick slab (middle left and right), a closer view of the fit (bottom left and right) and the whole tail (center left and right). In both (A and C) top panels, the one-disk thick slab of density is shown with six gp18M molecules fitted into the density with their domains I, II and III coloured blue, olive green and orange red, respectively. In the two-disk-thick slabs of density shown in (A) and (C), the tail tube density is colored red and two sequential disks of the tail sheath are colored blue and green. In the closer view of the fit, four disks of the tail sheath are shown, with disks one and three colored green and disks two and four colored blue. In (B), the arrangement of the gp18M domains is shown in the linear sequence (top), in the ribbon diagram of the crystal structure (left) and in the structure fitted into the piece of density map that corresponds to full-length gp18 (right).
The whole of the gp18 molecule is S-shaped and consists of four domains, three of which comprise gp18M. The fitting results suggest that the fourth C-terminal domain forms the inner part of the sheath. To determine if domain IV (whose atomic structure remains unknown) changes its orientation relative to the three other domains of gp18 during contraction, the whole gp18 subunit was masked out of the extended tail map based on the density connectivity and placed into the contracted sheath map (Figure 3). This showed, within the limits of the present resolution, that domain IV does not change its orientation relative to the other domains of gp18 during contraction, showing that the whole of the sheath protein moves as a rigid body despite the drastic change in its orientation and position.
Figure 3.
Relative position of two gp18 molecules belonging to the same helical strand in the extended and contracted tail sheath. (A) Four successive rings of electron density of the extended tail sheath with the densities of two gp18 molecules. (B) Superposition of the gp18 density extracted from the extended sheath onto the contracted sheath. The top panels in both (A) and (B) show the surface of the sheath, whereas the lower panel is a cut-away view showing the arrangement of the inner domains (brown). The cut-away plane is shown on a diagrammatic top view of the sheath with the direction of view indicated by an arrow. The gp18 molecule densities are colored as shown in the linear sequence diagram, with domains I, II, III and IV colored blue, green, orange and brown, respectively.
The fitting of the structure showed that the protease-resistant part of gp18M is exposed on the tail surface, whereas the N- and C- termini are positioned further towards the interior of the tail sheath (Figure 2A and C). The exposed and buried residues in each conformation of the sheath are in agreement with previous chemical modification of the exposed tyrosines (Takeda et al, 1990) and immunolabelling studies in showing that the protease-resistant part of the protein is accessible (Arisaka et al, 1990). Domain I of gp18 is protruding outwards from the tail and is not involved in inter-subunit contacts. The other three domains form the core of the tail sheath (Figure 2A and C), with domains III and IV being the most conserved parts of tail sheath proteins among T4-related bacteriophages (Supplementary Figure 1). Although the tail sheath proteins of other Myoviridae phages do not show any clear sequence homology to gp18, they appear to have similar helical parameters (Donelli et al, 1972; Cremers et al, 1977; Parker and Eiserling, 1983; Müller et al, 1994; Fokine et al, 2007) and function in a similar manner. Thus, in view of the general conservation of structural proteins in bacteriophages, it is most likely that most Myoviridae phages have a similar tail sheath structure.
Despite the fact that domain I has apparently no role in gp18–gp18 interactions, in the extended tail sheath, this domain binds to the baseplate (Figure 2A and Supplementary Figure 2A). Thus, one of the roles of domain I may be to initiate sheath assembly and contraction. Domain I also binds the long tail fibres when they are retracted. It was shown previously that three mutations in domain I inhibit fibre retraction (Takeda et al, 2004). These single mutations (Gly106Ser, Ser175Phe or Ala178Val) map to two loops close to the retracted tail fibre attachment site on the surface of the extended tail sheath (Supplementary Figure 2B), presumably abrogating binding of the tail fibres.
Contraction of the tail sheath
There are two ways in which the tail sheath contraction can be initiated. The first way can be induced by urea or pH change (Coombs and Arisaka, 1994), in which case the baseplate changes its conformation from dome- to star-shaped while remaining attached to the distal end of the tail sheath similarly to events in vivo. The second way is to induce tail contraction by heat (Arisaka et al, 1981) or by cationic detergent (To et al, 1969), in which case the baseplate stays in its dome-shaped conformation while being attached to the tail tube. Thus, in both cases, sheath contraction is initiated by change in the contacts with the baseplate. Unfortunately, the structures of the baseplate proteins in contact with the sheath are not known and hence the mechanism by which the sheath contraction is initiated has yet to be determined.
The tail sheath contraction can be divided into several steps. Previous studies of partially contracted sheath showed that subsequent to the rearrangement of the baseplate, the conformational changes of the sheath are propagated ‘upwards' starting from the disk of the gp18 subunits closest to the baseplate (Moody, 1973). The cryo-EM reconstructions showed that during contraction, the tail sheath pitch decreases from 40.6 to 16.4 Å and its diameter increases from 240 to 330 Å (Leiman et al, 2004; Kostyuchenko et al, 2005). Owing to the apparent change in the tail sheath appearance during contraction, it was suggested previously that the gp18 undergoes a large conformational rearrangement. However, the present results show that gp18 monomers remain rigid during contraction and move about 50 Å radially outwards while tilting ∼45° clockwise, viewed from outside the tail, about an approximately radial axis passing through the specific subunit. In the extended sheath, each gp18 subunit interacts with two neighbouring subunits within one helical strand and with two subunits on either side within a disk (Figure 4A). After disk expansion induced by the baseplate, the interactions between neighbouring subunits within a disk are broken, leaving only the interactions between the subunits within each helical strand (Figure 4B). As a result, the subunits from the disk above get inserted into the gaps formed in the disk below, increasing by about four times the contact area between gp18 molecules. During contraction, new contacts are formed and old ones are broken between the three domains of gp18 and the subunits in the three sequential disks (Figure 4). Previously, it was suggested that a part of gp18 structure should retain the initial connectivity within subunits in the same helix to keep the integrity of the whole structure (Moody, 1973; Leiman et al, 2004). Here, it is shown that the inner domain (domain IV) performs this function by retaining the initial connectivity between subunits within each helical strand (Figure 4). However, the outer part of gp18 forms new types of contacts during contraction, driving the rearrangement forward.
Figure 4.
Connectivity between subunits in the extended and contracted tail sheath. The subunits that form three of the six neighbouring helices (pink, A; blue, B; and green, C) within the sheath are shown as surface representations of (A) the extended and (B) the contracted sheath. The successive hexameric disks are numbered 1, 2, 3, 4 and 5, with the disk being closest to the baseplate numbered 1. On the left is shown a surface representation side view of the tail. Immediately next to it is shown the whole view with a closer view of the three neighbouring helices (in pink, blue and green). In the same column as the closer view of the helical arrangement is shown, the schematic diagram of the arrangement of the three outer domains using the same colour scheme with each gp18 molecule is being represented by a cross. Right next to this is shown the arrangement of the inner helices consisting only of domains IV. This domain retains the connectivity between neighbouring subunits within each helix in both (A) the extended and (B) the contracted sheath. At the right of each panel is shown the top view of one disk of the tail sheath in the (A) extended and (B) contracted conformations. The three outer domains that correspond to the atomic structure are shown in ribbon representation. Domain IV, whose structure has not yet been determined, is shown as density segmented from the cryo-EM map.
To obtain the intermediate positions of gp18 during the sheath contraction, the rotational and translational components of the transformation matrix that superimposes the gp18M structure of the extended sheath onto the contracted sheath has been divided into several steps. The wave-like manner of contraction has been generated by assuming that gp18 molecules from the disk above are one step behind in comparison with gp18 from the disk below. The domain IV, absent in the crystal structure, was added to the gp18 coordinates by converting the density grid points cut from the extended tail sheath to the coordinate file (see Materials and methods). Two movies showing the contraction of nine sequential disks of the sheath have been made: one showing the complete gp18 molecule (the three domains of the gp18M crystal structure and domain IV from the cryo-EM) (Supplementary Movie 1) and the other showing the inner structure made of domain IV alone (Supplementary Movie 2). The amino acids from the three domains of the gp18 molecule that are involved in interactions with four neighbouring subunits in the extended, intermediate and contracted conformations are summarized in Table I and more details are given in Supplementary Table I. In the intermediate state, the gp18 subunit used for the analysis remains partially in contact with the helical strand of the extended tail while also making new contacts to the neighbouring strands. As can be seen in Table I, the number of residues involved in interactions increases as contraction progresses. During the initial steps of contraction, the number of hydrophobic residues involved in contacts increases the most. However, during the final steps of contraction, there is a greater increase in the amount of charged and polar interactions, thus creating specific contacts that provide directionality for binding of a subunit to a subunit.
Table 1.
Summary of the surface residues of gp18 M involved in gp18–gp18 contacts
| Extended tail sheath | Intermediate state | Contracted tail sheath | |
|---|---|---|---|
| Number of residues involved in contacts with other gp18 molecules | 29 | 69 | 98 |
| Hydrophobic residues no. of residues, % of contact residues) | 10 residues, 34.5% | 34 residues, 49.3% | 41 residues, 41.8% |
| Charged residues (no. of residues, % of contact residues) | 13 residues, 44.8% | 20 residues, 29% | 34 residues, 34.7 % |
| Polar residues (no. of residues, % of contact residues) | 6 residues, 20.7% | 15 residues, 21.7% | 23 residues, 23.5% |
Polymerization properties of gp18
As was shown previously (Poglazov et al, 1999), most gp18 mutants with deletions of about 250 C-terminal residues can form polysheaths. Nevertheless, several mutants with substitutions of the C-terminal residue, including gp18M, lose their polymerization ability (Efimov et al, 2002). All of the deletion mutants that do not polymerize in solution differ by less than 10 C-terminal amino acids, whereas mutants that are either a few residues smaller or larger retain polymerization properties (Figure 5). In the crystal structure of gp18M, the last 27 residues (484–510) form a loop that interacts with a β-hairpin formed by residues 454–470 (Figure 1C). The residues covered by this loop and the β-hairpin are involved in inter-subunit contacts in both the extended and in the contracted sheath, although in different ways (Figure 6). It is, therefore, possible that this loop disrupts the important polymerization interface and/or fixes the β-hairpin in a specific position, preventing its interaction with another gp18 subunit. Thus, in the full-length gp18 and in the deletion mutants larger than gp18M, this 27-residue region (residues 484–510) might be a linker between domains III and IV and have a different structure and position, exposing the surface and not contacting the β-hairpin. This is supported by the fact that gp18 mutants with small internal deletions within the loop (residues 507–522) but with the intact C-terminal sequence retain their polymerization properties (Efimov et al, 2002). In gp18 mutants that are only a few residues shorter than gp18M, the shorter loop possibly fails to interact with the β-hairpin and/or fails to cover the important polymerization interface. It is also possible that C-terminal mutations in the deletion mutants have an important function in disrupting the inter-subunit contacts, therefore changing the protein polymerization properties. Likewise, the deletion mutant with 507 residues forms polymers, whereas another mutant of the same size (506Asn) does not.
Figure 5.
Analysis of polymerization properties of gp18 deletion mutants. The mutants that can polymerize (including the full-length protein) are shown in blue and those that do not polymerize are shown in red. The gp18M construct is indicated by an arrow. The region of the sequence that influences polymerization properties is shown in the red rectangle and corresponds to the C-terminal 27 residues of the gp18M structure. Internal deletions are shown as thin lines, with deleted residues being indicated on top. The C-terminal mutations are shown with the three-letter amino-acid code.
Figure 6.
Interactions between the gp18 subunits that involve β-hairpin (residues 454–470). Neighbouring gp18 subunits are shown for (A) extended and (B) contracted tail sheaths. One of the subunits is shown in transparent surface representation and both subunits are shown in ribbon representation. Gp18 domains colored as in linear sequence diagram with β-hairpin as well as the last 27-residue loop are shown in cyan. The interacting subunits correspond to subunits 1B and 2B of the extended tail sheath and subunits 1B and 3A for the contracted sheath according to the labelling in Figure 4.
The energy of contraction
The tail of T4 is a unique motor, designed to function only once. As was pointed out previously by Moody (1973), this irreversibility suggests that there is no requirement for chemical energy to return the system to its initial state in contrast to many cellular motors. Despite the fact that GTP binding was proposed in early studies (Serysheva et al, 1984, 1992), neither binding nor hydrolysis of ATP or GTP have been observed during assembly and contraction. Thus, it is not surprising that there is no known nucleotide binding fold in the partial gp18 structure presented in this work. Furthermore, it is unlikely that the yet to be determined C-terminal domain of gp18 has a nucleotide binding fold because it functions to keep the integrity of the tail sheath. The observations that contraction can be induced by various chemical and physical factors, such as heat, urea and pH change, suggest that the contracted sheath has a lower energy in comparison with the extended sheath (Coombs and Arisaka, 1994). Additionally, polysheaths that self-assemble from gp18 alone in vitro have helical parameters of the contracted state and never of the extended state (Moody, 1967). Thus, the tail has to be assembled into a stable, extended structure that has higher energy than the contracted conformation. Assembly of the extended tail sheath happens only in the presence of the baseplate–tail tube complex that determines the orientation of gp18 subunits, resulting in a complex that has a lower free energy per subunit than individual monomeric proteins. Additionally, the formation of the sheath in the extended conformation occurs about 50 times faster than the formation of polysheaths and, therefore is a preferred pathway due to a lower activation energy barrier (Arisaka et al, 1979; Tschopp et al, 1979). A similar situation occurs in, for instance, enveloped viruses when viral glycoproteins are assembled in a prefusion form that is thought to be of a higher energy state than the post-fusion conformation. The analysis of the tail sheath structures together with previous data suggests that the tail sheath contraction is driven by enthalpy gain from the increase in the inter-subunit binding energy.
Materials and methods
Protein expression, purification and crystallization
The deletion mutant gp18M was expressed and purified as described previously (Efimov et al, 2002). After extensive screening, only one crystallization condition was found: 12% PEG 20 000 buffered with 0.1 M MES (pH 6). Despite numerous optimization attempts, most crystals diffracted to 6 Å resolution at best. However, one crystal was found that diffracted to 3.5 Å resolution. A seleno-methionine (SeMet) derivative of gp18M was obtained using the method described by Ramakrishnan et al (1993). Unfortunately, it did not produce any diffracting crystals. Nevertheless, it was used to make a SeMet derivative of a smaller protease-resistant fragment, gp18PR, described previously by Arisaka et al (1990). Gp18PR was obtained by incubation of gp18M at 0.5 mg/ml, with trypsin at 1 μg/ml for 16 h at room temperature and subsequent ion-exchange chromatography. The crystallization condition of the SeMet derivative of gp18PR contained 1.2 M sodium acetate and 0.1 M imidazole. Mass-spectrometry analysis of the native and SeMet gp18PR showed that two Se atoms were present per protein molecule and that the protease-resistant fragment was larger than described previously (Arisaka et al, 1990). The subsequent structure determination confirmed this observation and showed that residues 86–361 were visible in the electron density map. This suggested that trypsin had cut the protein at Arg 365, rather than at Lys 316 as reported previously.
Data collection, structure determination and refinement
Gp18PR crystals were washed in cryoprotectant solution, containing mother liquor with the addition of 25% glycerol, for 20–30 s before flash-freezing at 100 K in the nitrogen stream. All screened gp18PR crystals had similar quality and belonged to spacegroup F432. Three wavelength MAD data sets were collected at the Advanced Proton Source, GM/CA, beam line 23 ID-D. Data sets were indexed, integrated and scaled using DENZO and SCALEPACK (Otwinowski and Minor, 1997). Heavy atoms' positions were determined with the program SHELXD (Usón and Sheldrick, 1999) using data to 2.8 Å resolution. The correlation coefficient between the anomalous differences dropped below 30% in the next resolution shell. Subsequently, phasing and density modification were performed in SHELXE (Usón and Sheldrick, 1999) using data to 2.2 Å resolution. The atomic model was built manually with the help of the program COOT (Emsley and Cowtan, 2004). The programs CNS (Brünger et al, 1998) and PHENIX (Afonine et al, 2005) were used to 2.2 and 1.8 Å resolution in initial and final stages of refinement, respectively (Table II and Supplementary Figure 3).
Table 2.
Data collection and refinement statistics
| Crystal gp18PR, SeMet derivative | Crystal gp18 M | |
|---|---|---|
| Data collection | ||
| Space group | F432 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 203.66 | 99.591 |
| 203.66 | 116.288 | |
| 203.66 | 433.759 | |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50 (1.8)a | 50 (3.5) |
| Rmerge | 6.4 (79.4) | 10.6 (29.1) |
| I/ΔI | 26.8 (2) | 11.9 (3.2) |
| Completeness (%) | 96.6 (95.5) | 87.1 (61.1) |
| Redundancy | 5.5 (4.4) | 3.7 (2.7) |
| Refinement | ||
| Resolution (Å) | 36 (1.8) | 50 (3.5) |
| No. of reflections | 55815 | 28369 |
| Rwork/Rfree | 18.98/21.94 | 26.54/29.86 |
| R.m.s. deviations | ||
| Bond length (Å) | 0.005 | 0.006 |
| Bond angle (deg) | 1.013 | 1.054 |
| aValues in parentheses are for highest-resolution shell. | ||
The best crystal of gp18M diffracted to 3.5 Å resolution and belonged to spacegroup C2221 with four molecules per asymmetric unit. The data set was collected at the Advanced Photon Source, GM/CA, beam line 23 ID-D. It was indexed, integrated and scaled using DENZO and SCALEPACK (Otwinowski and Minor, 1997). Out of 180 degrees of data collected, only the first 100 degrees were used due to significant radiation damage. The structure of gp18M was solved by the MR method using the program PHASER (Read, 2001), with gp18PR as a search model that constituted less than 12% of the unit cell content. The initial density calculated using phases from the MR solution was significantly improved by non-crystallographic symmetry averaging (NCS). It was possible to make a mask for the unknown domain and then to optimize NCS operators separately for the unknown and for the known parts of the gp18M structure using the RAVE program package from the Uppsala Software Factory (Kleywegt et al, 2001). Subsequently, the electron density was averaged and the solvent was flattened using the program DM (Cowtan, 1994). The programs CNS (Brünger et al, 1998) and PHENIX.REFINE (Afonine et al, 2005) were used in the initial and final stages of the refinement, respectively (Table II and Supplementary Figure 4).
Fitting of the crystal structures into the cryo-EM maps of the extended and contracted tail
The crystal structure of gp18M as a whole, as well as domains I and II combined (half of the gp18M molecule), were fitted into the cryo-EM density of the extended and contracted tail using the COLORES program from the SITUS package (Wriggers et al, 1999). The search was confined to the electron density corresponding to one disk of the tail sheath. The correlation coefficient corresponding to the best rotational fit at each grid point of the cryo-EM density was plotted as a three-dimensional ‘fit map'. The best fit within the asymmetric unit (a wedge of 60°) was 11σ above background for the extended tail and 10σ for the contracted tail, with the highest noise peak being about 6σ of the ‘fit map'. The r.m.s.d. between Cα positions when domains I, II and III were fitted together as compared with when domains I and II were used for fitting was 1.6 Å or less, showing the robustness of the fitting procedure.
The inter-subunit contact area in the initial and final states of contraction was calculated using the program AREAIMOL (Lee and Richards, 1971) assuming a 1.4-Å-radius probe. The residues that are in contact in the sheath were determined using the program CONTACT (from CCP4 program package by Tadeusz Skarzynski, Imperial College, London, 1.12.88).
A transformation matrix, which superimposes gp18M from the extended sheath onto the subunits in the contracted sheath, was calculated using LSQKAB (Kabsch, 1976). This matrix was used to create the contraction intermediates. The known helical parameters of the sheath were used to generate the intermediates for about one-third of the sheath structure (a total of 54 subunits). The rotational and translational components were divided into five steps, making the assumption that both rotation and translation of a subunit occur simultaneously. Once the intermediates were generated for one subunit, the intermediates for all other subunits were calculated using the helical parameters of the tail sheath. To create the contraction wave, which was observed in vitro by Moody (1973), the subunits were combined so that the closest disk to the baseplate changes first and each subsequent disk is a step behind. To assess what happens to the C-terminal domain absent in the gp18M structure, its density was cut out of the extended tail sheath using the program MAPROT (Stein et al, 1994) and a PDB file was created from the density by placing an atom at every grid point of the map using the program VOL2PDB from SITUS (Wriggers et al, 1999). Two movies were compiled from the calculated intermediate positions of gp18: one showing the whole of gp18 (Supplementary Movie 1) and the other showing only the inner structure composed of domain IV (Supplementary Movie 2).
Protein data bank accession numbers
The refined atomic models of gp18PR and gp18M have been deposited in the PDB with accession numbers 1FO8 and 1FOA, respectively. Additionally, fitted coordinates corresponding to one disk of the tail sheath (six gp18M molecules) were deposited in the PDB with accession numbers 1FOH and 1FOI for extended and contracted tail sheaths, respectively.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Information
Acknowledgments
We thank Siyang Sun, Ye Xiang and Anthony Battisti for helpful discussions and Sheryl Kelly for help in the preparation of the manuscript. We thank Paul Chipman, Valorie Bowman and Heather Holdaway for collecting some potentially useful cryo-EM data, although it was not used in this study. We thank the staff of beam line 23 (GM/CA) of the Advanced Photon Source for excellent support of our data collection. We thank ‘CCP4 school: from data processing to structure refinement and beyond', held in May 2008 at Argonne National Laboratory, for helpful discussions of refinement strategies and Pavel Afonine for advice on the use of the PHENIX.REFINE program. This work was supported by a National Science Foundation grant (MCB-0443899) to M.G.R., a Purdue Research Foundation grant to M.G.R. in support of A.A.A. and a Russian Fund for Basic Research grant (08-04-01260) to L.P.K.
Competing interests statement The authors declare that they have no competing financial interests.
References
- Ackermann H-W (2006) Classification of bacteriophages. In The Bacteriophages, Calendar R (ed), 2nd edn, pp 8–16. Oxford University Press: New York, NY [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Adams PD (2005) The Phenix refinement framework. CCP4 Newsletter. Number 42, Contribution 8 [Google Scholar]
- Arisaka F, Engel J, Horst K (1981) Contraction and dissociation of the bacteriophage T4 tail sheath induced by heat and urea. Prog Clin Biol Res 64: 365–379 [PubMed] [Google Scholar]
- Arisaka F, Takeda S, Funane K, Nishijima N, Ishii S (1990) Structural studies of the contractile tail sheath protein of bacteriophage T4. 2. Structural analyses of the tail sheath protein, gp18, by limited proteolysis, immunoblotting, and immunoelectron microscopy. Biochemistry 29: 5057–5062 [DOI] [PubMed] [Google Scholar]
- Arisaka F, Tschopp J, van Driel R, Engel J (1979) Reassembly of the bacteriophage T4 tail from the core-baseplate and the monomeric sheath protein P18: a co-operative association process. J Mol Biol 132: 369–386 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Cryst D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Coombs DH, Arisaka F (1994) T4 tail structure and function. In Molecular Biology of Bacteriophage T4, Karam JD (ed), pp 259–281. American Society for Microbiology: Washington, D.C. [Google Scholar]
- Cowtan KD (1994) ‘dm': an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31: 34–38 [Google Scholar]
- Cremers AFM, Schepman AMH, Visser MP, Mellema JE (1977) An analysis of the contracted sheath structure of bacteriophage Mu. Eur J Biochem 80: 393–400 [DOI] [PubMed] [Google Scholar]
- De Rosier DJ, Klug A (1968) Reconstruction of three dimensional structures from electron micrographs. Nature 217: 130–134 [DOI] [PubMed] [Google Scholar]
- Donelli G, Guglielmi F, Paoletti L (1972) Structure and physico-chemical properties of bacteriophage G: I. Arrangement of protein subunits and contraction process of tail sheath. J Mol Biol 71: 113–125 [DOI] [PubMed] [Google Scholar]
- Efimov VP, Kurochkina LP, Mesyanzhinov VV (2002) Engineering of bacteriophage T4 tail sheath protein. Biochemistry (Mosc) 67: 1366–1370 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Cryst D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG (2007) Cryo-EM study of the Pseudomonas bacteriophage fKZ. Structure 15: 1099–1104 [DOI] [PubMed] [Google Scholar]
- Goldberg E, Grinius L, Letellier L (1994) Recognition, attachment, and injection. In Molecular Biology of Bacteriophage T4, Karam JD (ed), pp 347–356. American Society for Microbiology: Washington, D.C. [Google Scholar]
- Holm L, Sander C (1996) Mapping the protein universe. Science 273: 595–602 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1976) A solution for the best rotation to relate two sets of vectors. Acta Cryst A 32: 922–923 [Google Scholar]
- Kemp P, Garcia LR, Molineux IJ (2005) Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340: 307–317 [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA (2001) Around O. In International Tables for Crystallograhy, Vol. F, Crystallography of Biological Macromolecules, Rossmann MG, Arnold E (eds), pp 353–356. Kluwer Academic Publishers: Dordrecht/Boston/London [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG (2005) The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol 12: 810–813 [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM (1971) The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55: 379–400 [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG (2004) Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118: 419–429 [DOI] [PubMed] [Google Scholar]
- Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG (2003) Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci 60: 2356–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MF (1967) Structure of the sheath of bacteriophage T4. I. Structure of the contracted sheath and polysheath. J Mol Biol 25: 167–200 [DOI] [PubMed] [Google Scholar]
- Moody MF (1973) Sheath of bacteriophage T4. III. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol 80: 613–635 [DOI] [PubMed] [Google Scholar]
- Moody MF, Makowski L (1981) X-ray diffraction study of tail-tubes from bacteriophage T2L. J Mol Biol 150: 217–244 [DOI] [PubMed] [Google Scholar]
- Müller M, Engel A, Aebi U (1994) Structural and physicochemical analysis of the contractive MM phage tail and comparison with the bacteriophage T4 tail. J Struct Biol 112: 11–31 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Parker ML, Eiserling FA (1983) Bacteriophage SPO1 structure and morphogenesis. I. Tail structure and length regulation. J Virol 46: 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plisson C, White HE, Auzat I, Zafarani A, Sao-Jose C, Lhuillier S, Tavares P, Orlova EV (2007) Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J 26: 3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poglazov BF, Efimov AV, Marco S, Carrascosa J, Kuznetsova TA, Aijrich LG, Kurochkina LP, Mesyanzhinov VV (1999) Polymerization of bacteriophage T4 tail sheath protein mutants truncated at the C-termini. J Struct Biol 127: 224–230 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–223 [DOI] [PubMed] [Google Scholar]
- Read RJ (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr Sect D 57: 1371–1382 [DOI] [PubMed] [Google Scholar]
- Serysheva II, Tourkin AI, Bartish IV, Poglazov BF (1992) GTPase activity of bacteriophage T4 sheath protein. J Mol Biol 223: 23–25 [DOI] [PubMed] [Google Scholar]
- Serysheva II, Tourkin AI, Venyaminov SY, Poglazov BF (1984) On the presence of guanosine phosphate in the tail of bacteriophage T4. J Mol Biol 179: 565–569 [DOI] [PubMed] [Google Scholar]
- Stein PE, Boodhoo A, Armstrong GD, Cockle SA, Klein MH, Read RJ (1994) The crystal structure of pertussis toxin. Structure 2: 45–57 [DOI] [PubMed] [Google Scholar]
- Takeda S, Arisaka F, Ishii S, Kyogoku Y (1990) Structural studies of the contractile tail sheath protein of bacteriophage T4. 1. Conformational change of the tail sheath upon contraction as probed by differential chemical modification. Biochemistry 29: 5050–5056 [DOI] [PubMed] [Google Scholar]
- Takeda Y, Suzuki M, Yamada T, Kageyama F, Arisaka F (2004) Mapping of functional sites on the primary structure of the contractile tail sheath protein of bacteriophage T4 by mutation analysis. Biochim Biophys Acta 1699: 163–171 [DOI] [PubMed] [Google Scholar]
- To CM, Kellenberger E, Eisenstark A (1969) Disassembly of T-even bacteriophage into structural parts and subunits. J Mol Biol 46: 493–511 [DOI] [PubMed] [Google Scholar]
- Tschopp J, Arisaka F, van Driel R, Engel J (1979) Purification, characterization, and reassembly of the bacteriophage T4D tail sheath protein P18. J Mol Biol 128: 247–258 [DOI] [PubMed] [Google Scholar]
- Usón I, Sheldrick GM (1999) Advances in direct methods for protein crystallography. Curr Opin Struct Biol 9: 643–648 [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA (1999) Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 125: 185–189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Information