Abstract
Dysregulation of growth and differentiation factor 5 (GDF-5) signalling, a member of the TGF-β superfamily, is strongly linked to skeletal malformation. GDF-5-mediated signal transduction involves both BMP type I receptors, BMPR-IA and BMPR-IB. However, mutations in either GDF-5 or BMPR-IB lead to similar phenotypes, indicating that in chondrogenesis GDF-5 signalling seems to be exclusively mediated through BMPR-IB. Here, we present structural insights into the GDF-5:BMPR-IB complex revealing how binding specificity for BMPR-IB is generated on a molecular level. In BMPR-IB, a loop within the ligand-binding epitope functions similar to a latch allowing high-affinity binding of GDF-5. In BMPR-IA, this latch is in a closed conformation leading to steric repulsion. The new structural data now provide also a molecular basis of how phenotypically relevant missense mutations in GDF-5 might impair receptor binding and activation.
Keywords: CDMP-1, protein recognition, protein specificity, skeletal malformation diseases, TGF-β superfamily
Introduction
Synovial joints are essential for the biomechanical function of the skeleton. As improper function, as observed in arthritic diseases, directly results in a severe loss of life quality, joint biology has been in focus of extensive research for years leading to an understanding of joint anatomy and histology as well as the biomechanical properties and roles of articular cartilage and other components in joint function and maintenance. However, little is known about how synovial joints acquire their structure in the developing embryo and in particular what factors are required for the differentiation of progenitor cells, which then give rise to each joint component (Pacifici et al, 2005). As a first sign of joint formation in the embryonic limb, an emergence of a mesenchymal interzone at each prospective joint site can be observed (Holder, 1977; Mitrovic, 1978). This interzone is a tripartite tissue structure composed of an intermediate cell layer and two outer cell layers with higher cell density. Interzone cells express a number of genes being involved in chondrogenesis such as Wnt-9a, Wnt-4, Noggin and growth and differentiation factor 5 (GDF-5) (Storm et al, 1994; Brunet et al, 1998; Hartmann and Tabin, 2001).
GDF-5, a member of the large TGF-β superfamily of secreted growth factors, shows chondrogenic activity and congenital GDF-5 mutations cause defects in digit, wrist and ankle joints in mice and humans (Storm et al, 1994; Thomas et al, 1997). The expression of GDF-5 is most strikingly limited to regions where joints will develop and is one of the earliest markers of joint formation (Storm and Kingsley, 1999). Similar to other TGF-β superfamily members, GDF-5 binds to and oligomerizes two types of membrane bound serine-threonine kinase receptors termed type I and II. Upon ligand binding, these complexes transduce signals by phosphorylating members of the SMAD family of transcription factors, which upon activation enter the nucleus and regulate transcription of responsive genes (Massague, 1996). Recent experiments have implicated two different type I receptors in skeletal patterning, BMPR-IA and BMPR-IB. Both receptors are expressed in dynamic patterns during normal development. In several limb structures, for example, in joint interzones and perichondrium, an overlapping expression of BMPR-IA and BMPR-IB is observed (Mishina et al, 1995; Zou et al, 1997; Baur et al, 2000). With regard to the BMPR-IA and BMPR-IB expression patterns, GDF-5 signal transduction should be accomplished by the interaction with both BMPR-IA and BMPR-IB (Chang et al, 1994; Zou et al, 1997). Null mutations in the bmpr-1b gene produce viable mice with defects in bone and joint formation that closely resemble those seen in mice missing GDF-5 (Storm and Kingsley, 1996; Yi et al, 2000), whereas bmpr-ia−/− mice are known to die early in embryogenesis (Mishina et al, 1995). However, a conditional knockout of BMPR-IA under the control of a GDF5-Cre driver bypasses embryonic lethality and produces viable mice with normally formed joints. But, after birth articular cartilage within the joints wears away in a process reminiscent to osteoarthritis, which points at the importance of this receptor in cartilage homoeostasis and repair (Rountree et al, 2004).
In the past, several single missense mutations in the mature part of the human GDF-5 have been described resulting in phenotypes such as brachydactyly A2 (BDA2), DuPan syndrome and symphalangism type I (SYM1) (for details, see Supplementary Table I). Interestingly, the majority of phenotypically relevant mutations occur within a central loop (pre-helix loop) of the so-called wrist epitope of GDF-5 that represents the binding site for BMP type I receptors. The corresponding pre-helix loop in BMP-2 harbours the main binding determinants for type I receptor interaction. In contrast to BMP-2, which binds BMPR-IA and BMPR-IB with almost identical affinities (KD∼1–3 nM), GDF-5 binds BMPR-IB in vitro with about 10- to 20-fold higher affinity (KD∼1–2 nM) as compared with BMPR-IA (KD∼15–20 nM). A mutagenesis study revealed that a single residue in GDF-5 located in the pre-helix loop, Arg57, solely determines the binding specificity for the BMP type I receptor IB (Nickel et al, 2005).
To date, neither the structure of GDF-5 bound to BMPR-IA nor to BMPR-IB has been reported. Only structure data for the unbound GDF-5 (Nickel et al, 2005; Schreuder et al, 2005) and other TGF-β members such as BMP-2 (Kirsch et al, 2000b; Keller et al, 2004; Allendorph et al, 2006; Weber et al, 2007), BMP-7 (Greenwald et al, 2003), activin-A (Thompson et al, 2003) or TGF-β3 (Hart et al, 2002; Groppe et al, 2008) in complex with either receptor subtype are currently available. A theoretical model for GDF-5 ligand–receptor complex based on these data has, however, failed to explain type I receptor specificity of GDF-5 (Nickel et al, 2005). Here, we present the crystal structure of GDF-5 bound to BMPR-IB allowing the deduction of mechanisms by which type I receptor specificity is encoded on a molecular level. Furthermore, these structural data allow for the first time an understanding of how phenotypically relevant missense mutations found in GDF-5 impair receptor binding and activation.
Results
Architecture of the complex of GDF-5 and its high-affinity type I receptor BMPR-IB
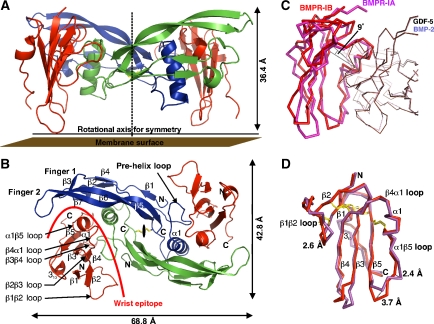
The crystal structure of the binary complex GDF-5 bound to the extracellular domain of BMPR-IB was determined to 2.1 Å resolution. The final structure model was obtained by refining native data (Rmerge is 7.1% for all reflections and 35.4% for those in the highest resolution shell) and exhibits an Rfree of 25.6% and an Rcryst of 21.5% (for further processing and refinement statistics, see Table I). The asymmetric unit contains one GDF-5 monomer with one BMPR-IB ectodomain bound, thus the biological assembly is formed by a two-fold crystallographic symmetry axis resulting in a fully symmetrical dimer assembly. The GDF-5 dimer exhibits a butterfly-shaped architecture, with a central core formed by the cystine-knot flanked by the two α-helices from each monomer. Two β-sheets comprised of four strands each form the two fingers per monomeric GDF-5 subunit. Four receptor-binding sites exist in the GDF-5 dimer, the type II receptor sites (knuckle) are located at the back of the two fingers, whereas the type I receptor-binding sites (wrist) are located in the cleft between the helix α1 of one monomer and the front side of the fingers 1 and 2 of the other monomer. The two BMPR-IB molecules in the complex bind to the wrist epitopes of GDF-5 (Figure 1), resembling a similar ligand–receptor assembly as also found for BMP-2 when bound to BMPR-IA (Kirsch et al, 2000b; Keller et al, 2004). However, closer inspection reveals clear differences, with the BMPR-IB moved upward by almost 2 Å compared with the complex BMP-2:BMPR-IA (Figure 1). This coincides with a change in the tilt angle of about 9° when comparing a single receptor molecule (BMPR-IB versus BMPR-IA) in the binary ligand–receptor complexes of GDF-5 (this study) and BMP-2 (PDB entry 1REW) (Figure 1). The change becomes most apparent when the Cα atoms of both dimeric ligands GDF-5 and BMP-2 are superimposed (residues 12–69, 75–114 of BMP-2 and 17–74, 81–120 of GDF-5) yielding an r.m.s.d. of only 0.86 Å, indicating that the structures of the ligands are highly similar. In contrast, the two receptor ectodomains of BMPR-IA and BMPR-IB are clearly differently placed in the wrist epitope of both superimposed complexes. A line through the Cα atoms of Phe66 (Phe85 in BMPR-IA), which marks the centre of rotation and Cys82 of BMPR-IB and the equivalent Cys101 in BMPR-IA in the central β-sheet shows that a single BMPR-IB rotates upward by an angle of 9°. Although this change in location and orientation for BMPR-IB in the wrist epitope of GDF-5 (compared with BMP-2:BMPR-IA) is far less pronounced compared with the differences found for the TGF-β type I receptor in the TGF-β3:TβR-II:TβR-I complex (Groppe et al, 2008), it clearly shows that the location and orientation of the receptor ectodomains in the ligand-binding sites of different BMPs can vary.
Table 1.
Data collection and refinement statistics for the GDF-5:BMPR-IB complex structure
| MAD dataa | Native dataa | |||
|---|---|---|---|---|
| Processing | ||||
| Space group | P42212 | P42212 | ||
| Unit cell | a=b=76.62, c=82.12 | a=b=76.46, c=82.78 | ||
| α=β=γ=90° | α=β=γ=90° | |||
| Wavelength (Å) | 0.97979 (inflection) | 0.97962 (peak) | 0.90789 (remote) | 1.10485 (native) |
| Resolution (Å) | 36.32–2.90 (3.00–2.90) | 38.31–2.60 (2.69–2.60) | 36.26–2.90 (3.00–2.90) | 34.19–2.10 (2.18–2.10) |
| Rmerge | 15.7 (49.2) | 10.0 (40.8) | 12.0 (39.0) | 7.1 (35.4) |
| I/σI | 6.2 (2.5) | 9.1 (3.5) | 8.6 (3.7) | 14.7 (5.3) |
| Completeness (%) | 99.8 (100.0) | 99.9 (100.0) | 99.9 (100.0) | 99.5 (100) |
| Redundancy | 6.69 (6.93) | 6.75 (6.85) | 6.1 (6.3) | 9.7 (9.9) |
| Refinement | ||||
| Resolution (Å) | 34.19–2.10 | |||
| No. of reflections | 14 062 | |||
| Rcryst/Rfree (%) | 21.5 (28.9)/25.6 (24.2) | |||
| No. of atoms | ||||
| Protein | 1484 | |||
| Water | 52 | |||
| B-factors | ||||
| Protein (Å2) | 71.2 | |||
| Water (Å2) | 60.9 | |||
| r.m.s.d. | ||||
| Bond length (Å) | 0.014 | |||
| Bond angles (deg) | 1.340 | |||
| Values in parentheses are for the highest resolution shell. | ||||
| aOne crystal was used to collect the diffraction data. | ||||
Figure 1.
Architecture of the complex of GDF-5 bound to BMPR-IB. (A) Ribbon representation of the full tetrameric complex of GDF-5 dimer (in blue and green) bound to the extracellular domains of two BMPR-IB molecules (red). A stippled line indicates the crystallographic two-fold axis. (B) As in (A), but viewed from the top. (C) The complex structures of GDF-5:BMPR-IB and BMP-2:BMPR-IA (PDB 1REW) were structurally aligned using the Cα atoms of both ligand dimers and the program Quanta2006. The ligand dimer superposition exhibits an r.m.s.d. of 0.86 Å (Cα of GDF-5: 17–74, 80–120 versus BMP-2: 12–74, 75–114). The ligand superposition clearly reveals that BMPR-IA and BMPR-IB are shifted in both complexes up to 2 Å. Further inspection shows that the BMPR-IB molecule (red) is tilted by 9° (angle between Cys82(BMPR-IB)–Phe66(BMPR-IB)–Cys101(BMPR-IA), see line) towards finger 2 of GDF-5 compared with the orientation of BMPR-IA (magenta) in complex with BMP-2. Residue Phe66 (Phe85 in BMPR-IA) presents the centre of rotation. (D) Despite the reorientation the core structures of both type I receptors are identical (r.m.s.d. 0.7 Å for β-sheet core without β1β2, β3β4 and α1β5 loops), only the β1β2 and α1β5 loops differ significantly.
The structure of the binding loop of BMPR-IB differs from that of BMPR-IA
Our complex structure GDF-5:BMPR-IB now yields data for a BMP type I receptor ectodomain other than BMPR-IA and thus allows to detect structural differences and variability among BMP type I receptors. The ectodomain of BMPR-IB shares about 50% identity on amino-acid sequence level with BMPR-IA (Supplementary Figure 1). Therefore, many of the secondary structure elements and the tertiary fold are conserved between BMPR-IB and BMPR-IA (Figure 1). However, a detailed comparison of the structures of both receptor ectodomains reveals some structural differences between BMPR-IB and BMPR-IA (PDB entry 1REW). The structural core comprising five β-strands superimposes well showing an r.m.s.d. of 0.7 Å, but considering all Cα in the ectodomains the r.m.s.d. rises to 2.2 Å, showing that the loop sections differ significantly between BMPR-IB and BMPR-IA. As binding and recognition by GDF-5 are mainly mediated through the β1β2 and the α1β5 loops of BMPR-IB this is of important biological consequence. Taken together, both loops contribute almost 80% of the buried surface area of the BMPR-IB ectodomain in the complex. These loops show large structural differences between BMPR-IB and BMPR-IA with Cα positions being shifted up to 3 Å in the β1β2 loop and up to 5 Å in the α1β5 loop. Thus, the binding determinants of the GDF-5:BMPR-IB and BMP-2:BMPR-IA (PDB entry 1REW) complexes likely differ not only due to the different orientations of the type I receptor ectodomains but also due to the differences present in the binding loops of BMPR-IB and BMPR-IA. Notably, the length of the α-helix, which carries the hot spot of binding for the BMP-2:BMPR-IA interaction, also varies between BMPR-IB and BMPR-IA. In BMPR-IA, the α-helix comprises residues Ser83 to Lys88 and hence has a length of more than 1.6 turns. In BMPR-IB, the α-helix is two residues shorter (Ser64 to Gln67) and consists of just one turn.
The GDF-5–type I receptor interaction in the GDF-5:BMPR-IB complex
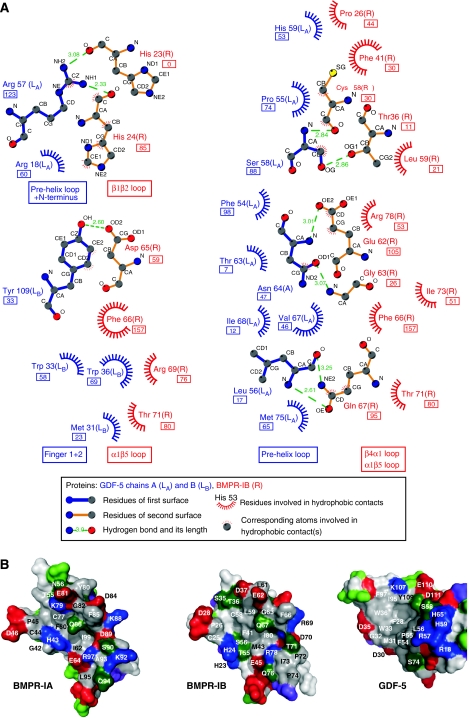
About 1040 Å2 solvent accessible surface of the BMP type IB receptor (for one receptor ectodomain of the dimeric assembly) and 1100 Å2 solvent accessible surface of GDF-5 (per type I receptor-binding site) are buried upon complex formation. Thus, considering the dimeric assembly with two BMPR-IB and two GDF-5 wrist epitopes involved, about 4280 Å2 of the protein surfaces are buried upon binding of GDF-5 to two BMPR-IB. Here, 22 residues of BMPR-IB (per molecule) and 21 residues of GDF-5 (per monomeric subunit) mark the contact interface. Of the 22 contact residues of BMPR-IB and the 21 contact residues of GDF-5 in the GDF-5:BMPR-IB interface, 15 are conserved with BMPR-IA and 13 are conserved with BMP-2 in the BMP-2:BMPR-IA interface (Figure 2). Nine intermolecular hydrogen bonds (H-bonds) are observed in the GDF-5:BMPR-IB contact (Supplementary Table II). Five of these involve residues in helix α1 and the α1β5 loop of BMPR-IB, suggesting that these elements are highly important for ligand recognition and binding. Interestingly, the bi-dentate H-bond between the conserved glutamine in helix α1 of the type I receptor (BMPR-IB:Gln67; BMPR-IA:Gln86) and the main chain polar groups of a conserved leucine in the pre-helix loop of the ligand (GDF-5 Leu56; BMP-2 Leu51) is also present in the GDF-5:BMPR-IB contact. However, whether this H-bond, similar as for the BMP-2:BMPR-IA interaction (Keller et al, 2004), presents the hot spot of binding for the GDF-5:BMPR-IB complex cannot be told from the structure.
Figure 2.
Interface of the GDF-5:BMPR-IB complex. (A) Ligplot (Wallace et al, 1995) analysis of the GDF-5:BMPR-IB interface. H-bonds are indicated as stippled lines with the residues involved shown as ball-and-stick models. Hydrophobic contacts are presented as spheres; the buried surface area of each contact residue is given in Å2 (boxed values below residues). Only hydrophobic contacts with buried surface areas ⩾5 Å2 are shown. (B) ‘Open book' view of the GDF-5:BMPR-IB complex with BMPR-IB rotated out of the interface by a 120° rotation in the y axis. The surface is colour coded by amino-acid polarity; red marks negatively charged residues, blue positively charged residues, green indicates polar uncharged amino acids and grey colour represents hydrophobic amino acids. Contact residues are indicated by residue number and amino-acid type in one-letter code. The contact surface of BMPR-IA in the BMP-2:BMPR-IA complex (PDB entry 1REW) is given for comparison.
Therefore, Gln67 and Phe66 in BMPR-IB were mutated to alanine, and their binding to GDF-5 and BMP-2, was tested by SPR. Surprisingly, the surmised hot spot of binding Gln67 showed only slightly decreased affinities for GDF-5 (6.3-fold) and BMP-2 (4.5-fold), respectively (Table II). Thus, in contrast to BMPR-IA, where the mutation BMPR-IAQ86A leads to almost 100-fold loss in affinity for BMP-2 (Keller et al, 2004), the conserved glutamine does not represent a hot spot of binding for BMPR-IB. Exchange of Phe66 in BMPR-IB by alanine, however, almost completely abolished binding to GDF-5 and BMP-2. Hatta et al (2000) observed that the affinity of the BMPR-IA variant F85A is decreased for BMP-2 only 15-fold (ΔΔG=1.5 kcal mol−1), suggesting that Phe85 is not a hot spot of binding in the BMP-2:BMPR-IA interaction. Thus, the conserved phenylalanine is crucial only for binding of BMPR-IB to BMPs, whereas the conserved glutamine seems important only for binding of BMPR-IA to the ligands. These findings corroborate our hypothesis that recognition and binding of BMPR-IB to BMPs differ from BMPR-IA.
Table 2.
Binding affinities of GDF-5 and BMP-2 to immobilized BMPR-IB variants
| GDF-5 | BMP-2 | |||
|---|---|---|---|---|
| KD (nM)a | ΔΔG (kcal mol−1)b | KD (nM)a | ΔΔG (kcal mol−1)b | |
| BMPR-IB | 1.3±0.55 | — | 4.8±1.80 | — |
| F66A | ⩾1000c | ⩾4.0 | n.b.d | ⩾4.5 |
| Q67A | 8.2±2.98 (6.3 × ) | 1.1 | 21.7±11.31 (4.5 × ) | 0.9 |
| H22S/H23G | 4.1±1.88 (3.2 × ) | 0.7 | 7.2±1.21 (1.5 × ) | 0.2 |
| aThe apparent binding constant KD was derived from calculating KD=koff/kon. Numbers in parentheses represent the relative change compared with wild-type BMPR-IB. | ||||
| bCalculated using ΔΔG=(−RTlnKD)wt−(−RTlnKD)var with R=1.98 cal mol−1 K−1 and T=293.15 K. Values ⩾2.0 kcal mol−1 identify a hot spot of binding. | ||||
| cThe apparent KD was estimated from the dose dependency of equilibrium binding and presents the lower limit due to technical limitations of the BIAcore2000 system. | ||||
| dNo binding above background levels could be detected, from the highest analyte concentration applicable in the analysis, the binding affinity was estimated to be ⩾10 μM. | ||||
GDF-5 passes through an induced fit upon complex formation
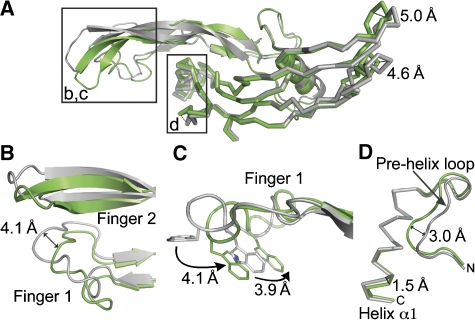
Comparison of free GDF-5 and GDF-5 bound to BMPR-IB shows that GDF-5 passes through an induced fit upon complex formation. Both fingers of GDF-5 move towards BMPR-IB upon binding, indicating that the opening of the wrist epitope is wider in the free form and upon BMPR-IB binding the fingers ‘wrap around' BMPR-IB to make packing tighter (Figure 3).
Figure 3.
Structural rearrangements in GDF-5 upon complex formation. (A) Superposition of free (grey, PDB entry 1WAQ) and GDF-5 bound to BMPR-IB (green) showing the structural rearrangements upon receptor binding. Regions of interest are highlighted in b, c and d. Distances between Cα positions are indicated. (B) BMPR-IB binding leads to shifts of up to 4 Å in fingers 1 and 2 of GDF-5. (C) The tryptophans 33 and 36 of GDF-5 change their side chain conformation upon type I receptor binding. (D) The C-terminal end of the α-helix moves towards the β-sheet of GDF-5 by 1.5 Å; also the pre-helix loop undergoes an induced fit upon BMPR-IB binding.
The finger 2 of GDF-5 moves as a rigid body with the side chains being pre-oriented in the free form. In contrast, in finger 1 of GDF-5 several side chains move significantly. In unbound GDF-5, Trp36 is pointing away from the receptor-binding epitope. In the complex, the Trp36 points towards the receptor, with the side chain atoms moving by almost 8 Å (Figure 3). These large rearrangements in the ligand are not observed in the BMP-2:BMPR-IA complex formation. In BMP-2, the movements in fingers 1 and 2 are less than 2 Å with the side chains being pre-oriented before receptor binding. The GDF-5 pre-helix loop also rearranges upon BMPR-IB binding. The changes in Cα position are comparable to those in BMP-2 upon binding to BMPR-IA (free BMP-2: 3BMP; bound BMP-2: 1REW).
On the basis of its homology to BMPR-IA, we also think that BMPR-IB passes through an induced fit mechanism upon ligand binding. NMR studies on the extracellular domain of free BMPR-IA showed that the β1β2- and especially the β4β5- loops are highly flexible and disordered in solution (Klages et al, 2008). Helix α1 is absent in free BMPR-IA (PDB entry 2K3G). As the helix of bound BMPR-IB is even shorter compared with BMPR-IA and thus probably less stable, this loop segment is likely similarly flexible in free BMPR-IB. Thus, both complex partners bind by an induced fit mechanism, which possibly represents a molecular mechanism to generate specificity as well as promiscuity.
The BMPR-IB specificity of GDF-5 is based on a spring-loaded latch in BMPR-IB
In contrast to BMP-2, GDF-5 is described to have a pronounced type I receptor specificity in vivo (Nishitoh et al, 1996). In vitro, type I receptor discrimination of GDF-5 seems less dramatic with respect to affinities for BMPR-IB (KD 1.3 nM) and BMPR-IA (KD 16.2 nM), showing that only a factor of 10–20 is sufficient for discrimination between BMPR-IB and BMPR-IA (Nickel et al, 2005). However, even this supposedly small difference is of great physiological significance for GDF-5 function in vivo as can be seen from the R57L mutation (R438L pro-protein numbering). The mutant GDF-5R57L exhibits an enhanced affinity for BMPR-IA (KD 4.2 nM) and nearly unaltered binding to BMPR-IB (KD 0.7 nM) (Table III). This correlates with the mutation of Arg57 to alanine that abrogates type I receptor specificity of GDF-5 completely (Nickel et al, 2005). Therefore, the mechanism encoding for BMPR-IB specificity should be in close proximity of GDF-5 Arg57. In the complex structure, GDF-5 Arg57 is completely hidden inside the ligand–receptor interface and shares contact with several residues of BMPR-IB. This is in contrast to the modelling studies, that suggested Arg57 pointing away from the interface out into the solvent (Nickel et al, 2005).
Table 3.
Binding affinities (KD (nM)) of GDF-5 variants to immobilized BMPR-IAec, -IBec and -IIec
| BMPR-IAa | BMPR-IBa | BMPR-IIb | |
|---|---|---|---|
| GDF-5 | 16.2±6.38 | 1.3±0.62 | 65.8±4.2 |
| GDF-5 R57L | 4.2±1.51 | 0.7±0.16 | 55.9±3.8 |
| GDF-5 R57A | 2.0±0.68 | 0.6±0.19 | 72.4±8.4 |
| GDF-5 L60P | n.b.c | 42.3±7.32 | 31.9±5.9 |
| GDF-5 ΔL56+S58T+H59L | ⩾1000d | ⩾1000d | 73.0±5.4 |
| aThe apparent binding constant KD was derived from calculating KD=koff/kon. | |||
| bKD values were evaluated from the dose dependency of equilibrium binding. | |||
| cNo binding above background levels could be detected. | |||
| dThe values for the apparent KD represent the lower limit estimated from the highest analyte concentration used in the SPR analysis. | |||
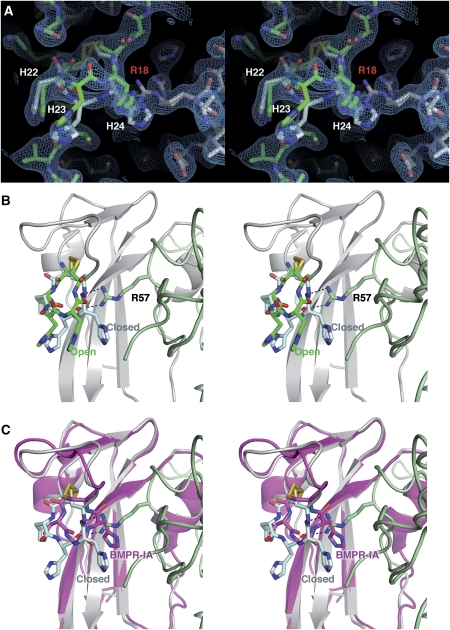
During refinement, two alternative conformations were detected for the β1β2 loop of BMPR-IB. As there is only one receptor ectodomain in the asymmetric unit—the full tetrameric GDF-5:(BMPR-IB)2 complex is formed by a two-fold crystallographic axis—both conformations are observed in the same BMPR-IB molecule (Figure 4). In the ‘closed' conformer, the β1β2 loop of BMPR-IB contacts Arg57, with the Arg side chain being clamped between the aromatic ring of Phe41 and the backbone of His23 and His24 of BMPR-IB. In this conformer, Arg57 is shielded from solvent and forms two H-bonds with the backbone carbonyl of His23 and His24 of BMPR-IB (see also Supplementary Figure 2). The second conformer presents an ‘open' conformation, in which the backbone of this section of the β1β2 loop moves away from Arg57 towards the solvent. This movement is due to a 180° flip in the Psi torsion angle of His22 resulting in an altered backbone route for BMPR-IB His23 and His24 (Supplementary Figure 3). The Cα atoms of the latter histidines relocate by 2.3 Å, possibly allowing water to fill the cleft nearby GDF-5 Arg57. The side chain conformation of Arg57 is unchanged in both conformations. As at the resolution of 2.1 Å, it is not reasonable to do occupancy refinement, the relative populations for both conformers can be estimated only from the electron density, suggesting that both conformations are roughly equally populated.
Figure 4.
A spring-loaded latch in BMPR-IB for high-affinity binding to GDF-5. (A) Stereoview of the two alternative conformations of the BMPR-IB β1β2 loop in the GDF-5:BMPR-IB complex. The electron density is contoured at 0.8σ. The open conformation is shown with the C atoms coloured in green and in the closed conformation the C atoms are shown in cyan. Arg18 of GDF-5 (grey), which also adopts two alternative conformations, is labelled in red. (B) Scheme (stereoview) of the two alternative β1β2 loop conformations (colour code for BMPR-IB as in (A), GDF-5 is in pale green) and their interaction with the specificity-determining residue of GDF-5 Arg57. In the closed conformation, two H-bonds are formed between the β1β2 loop and Arg57 of GDF-5. (C) Docking of BMPR-IA (magenta C atoms) onto BMPR-IB reveals that the BMPR-IA β1β2 loop will cause a steric clash with GDF-5 Arg57, if same conformation for the BMPR-IA β1β2 loop as in the complex BMP-2:BMPR-IA is assumed.
The local conformational change in the β1β2 loop possibly results from fixing the segment His22 to His24 in between the disulphide bond Cys21–Cys25. In the resulting pre-stressed loop, resembling a pentameric cyclic peptide, backbone torsion angle space in the N-terminal section of the β1β2 loop is limited to a few allowed states. In BMPR-IA, His23 is replaced by a glycine. This exchange probably removes the tension on the backbone between the conserved disulphide. Indeed, BMPR-IA Gly42 (His23 BMPR-IB) exhibits backbone torsion angles that are not amenable to non-glycine residues (Supplementary Figure 3). Thus, docking of the BMPR-IA ectodomain onto the GDF-5:BMPR-IB complex shows that the BMPR-IA β1β2 loop (Cys40–Cys44) would move towards the pre-helix loop of GDF-5 (using the above terminology, the β1β2 loop would then adopt a locked conformation). This cleft is, however, occupied by Arg57 leading to a steric clash and possibly explaining the lower binding affinity of GDF-5 for BMPR-IA (Figure 4).
Therefore, discrimination between BMPR-IB and BMPR-IA by GDF-5 might be based on a molecular latch mechanism, in which a receptor loop is ‘actively' moved away from a bulky residue in GDF-5; the type I receptor with lower affinity—BMPR-IA—has a mutation that removes the spring from this latch. Consequently, substitution of Arg57 in GDF-5 by smaller, less bulky residues increases the affinity to BMPR-IA by removing the steric clash. The fact that the GDF-5 mutations R57L and R57A do not lower the affinity for BMPR-IB (Nickel et al, 2005; Seemann et al, 2005) (Table III) despite the existence of two H-bonds between ligand and receptor can also be explained with the presence of the two conformations. Only one conformer allows the formation of the H-bonds, whereas the other conformer is open and water can fill in the cleft between Arg57 and the receptor loop. Thus, these two H-bonds are energetically silent due to entropy loss required for fixing the loop and the water molecules competing for intermolecular H-bond formation.
To test our hypothesis whether His23 controls the spring-lock mechanism, we have prepared the BMPR-IB variant H22S/H23G, in which the β1β2 loop between the two conserved cysteine residues reflects the situation found in BMPR-IA. However, SPR analysis of the BMPR-IB variant showed only a 3- to 4-fold reduced binding affinity for GDF-5 (KD 4.1 nM) and not the expected 10- to 20-fold decrease (Table II). This suggests that additional factors might contribute to the BMPR-IB specificity of GDF-5; one possible source could be the difference in the tilt angle. The differences in the tilt angle of the type I receptor might also influence the two conformations as the contacts between the β1β2 loop and the ligand differ through the different tilt angle thereby influencing the conformation of the latch.
Phenotypic mutations alter binding affinity of GDF-5 to BMP type I receptors
To date, eight missense mutations in the mature part of GDF-5 have been described leading to skeletal malformations such as brachydactyly, symphalangism or chondrodysplasia (Thomas et al, 1997; Akarsu et al, 1999; Everman et al, 2002; Seemann et al, 2005; Szczaluba et al, 2005; Wang et al, 2006). Exchanges of or substitution by cysteines were considered to cause protein misfolding (Thomas et al, 1997; Everman et al, 2002). The other mutations seem to be functional amino-acid exchanges and thus raise the question about their molecular mechanism in causing the disease. As no structural data are available for the GDF-5:type II receptor interaction, we focused on mutations affecting GDF-5:type I receptor interaction. Of the mutants described, three cluster in the type I receptor epitope, R57L (R438L pro-protein numbering), ΔL56/S58T/H59L (ΔL437/S439T/H440L pro-protein numbering) and L60P (L441P pro-protein numbering) (Supplementary Table I).
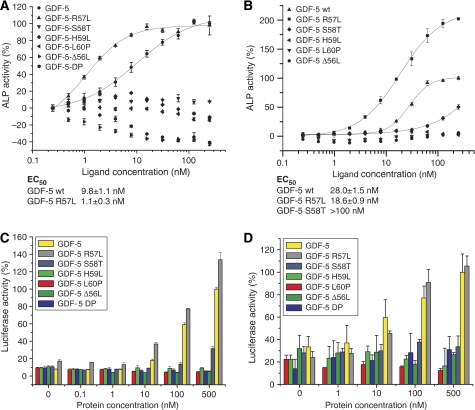
For two variants, GDF-5L60P and GDF-5ΔL56/S58T/H59L (in the following GDF-5DP), type I receptor binding was either abrogated (GDF-5DP) or greatly decreased (GDF-5L60P) (Table III). No induction of alkaline phosphatase (ALP) expression could be observed in cell-based assays (Figure 5). Leu60 is also conserved in BMP-2 and exchange for proline does not seem to cause a steric clash internally or in the contact with BMPR-IB. Thus, the most likely explanation for the loss of binding is in a local structural rearrangement of the preceding pre-helix loop of GDF-5, which is involved in several H-bonds with BMPR-IB. The mutation GDF-5DP was recently described in patients suffering from DuPan syndrome (Szczaluba et al, 2005) and leads to a complete loss of binding to BMPR-IA and BMPR-IB (Table III). As type II receptor binding seems to be not affected by the mutation, structural rearrangements are likely limited to the type I receptor epitope.
Figure 5.
Biological activities of phenotypically relevant GDF-5 variants. (A) Induction of ALP expression in RobC26 cells. GDF-5 exhibits an EC50 of about 10 nM, whereas R57L requires a 10-fold lower concentration for half-maximal stimulation. All other variants showed no ALP induction in RobC26 cells. (B) As in (A), but using ATDC5 cells that lack BMPR-IB receptor. The variant GDF-5R57L exhibits an ∼2-fold lower EC50 value correlating nicely with the two-fold increased affinity for BMPR-IA. The variant S58T, which is unable to induce ALP expression in RobC26 cells shows a residual activity in ATDC5 cells. (C) Reporter gene activation in C3H10T1/2 cells stably transfected with a p(BRE2)-Luc reporter construct. GDF-5 and its variant R57L with an increased affinity for BMPR-IA show a dose-dependent activation of the SMAD1/5/8 pathway. As in the ALP expression studies, all other variants that have either a decreased type I receptor affinity or exhibit an accelerated complex dissociation rate are inactive in SMAD activation. (D) As in (C), but using ATDC5 cells that were transiently transfected with the p(BRE2)-Luc reporter construct.
In contrast, exchange of Arg57 by leucine results in an enhanced affinity of GDF-5R57L to BMPR-IA (four-fold), whereas the affinity for BMPR-IB is almost unaltered (⩽2-fold) (Seemann et al, 2005). In ALP assays, GDF-5R57L—similar to GDF-5R57A (Nickel et al, 2005)—exhibits a lower concentration required for 50% stimulation (EC50) in both ATDC5 (Seemann et al, 2005) and RobC26 cells (Figure 5). In ATDC5 cells, which lack BMPR-IB and signalling thus utilizes the BMPR-IA, the EC50 is decreased from ∼30 nM for GDF-5 to 20 nM. In RobC26 cells, in which GDF-5 reportedly signals through BMPR-IB (Nishitoh et al, 1996; Erlacher et al, 1998), a larger shift is observed in the EC50 from 10 nM for GDF-5 to 1 nM for GDF-5R57L.
Additionally, we conducted reporter gene assays in ATDC5 cells and in the mouse fibroblast cell line C3H10T1/2 to directly measure SMAD1/5/8 activation. Consistent with the ALP assays, luciferase reporter activity is diminished for GDF-5L60P and GDF-5DP in both cell lines. Furthermore, the specificity impaired variant GDF-5R57L exhibits an enhanced activity in C3H10T1/2 cells and requires lower concentrations for half-maximal stimulation in ATDC5 cells (Figure 5).
Discussion
Mutations in either GDF-5 or BMPR-IB leading to skeletal malformation diseases such as brachydactyly or symphalangism suggest that both proteins are functionally tightly coupled in vivo (Lehmann et al, 2003; Seemann et al, 2005). Understanding how GDF-5 specifically interacts with BMPR-IB will thus yield important insights into the molecular mechanisms underlying those diseases. Although the complexes of GDF-5:BMPR-IB and BMP-2:BMPR-IA seem similar, numerous differences are observed. First, several regions in GDF-5 show considerable conformational rearrangements upon binding to BMPR-IB, whereas in BMP-2 a much smaller induced fit is observed only in a very limited area (Kirsch et al, 2000b; Keller et al, 2004). As—based on the structure of free BMPR-IA (Klages et al, 2008)—also BMPR-IB probably passes through an induced fit upon complex formation, the epitopes of both interaction partners seem ‘soft' and the final interface is developed when the complex is formed. The rearrangements seen in the GDF-5:BMPR-IB complex formation are on an intermediate level compared with the large changes in the ligand architecture observed for the TGF-β and activin-A ligand–receptor complexes on the one hand (Hart et al, 2002; Thompson et al, 2003) and the very rigid ligand architecture found in BMP-2/BMP-7 complexes on the other hand, in which the BMP ligands function as rather rigid clamps bringing type I and II receptors together (Kirsch et al, 2000b; Greenwald et al, 2003; Keller et al, 2004; Allendorph et al, 2006; Weber et al, 2007).
Second, the orientation of the BMPR-IB ectodomain differs from that of BMPR-IA in complex with BMP-2. Although the tilting of BMPR-IB in complex with GDF-5 compared with BMPR-IA in the BMP-2:BMPR-IA complex seems small (9°), the relative orientation of the type I and II receptor kinase domains could change presuming that the complete receptor is ‘rigid' and movements of the extracellular domain can propagate to the intracellular kinase domain. These small orientation differences now observed between different ligand–receptor complexes might explain how ligand-specific signals could be delivered by different BMP ligands, although the receptor composition of the complexes is identical. Koshland described different activation mechanisms (such as piston, rotation, see–saw and others) differing from the classical ligand-induced receptor oligomerization scheme, which allow single transmembrane receptors to activate and modulate intracellular signalling cascades (Ottemann et al, 1999). Thus, orientation differences on the outside can be relayed through a single transmembrane segment and influence the inside signalling cascade.
Another difference between GDF-5 and BMP-2 is in the way type I receptor specificity and promiscuity are achieved. In BMP-2 interactions with type I receptors, side chain and backbone flexibility in the ligand and the receptor ‘encode' for promiscuous binding by allowing adapting their surfaces to differing surface geometries. In the GDF-5:BMPR-IB complex investigated here, the flexibility of a loop can generate type I receptor specificity instead. The spring-loaded latch implemented in the BMPR-IB β1β2 loop seems required for high-affinity binding to GDF-5 by preventing a possible steric clash of this loop with the bulky Arg57 in GDF-5. In BMPR-IA, the ‘spring' is removed therefore resulting in an altered, unfavourable conformation of this loop. The rearrangement of the receptor loop upon GDF-5:BMPR-IA interaction requires energy and thus binding affinity of GDF-5 for BMPR-IA is reduced. Trials to directly manipulate the latch by removal of the ‘spring' residue His23 and replacing it with the corresponding Gly residue of BMPR-IA did, however, not show the maximal effect. One possible explanation might be in the differing orientation of both type I receptors in complexes with either GDF-5 or BMP-2, which could possibly influence the latch mechanism indirectly. Other mechanisms can also provide binding specificity in the TGF-β ligand–receptor interaction. For BMP-6, N-glycosylation of Asn73 in BMP-6 is required for binding to the activin type I receptor (ActR-I), whereas for binding to BMPR-IA or BMPR-IB presence of the carbohydrate moiety is dispensable (Saremba et al, 2008). In BMP-2, activation of a silent H-bond in the type II receptor epitope confers high binding affinity specifically to ActR-IIB, whereas binding to BMPR-II and ActR-II is not or only partially affected (Weber et al, 2007). Thus, the molecular mechanisms to generate receptor specificity to either type I or II BMP receptors are diverse throughout the BMP/TGF-β family.
Finally, two single missense mutations, S58T and H59L, in GDF-5 exhibit a surprising relationship between BMP type I receptor-binding affinity and biological activity. Both mutations render the variants inactive in ATDC5-based assays, although their equilibrium binding constants for BMPR-IA are basically unaltered (Figure 5; Supplementary Table III; Supplementary Figure 4). The unchanged equilibrium-binding constants are a result from a compensatory change in association and dissociation kinetics. Thus, the loss of ALP induction for these two GDF-5 mutants might be related to their shortened complex lifetime due to the increased dissociation rate. Whether a certain complex lifetime is required to allow efficient activation of the intracellular kinases is currently not clear; however, a similar effect has been described for human growth hormone (hGH). Mutations that increase the dissociation rate of hGH from its ‘capturing' receptor and hence lower the complex lifetime render hGH inactive (Pearce et al, 1999). Thus, further investigations are required, but the interesting signalling properties of these two mutants make them valuable tools to study the receptor activation mechanism of GDF-5.
Materials and methods
Recombinant protein expression and purification
The mature part of human GDF-5 and variants thereof were expressed in Escherichia coli and purified from inclusion bodies as described before (Nickel et al, 2005). For selenomethionine (Se-Met) labelling of GDF-5, Met-auxotroph E. coli B834 (DE3) cells (Novagen) carrying the expression plasmid were grown on M9 minimal medium supplemented with 50 mg l−1 (D,L)-Se-Met (Sigma). Similar to BMPR-IA, the BMPR-IB ectodomain was expressed in E. coli as a thioredoxin fusion protein (Kirsch et al, 2000b). The cells were disrupted by sonication and, after centrifugation, the supernatant was purified using metal ion affinity chromatography (Ni2+-NTA; Qiagen Inc.). After thrombin cleavage, thioredoxin and BMPR-IB proteins were separated by gel filtration chromatography. Monomeric BMPR-IB was collected and further purified by a second metal ion affinity chromatography step. Active BMPR-IB was finally obtained from affinity chromatography employing resin with immobilized BMP-2. Protein purity and homogeneity were analysed by SDS–PAGE and ESI FT-ICR mass spectrometry.
Complex preparation and crystallization
Complex purification and crystallization followed protocols similar to procedures published for BMP-2:BMPR-IA (Kirsch et al, 2000b). Details of the preparation and crystallization will be published elsewhere. Single crystals of approximately 200 μm × 100 μm × 100 μm in size consisting of two BMPR-IB molecules bound to wild-type and Se-Met-labelled GDF-5 dimer, respectively, grew in hanging drops over a reservoir buffer containing 0.1 M sodium acetate pH 5.25, 50% PPG400, 30 mM MgSO4 within 5 days at 21°C.
Data acquisition and structure analysis
Multiple anomalous dispersion (MAD) data (inflection, peak and remote) were collected from a single crystal at 100 K at the beamline BL14.2 at the Protein Structure Factory (BESSY, Berlin, Germany) from a 90° sweep (1° per frame) with a resolution of 2.9 Å. Native data were obtained from a single crystal at 100 K at the beamline X06SA at the Swiss Light Source (Paul-Scherrer-Institute, Villigen, Switzerland) by 125° rotation (1° per frame) with a resolution of 2.1 Å. Data were processed using CrystalClear 1.3.6 (Rigaku). Selenium positions were determined using SHELX and refined with SHARP/AutoSHARP version 2.2.0. One crystal component was defined for the low-resolution MAD data, a second crystal compound was defined for the high-resolution native data yielding a ‘MAD+native' scenario in SHARP. The peak data set gave the strongest signal yielding values for RCullis of 0.66, phasing power of 1.8 and r.m.s. lack-of-closure of 1.1 (resolution 38.3–4.6 Å). The figure-of-merit after density modification was 0.83. Automated tracing of the electron density using ARP/wARP yielded fragments, which could be assigned to structures of GDF-5 (PDB entry 1WAQ) and BMPR-IA (PDB entry 1REW) providing an initial model of the binary complex. After several rounds of rebuilding using the XBuild/Autofit tool of Quanta2006 and refinement using Refmac5.0.2, only the native high-resolution data (2.1 Å) were used for further improvement of the structure. The Procheck analysis shows that 138 (85.2%) of all residues reside in most-favoured regions. In total, 21 (13.0%), 2 (1.2%) and 1 (0.6%) of a total of 162 residues reside in additionally allowed, generously allowed and disallowed regions, respectively. Detailed processing and refinement statistics are given in Table I. Supplementary Figure 5 provides an insight into the quality of the electron density map.
Interaction analysis
Biosensor-based interaction analysis was performed as described previously (Kirsch et al, 2000a; Nickel et al, 2005). Receptor ectodomains were biotinylated and immobilized onto a streptavidin-coated CM5 sensor chip at a density of about 200 RU. Thus, binding affinities for the so-called 1:2 interaction, in which one ligand can bind simultaneously to two receptors on the biosensor surface, were obtained. GDF-5 or variants thereof was perfused over the sensor chip using six different analyte concentrations (10–120 nM). The sensorgrams were evaluated using the software BIAevaluation version 2.2.4. Bulk face effects were corrected by subtracting a control flow cell (FC1) sensorgram from all other sensorgrams and the data were fitted to a 1:1 Langmuir-type interaction. To obtain dissociation constants (KD) from the kinetic rate constants for complex formation (kon) and dissociation (koff), the last 20% of the association phase before reaching equilibrium and the data points of the association phase exhibiting a linear slope for the derivative ln(abs(∂(RU)/∂t)) were used in the analysis. This ensures that mass transport limitations and rebinding effects are excluded in the analysis. The χ2 statistics for this kind of data analysis yields χ2 values close to the noise level of the SPR system (0.5 RU). Owing to the experimental setup, all our equilibrium binding constants obtained by SPR are termed apparent KD values, indicating that the absolute values might differ from values obtained by other methods. However, in a direct comparison of values obtained under identical conditions a difference of more than two-fold is considered significant. Mean values plus/minus standard deviations for kon, koff and KD are indicated in Tables II and III and Supplementary Table III.
ALP assays
The teratocarcinoma AT508-derived cell line ATDC5 (RIKEN, no. RCB0565) was cultured in DMEM/HAMs F12 (1:1 v/v) medium containing 5% (v/v) fetal calf serum (FCS) and antibiotics (100 U ml−1 of penicillin G and 100 mg ml−1 of streptomycin). The osteoblastic cell line RobC26 isolated from neonatal rat calvariae (gift from Akira Yamaguchi) was cultured in α-MEM containing 10% (v/v) FCS and antibiotics. For ALP assays, the cells were serum-starved (2% FCS) and exposed to ligands for 72 h in 96-well microplates. After cell lysis, ALP activity was measured by p-nitrophenylphosphate conversion using an ELISA reader at 405 nm.
Reporter gene assays
The fibroblastic cell line C3H10T1/2 stably transfected with the BMP-responsive p(BRE2)-Luc reporter construct (gift from Peter ten Dijke) was cultured in DMEM containing 10% (v/v) FCS, 100 U ml−1 penicillin G, 100 mg ml−1 streptomycin and 200 μg ml−1 G418 sulphate. ATDC5 cells were cultured as described and transfected with 100 ng p(BRE2)-Luc and 400 ng pcDNA3.1 (empty vector) using lipofectamine and plus reagent (Invitrogen) according to the manufacturer's protocol. Cells were serum-starved (ATDC5: 2% FCS; C3H10T1/2: 0.1% FCS) and stimulated with ligands for 72 h in 96-well microplates. β-Galactosidase activity was determined by o-nitrophenyl galactopyranoside conversion at 405 nm. Luciferase activity was determined using the Promega luciferase assay kit.
Coordinate deposition
The atomic coordinates and structure factors for the structure of the GDF-5:BMPR-IB complex have been deposited with the Protein Data Bank (accession code 3EVS).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information
Acknowledgments
We thank B Midloch for excellent assistance and W Schmitz for mass spectrometry analysis. We gratefully acknowledge C Schulze-Briese, T Tomizaki (SLS), U Mueller and J Schulze (BESSY) for assistance during data acquisition and acknowledge the access to the beamlines X06SA and BL14.2 at the Swiss Light Source (SLS), Switzerland, and BESSY, Germany, respectively. We also thank C Kisker and H Schindelin for generous support and access to biophysics instrumentation facility at the Rudolf-Virchow Center, Würzburg. This project was supported by the Deutsche Forschungsgemeinschaft (DFG), SFB 487 TP B2. Author contributions: AK performed protein preparation, crystallization and structure analysis; JN performed the in vitro interaction studies and ALP expression assays; AS performed the reporter gene assays; JN, WS and TDM designed and supervised the experiments and AK, JN and TDM wrote the paper.
References
- Akarsu AN, Rezaie T, Demirtas M, Farhud DD, Sarfarazi M (1999) Multiple synostosis type 2 (SYNS2) maps to 20q11.2 and caused by a missense mutation in the growth/differentiation factor 5 (GDF5). In ASHG Annual Meeting, Vol. 1569, 19–23 October 1999. San Francisco, CA, USA [Google Scholar]
- Allendorph GP, Vale WW, Choe S (2006) Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci USA 103: 7643–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur ST, Mai JJ, Dymecki SM (2000) Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development 127: 605–619 [DOI] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM (1998) Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455–1457 [DOI] [PubMed] [Google Scholar]
- Chang SC, Hoang B, Thomas JT, Vukicevic S, Luyten FP, Ryba NJ, Kozak CA, Reddi AH, Moos M Jr (1994) Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J Biol Chem 269: 28227–28234 [PubMed] [Google Scholar]
- Erlacher L, McCartney J, Piek E, ten Dijke P, Yanagishita M, Oppermann H, Luyten FP (1998) Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J Bone Miner Res 13: 383–392 [DOI] [PubMed] [Google Scholar]
- Everman DB, Bartels CF, Yang Y, Yanamandra N, Goodman FR, Mendoza-Londono JR, Savarirayan R, White SM, Graham JM Jr, Gale RP, Svarch E, Newman WG, Kleckers AR, Francomano CA, Govindaiah V, Singh L, Morrison S, Thomas JT, Warman ML (2002) The mutational spectrum of brachydactyly type C. Am J Med Genet 112: 291–296 [DOI] [PubMed] [Google Scholar]
- Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S (2003) The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 11: 605–617 [DOI] [PubMed] [Google Scholar]
- Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP (2008) Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 29: 157–168 [DOI] [PubMed] [Google Scholar]
- Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP (2002) Crystal structure of the human TbetaR2 ectodomain–TGF-beta3 complex. Nat Struct Biol 9: 203–208 [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ (2001) Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell 104: 341–351 [DOI] [PubMed] [Google Scholar]
- Hatta T, Konishi H, Katoh E, Natsume T, Ueno N, Kobayashi Y, Yamazaki T (2000) Identification of the ligand-binding site of the BMP type IA receptor for BMP-4. Biopolymers 55: 399–406 [DOI] [PubMed] [Google Scholar]
- Holder N (1977) An experimental investigation into the early development of the chick elbow joint. J Embryol Exp Morphol 39: 115–127 [PubMed] [Google Scholar]
- Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD (2004) Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol 11: 481–488 [DOI] [PubMed] [Google Scholar]
- Kirsch T, Nickel J, Sebald W (2000a) BMP-2 antagonists emerge from alterations in the low-affinity binding epitope for receptor BMPR-II. EMBO J 19: 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch T, Sebald W, Dreyer MK (2000b) Crystal structure of the BMP-2–BRIA ectodomain complex. Nat Struct Biol 7: 492–496 [DOI] [PubMed] [Google Scholar]
- Klages J, Kotzsch A, Coles M, Sebald W, Nickel J, Muller T, Kessler H (2008) The solution structure of BMPR-IA reveals a local disorder-to-order transition upon BMP-2 binding. Biochemistry 47: 11930–11939 [DOI] [PubMed] [Google Scholar]
- Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik KH, Muller D, Knaus P, Nurnberg P, Mundlos S (2003) Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci USA 100: 12277–12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J (1996) TGFbeta signaling: receptors, transducers, and Mad proteins. Cell 85: 947–950 [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, Behringer RR (1995) Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev 9: 3027–3037 [DOI] [PubMed] [Google Scholar]
- Mitrovic D (1978) Development of the diarthrodial joints in the rat embryo. Am J Anat 151: 475–485 [DOI] [PubMed] [Google Scholar]
- Nickel J, Kotzsch A, Sebald W, Mueller TD (2005) A single residue of GDF-5 defines binding specificity to BMP receptor IB. J Mol Biol 349: 933–947 [DOI] [PubMed] [Google Scholar]
- Nishitoh H, Ichijo H, Kimura M, Matsumoto T, Makishima F, Yamaguchi A, Yamashita H, Enomoto S, Miyazono K (1996) Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J Biol Chem 271: 21345–21352 [DOI] [PubMed] [Google Scholar]
- Ottemann KM, Xiao W, Shin YK, Koshland DE Jr (1999) A piston model for transmembrane signaling of the aspartate receptor. Science 285: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Iwamoto M (2005) Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res C Embryo Today 75: 237–248 [DOI] [PubMed] [Google Scholar]
- Pearce KH Jr, Cunningham BC, Fuh G, Teeri T, Wells JA (1999) Growth hormone binding affinity for its receptor surpasses the requirements for cellular activity. Biochemistry 38: 81–89 [DOI] [PubMed] [Google Scholar]
- Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, Kingsley DM (2004) BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol 2: e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saremba S, Nickel J, Seher A, Kotzsch A, Sebald W, Mueller TD (2008) Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J 275: 172–183 [DOI] [PubMed] [Google Scholar]
- Schreuder H, Liesum A, Pohl J, Kruse M, Koyama M (2005) Crystal structure of recombinant human growth and differentiation factor 5: evidence for interaction of the type I and type II receptor-binding sites. Biochem Biophys Res Commun 329: 1076–1086 [DOI] [PubMed] [Google Scholar]
- Seemann P, Schwappacher R, Kjaer KW, Krakow D, Lehmann K, Dawson K, Stricker S, Pohl J, Ploger F, Staub E, Nickel J, Sebald W, Knaus P, Mundlos S (2005) Activating and deactivating mutations in the receptor interaction site of GDF5 cause symphalangism or brachydactyly type A2. J Clin Invest 115: 2373–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ (1994) Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature 368: 639–643 [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM (1996) Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development 122: 3969–3979 [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM (1999) GDF5 coordinates bone and joint formation during digit development. Dev Biol 209: 11–27 [DOI] [PubMed] [Google Scholar]
- Szczaluba K, Hilbert K, Obersztyn E, Zabel B, Mazurczak T, Kozlowski K (2005) Du Pan syndrome phenotype caused by heterozygous pathogenic mutations in CDMP1 gene. Am J Med Genet A 138: 379–383 [DOI] [PubMed] [Google Scholar]
- Thomas JT, Kilpatrick MW, Lin K, Erlacher L, Lembessis P, Costa T, Tsipouras P, Luyten FP (1997) Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat Genet 17: 58–64 [DOI] [PubMed] [Google Scholar]
- Thompson TB, Woodruff TK, Jardetzky TS (2003) Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand–receptor interactions. EMBO J 22: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Wang X, Xiao F, Yang Q, Liang B, Tang Z, Jiang L, Zhu Q, Chang W, Jiang J, Jiang C, Ren X, Liu JY, Wang QK, Liu M (2006) A novel mutation in GDF5 causes autosomal dominant symphalangism in two Chinese families. Am J Med Genet A 140: 1846–1853 [DOI] [PubMed] [Google Scholar]
- Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, Mueller TD (2007) A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM (2000) The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development 127: 621–630 [DOI] [PubMed] [Google Scholar]
- Zou H, Wieser R, Massague J, Niswander L (1997) Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes Dev 11: 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information