Abstract
The spindle assembly checkpoint monitors microtubule attachment to kinetochores and tension across sister kinetochores to ensure accurate division of chromosomes between daughter cells. Cytoplasmic dynein functions in the checkpoint, apparently by moving critical checkpoint components off kinetochores. The dynein subunit required for this function is unknown. Here we show that human cells depleted of dynein light intermediate chain 1 (LIC1) delay in metaphase with increased interkinetochore distances; dynein remains intact, localised and functional. The checkpoint proteins Mad1/2 and Zw10 localise to kinetochores under full tension, whereas BubR1 is diminished at kinetochores. Metaphase delay and increased interkinetochore distances are suppressed by depletion of Mad1, Mad2 or BubR1 or by re-expression of wtLIC1 or a Cdk1 site phosphomimetic LIC1 mutant, but not Cdk1-phosphorylation-deficient LIC1. When the checkpoint is activated by microtubule depolymerisation, Mad1/2 and BubR1 localise to kinetochores. We conclude that a Cdk1 phosphorylated form of LIC1 is required to remove Mad1/2 and Zw10 but not BubR1 from kinetochores during spindle assembly checkpoint silencing.
Keywords: cdk1, dynein, Mad2, mitosis, spindle assembly checkpoint
Introduction
To maintain genetic stability, cells must divide their replicated chromosomes equally between two daughter cells. Segregation of replicated chromatids occurs as cells progress from metaphase to anaphase, a transition monitored by the spindle assembly checkpoint (SAC) (Hoyt et al, 1991; McIntosh, 1991; Rieder et al, 1994; Zhou et al, 2002). The SAC monitors two parameters: microtubule attachment to kinetochores and tension across sister kinetochores (Rieder et al, 1994; Zhou et al, 2002). It is difficult to separate these parameters and to determine whether they provide two independent inputs to the SAC because they are structurally dependent: attachment is both needed for and actively strengthened with tension (King and Nicklas, 2000). What is clear is that the SAC ensures correct orientation of sister chromatids (i.e., attached and under tension) for proper segregation on anaphase onset.
The precise structural and molecular underpinnings of the SAC are still unresolved. The initial proposal—that an unattached kinetochore produces a ‘wait anaphase' signal (McIntosh, 1991)—is supported by studies in which laser ablation of the last unaligned kinetochore allows cells to progress to anaphase (Rieder et al, 1995). The main molecular components required for the checkpoint, the Mad and Bub protein families, were originally shown to be required for the wait anaphase signal in yeast and later in humans (Hoyt et al, 1991; Meraldi et al, 2004). Two members of these families, Mad2 and BubR1, function directly in the SAC by preventing degradation of cyclin B and securins and the onset of anaphase (Shah and Cleveland, 2000; Hoyt, 2001). Both localise to kinetochores and serve as indicators of an active ‘wait anaphase' signal (Waters et al, 1998; Howell et al, 2000; Hoffman et al, 2001) and both move off kinetochores before anaphase progression (Howell et al, 2004; Mayer et al, 2006; Zhang et al, 2007). Mad2 localises to kinetochores through Mad1 and moves off kinetochores along microtubules upon adequate microtubule attachment. Zw10, a member of the RZZ complex (Rod–Zw10–Zwilch) is implicated in anchoring of Mad1/2 to the kinetochore and appears to also move off the kinetochore with Mad2 (Buffin et al, 2005). BubR1 moves off kinetochores when tension across sister kinetochores is established (Chan et al, 1999; Hoffman et al, 2001; Skoufias et al, 2001). Thus, when both checkpoint criteria are fulfilled (attachment and tension), Mad2 and BubR1 are removed from kinetochores.
Although both Mad2 and BubR1 are recruited to kinetochores during SAC activation, their individual roles in the checkpoint are unclear. To date, there is no condition that activates the checkpoint in which one protein is absent from all kinetochores whereas the other is present on at least one; both are present on at least some kinetochores in cells with an active SAC. For example, both proteins localise to unattached kinetochores when microtubules are depolymerised, and BubR1 localises to all kinetochores and Mad2 to a few when microtubules are stabilised by taxol (i.e., attachment but no tension) (Waters et al, 1998). Additional experiments will be required to determine the separate roles of Mad2 and BubR1 in the SAC.
Cdk1(Cdc2)–cyclin B1 is the primary cyclin-dependent kinase complex in mitosis. It controls mitotic timing and ensures against premature anaphase onset, before sister chromatid segregation (Murray et al, 1989; Gorbsky et al, 1998; Chang et al, 2003; D'Angiolella et al, 2003). Anaphase onset is triggered only when cdk1 activity is downregulated through cyclin B1 destruction (Minshull et al, 1989; Clute and Pines, 1999; Chang et al, 2003; Reed, 2003). Cdk1 also plays an apparently antagonistic mitotic role by setting up an environment that leads to its own inactivation. Cdk1 provides Mad2/BubR1 with control over the timing of APC/C activation, and thus, the events of anaphase onset including cyclin B1 and securin degradation, cdk1 inactivation and chromatid separation (D'Angiolella et al, 2003). To accomplish this, cdk1 promotes binding of Cdc20, the APC/C activator, to Mad2 and BubR1 (D'Angiolella et al, 2003), providing Mad2/BubR1 with the capacity to inhibit the Cdc20-dependent activation of proteasome degradation and anaphase onset.
The multiprotein cytoplasmic dynein complex is a minus end directed microtubule motor involved in the SAC as well as other mitotic and interphase functions such as Golgi complex positioning, vesicle trafficking, spindle organisation and chromosome alignment (Corthesy-Theulaz et al, 1992; Merdes et al, 2000; Pfister, 2005). Dynein is comprised of six subunit families in animal cells including heavy, intermediate, light intermediate and three different light chain families (Pfister et al, 2006). It is required to move Mad2 and BubR1 off kinetochores in metaphase, thus releasing the SAC and permitting chromosome segregation (Hoffman et al, 2001). The Rab6A' GTPase facilitates dynein-mediated movement of Mad2 and BubR1 off kinetochores through its interaction with the p150Glued subunit of the dynein-modulating dynactin complex (Miserey-Lenkei et al, 2006). Dynein light intermediate chains (LICs) are required for mitosis in Caenorhabditis elegans (Yoder and Han, 2001) and become phosphorylated during animal cell division by cdk1–cyclin B1 (Dell et al, 2000; Addinall et al, 2001). This phosphorylation has been correlated with loss of the motor's interaction with vesicles (Addinall et al, 2001), although neither the mitotic role of these phosphorylation events nor the precise function of dynein LICs in mitosis are understood.
In this manuscript, we identify human LIC1 (DYNC1LI1) as a novel contributor to the SAC. LIC1 depletion induces a Mad1-, Mad2- and BubR1-dependent metaphase arrest that is suppressed by re-expressing a cdk1 site phosphomimetic form of LIC1, but not a nonphosphorylatable form.
Results
siRNA-mediated depletion of LIC1 induces a delay in metaphase
Human LIC1 (hLIC1) function was investigated by treating HeLa cells with small interfering RNAs (siRNAs) targeting hLIC1 or GFP (control). Protein levels were detected by immunoblotting using an antibody raised to a peptide sequence in hLIC1 that reacted with LIC1 but not LIC2 (Supplementary Figure S1). By 24–72 h after transfection, LIC1 protein levels were significantly reduced in HeLa cells (Figure 1A, also 3A, 4A, 5A, C).
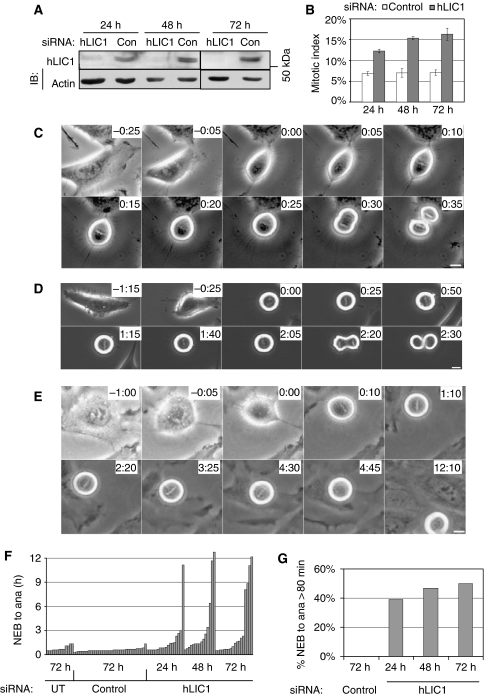
Figure 1.
LIC1-depleted cells delay in mitosis. (A) Immunoblot showing hLIC1 levels after treatment with control or hLIC1 siRNA for the indicated times. Actin, loading control; IB, immunoblot. (B) Mitotic index for control or hLIC1 siRNA-treated cells for indicated times (500–1000 cells were counted per bar, from three experiments). (C–E) Phase contrast images from time-lapse series. Control siRNA-treated Hela cell (C); hLIC1 siRNA-treated cell (D); hLIC1 siRNA-treated cell that did not initiate anaphase after 12 h but lost its metaphase plate (E, 4:30–4:45). 0:00 (h:min), bars, 10 μm. (F) Timing of individual cells (each bar) from NEB to anaphase onset (ana) under the indicated conditions. UT, untreated. (G) The percentage of mitotic cells that took >80 min (the longest control time) from NEB to ana (control: n=35 cells/bar, hLIC1: n=13–18 cells/bar) at the indicated times.
Depletion of hLIC1 led to an increase in mitotic HeLa cells (Figure 1B). Phase contrast time-lapse microscopy of hLIC1-depleted cells revealed a prolonged delay in mitosis (compare Figure 1C with D and E and Supplementary Video 1 with 2 and 3). Quantitative analysis showed that hLIC1-depleted cells were delayed between nuclear envelope breakdown (NEB) and anaphase onset (Figure 1F and G). Control siRNA-treated cells spent up to 80 min from NEB to anaphase onset (average 34 min at 72 h, n=35), whereas approximately 50% of hLIC1-depleted cells spent >80 min (average 184 min at 72 h, n=18) (Figure 1G). Of the LIC1 siRNA-treated cells, those delayed in mitosis showed lower LIC1 levels (up to seven-fold lower) than nondelayed cells (n=21 cells total) by immunostaining, consistent with a LIC1-dependent change in cell cycle progression. A mitotic delay was also observed in diploid hTERT-BJ1 cells and SAOS cells by time-lapse microscopy (Supplementary Figure S2) albeit weaker due to decreased protein depletion in these cell lines.
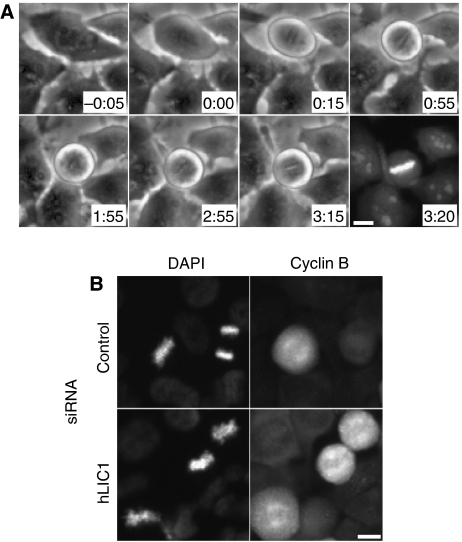
Analysis of chromosomes by phase contrast microscopy showed that cells were delayed in metaphase with normally aligned chromosomes (Figures 1D, E and 2A). Metaphase arrest was confirmed by immunofluorescence microscopy. Cells imaged for more than 100 min in mitosis and then stained with a vital DNA dye (Syto 13) showed normal metaphase chromosome alignment (Figure 2A, last panel; Supplementary Video 4). Time-lapse imaging confirmed that hLIC1 siRNA-treated Hela cells spent >2 h in metaphase as compared with approximately 30 min for control cells; the time spent in prophase or anaphase was comparable for control and LIC1 siRNA-treated cells (not shown). Moreover, all metaphase cells treated with LIC1 siRNA, including those delayed in mitosis, immunostained brightly for mitotic cyclin B, consistent with their inability to degrade this cyclin and enter anaphase (Figure 2B); in anaphase, the average cyclin B levels dropped from metaphase levels for both control siRNA and hLIC1 siRNA treatment (not shown). After prolonged metaphase arrest, some cells entered anaphase (Figure 1D, 2:20), whereas others lost the metaphase configuration but remained round (Figure 1E, 4:30–12:10), suggesting that they remained in mitosis with misaligned chromosomes.
Figure 2.
LIC1-depleted cells delay in metaphase. (A) A LIC1-depleted HeLa cell forms a metaphase plate approximately 15 min after NEB (0:00) then delays in metaphase for approximately 3 h. Staining with the viable DNA dye Syto13 (3:20). Bar, 10 μm. (B) Cyclin B immunofluorescence staining in cells treated with indicated siRNAs.
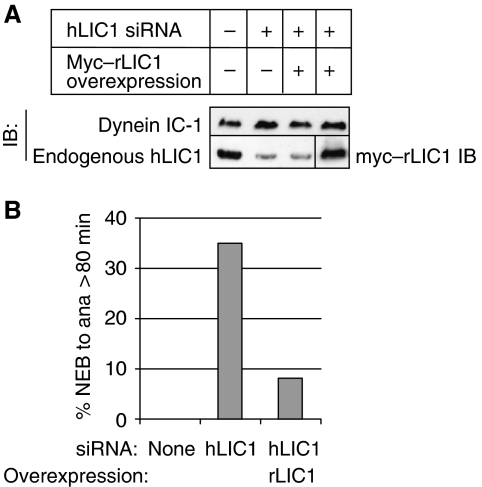
The metaphase delay and increased mitotic index observed in LIC1-depleted cells was reversed by expression of the rat homologue of hLIC1 (rLIC1), which was not targeted by the hLIC1 siRNA (Figure 3A and B). Thus, the mitotic phenotype was specific for LIC1 depletion and was not an off-target effect of the LIC1 siRNA. Taken together, these results show that LIC1 depletion induces a metaphase delay in HeLa cells.
Figure 3.
hLIC1-associated mitotic delay is suppressed by rLIC1 overexpression. (A) Immunoblot showing myc–rLIC1 overexpression in hLIC1-depleted cells. IC-1, cytoplasmic dynein 1 intermediate chain 1 (DYNC1I1). (B) The percentage of cells analysed by time-lapse imaging that exceeded 80 min from NEB to anaphase under the indicated conditions (n=13–17 cells/condition).
Cytoplasmic dynein levels, complex integrity, localisation and function are unaltered in LIC1-depleted cells
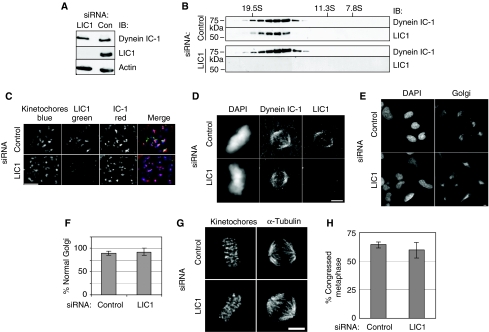
To test whether the mitotic delay induced by LIC1 depletion resulted from perturbation of cytoplasmic dynein, we examined several features of the motor. We found that level of the core intermediate chain of dynein was unaltered in LIC1-depleted cells, suggesting that the dynein complex was not destabilised or degraded by loss of LIC1 (Figure 4A). The dynein complex sedimented in sucrose gradients to the same position as controls (Figure 4B), showing that complex integrity was not significantly altered. Dynein localised normally to unattached kinetochores in nocodazole-treated cells (Figure 4C, 50/50 control and 52/52 LIC1 siRNA-treated cells) and to spindle poles in metaphase cells (Figure 4D, 50/50 control and 50/50 LIC1 siRNA-treated cells). Neither Golgi complex organisation (Figure 4E and F) (Corthesy-Theulaz et al, 1992) nor spindle pole focusing was altered in LIC1-depleted cells (Figure 4G) (Merdes et al, 2000); 29/29 control and 72/72 LIC1 siRNA-treated cells showed well-focused poles. Finally, chromosome congression in metaphase-arrested cells (2 h of MG132 treatment) (Yang et al, 2007) was comparable between control and LIC1-depleted cells (Figure 4H). Taken together, these data show that the mitotic delay observed on LIC1 depletion was not due to cytoplasmic dynein disruption, mislocalisation or functional abrogation. They also show that LIC1 is not required for localisation of dynein to kinetochores or other cellular sites.
Figure 4.
hLIC1 depletion does not alter dynein localisation, integrity or function. (A) Immunoblot of lysates from cells depleted of hLIC1 showing no significant change in dynein IC-1 levels. (B) hLIC1 co-sediments with dynein IC in sucrose gradients in control and LIC1-depleted cells. IB, immunoblot. S-value markers, above. (C) Dynein IC-1 localises to kinetochores (CREST) in nocodazole-treated hLIC1-depleted cells. (D) Dynein IC-1 localises to the spindle in control and hLIC1-depleted cells. (E) Golgi complex images and (F) percentage focused Golgi in control and LIC1-depleted cells (control: n=129 cells; LIC1: n=292 cells from three experiments). (G) Images of control/LIC1 siRNA-treated metaphase cells with focused spindle poles. (H) Percentage congressed mitotics (metaphase) after 2 h MG132 treatment (130–150 mitotics counted per experiment from three experiments). All scale bars=2 μM.
The mitotic arrest induced by LIC1 depletion requires Mad1, Mad2 and BubR1
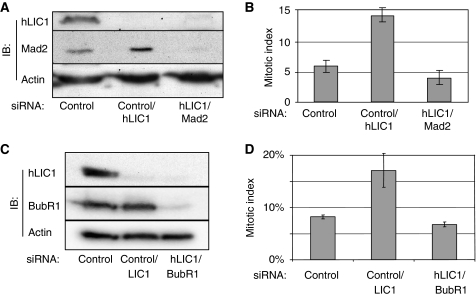
To test whether the mitotic delay induced by LIC1 depletion was due to activation of the SAC, we examined the role of the SAC proteins Mad1 and Mad2 (Hoyt et al, 1991; Chen et al, 1996, 1998; Waters et al, 1998; Meraldi et al, 2004). The increase in mitotic index in LIC1-depleted cells was suppressed in cells co-depleted of LIC1 and Mad2 (Figure 5A and B). Mad1 is involved in anchoring Mad2 at unattached kinetochores and enhances the ability of Mad2 to produce the ‘wait anaphase' signal (Chen et al, 1998; Meraldi et al, 2004; De Antoni et al, 2005). Mad1 depletion also suppressed the increased mitotic index induced by LIC1 depletion (Supplementary Figure S3A and B). BubR1 is associated with tension across sister kinetochores; depletion of BubR1 overcomes the SAC activated by microtubule depolymerisation (Chan et al, 1999; Hoffman et al, 2001; Skoufias et al, 2001). We found that co-depletion of LIC1 and BubR1 also suppressed the delay in mitosis (Figure 5C and D). These data show that the LIC1-dependent cell cycle delay requires Mad1, Mad2 and BubR1 and suggest that kinetochore-associated Mad2 is required for this phenotype.
Figure 5.
Mitotic arrest induced by LIC1 depletion requires Mad2 and BubR1. (A, C) Immunoblots of hMad2 (A), or BubR1 (C) and LIC1 (A, C) after treatment of siRNAs. (B, D) Mitotic indices after indicated siRNA treatments (n≥1200 cells from three experiments).
Structural organisation of SAC proteins at the kinetochore is normal in nocodazole-treated LIC1-depleted cells
When kinetochore-microtubule attachments are eliminated by microtubule depolymerisation, the two central players of the SAC, Mad2 and BubR1, localise to the unattached kinetochores (Chen et al, 1996; Taylor et al, 1998; Waters et al, 1998). In LIC1-depleted cells with depolymerised microtubules, recruitment of Mad1 (Figure 6A) or Mad2 (data not shown) to unattached kinetochores was not detectably different from control cells (Figure 6A, 50/50 control and 53/53 LIC1-depleted cells had Mad1 at kinetochores). Similar results were obtained with BubR1 (Figure 6B, 51/51 control and 50/50 LIC1-depleted cells); these cells also had Mad1 (data not shown) and dynein (Figure 4) on kinetochores. These data show that LIC1 depletion does not detectably alter the recruitment and molecular organisation of SAC proteins at the kinetochore.
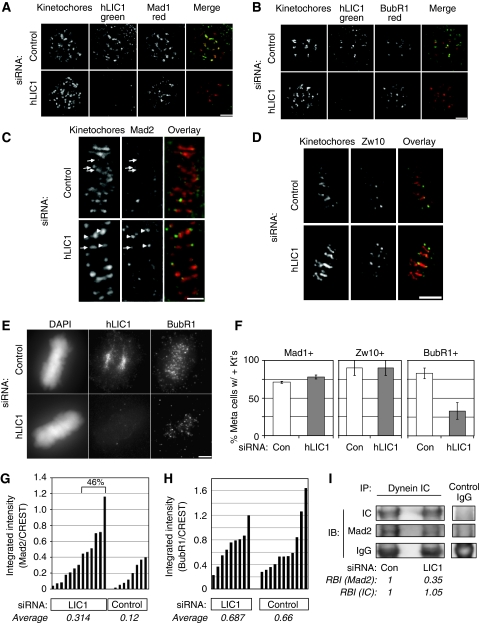
Figure 6.
Mad2, Mad1 and Zw10 but not BubR1 are inefficiently removed from metaphase kinetochores in LIC1-depleted cells. (A, B) Mad1 (A) and BubR1 (B) localise to kinetochores in hLIC1-depleted, nocodazole-treated cells just as in control. Scale bars in A–E, 2 μm. Kinetochore, CREST stain. Note, Mad1 or Mad2 (outer plate/fibrous corona proteins) do not completely overlap with CREST (inner plate) in stretched kinetochores. (C–E) Metaphase cells showing Mad2 (C, arrows—low levels; arrowheads—higher levels, Zw10 (D) or BubR1 (E) at kinetochores after the indicated siRNA treatments (F) The percentage of metaphase cells with Mad1-, Zw10- or BubR1-positive kinetochores (n=100 metaphase cells; bars, averages±s.d. from two experiments). Kt's, kinetochores. (G) Kinetochore Mad2 levels normalised to CREST staining in LIC1 depleted and control cells. LIC1 siRNA (left)—n=100 kinetochores from 13 cells; control siRNA—n=50 kinetochores from eight cells; from two experiments. LIC1-depleted cells (46%) had greater kinetochore Mad2 levels than the highest control cell. (H) Kinetochore BubR1 levels normalised to CREST; n=6 kinetochores each from 11 control cells and 9 LIC1 siRNA-treated cells analysed. Bars in G, H represent individual cells. Average intensity (G, H) is the mean fluorescence intensity from all quantified kinetochores. (I). Less Mad2 binds dynein in LIC1-depleted mitotic lysates. RBI, relative band intensity (normalised to respective control lane band intensity). IP, immunoprecipitation; IB, immunoblot. Representative of two identical experiments.
Mad1, Mad2 and Zw10, but not BubR1 are retained on kinetochores in LIC1-depleted metaphase cells
Although kinetochore recruitment of SAC proteins is normal, removal of some of these proteins is selectively impeded on LIC1 depletion. Quantification of Mad2 fluorescence intensity at metaphase kinetochores shows on average 2.6-fold higher retention of Mad2 in LIC1 siRNA-treated cells compared with control cells (Figure 6G, average intensities), with nearly half the LIC1 siRNA-treated cells (46%) showing higher kinetochore Mad2 levels than the highest control cell. However, a similar analysis for metaphase kinetochore BubR1 levels shows no change on LIC1 depletion (Figure 6H). We corroborated this observation by arresting LIC1-depleted cells and control cells in metaphase with the proteasome inhibitor MG132, which should allow efficient streaming of SAC proteins. Under these conditions, Mad1 (like Mad2) accumulates to about 2.8-fold higher levels on the visibly positive kinetochores in LIC1-depleted cells (Supplementary Figure S5) and, thus, appears to be inefficiently removed, whereas BubR1 kinetochore levels on visibly positive kinetochores are similar in LIC1 siRNA-treated and control cells (Supplementary Figure S6).
Counting the fraction of metaphase cells with visibly positive kinetochores (Figure 6F) also supports the earlier observations. The percentage of cells with Mad1- and Zw10-positive kinetochores is similar in LIC1-depleted and control cells; as approximately 50% of the LIC1-depleted cells were delayed in metaphase (Figures 1 and 2), this shows that Mad1 and Zw10 are retained on kinetochores in delayed cells. Zw10, a member of the RZZ complex (Rod–Zw10–Zwilch), is implicated in anchoring of Mad1/2 to the kinetochore (Buffin et al, 2005). The prolonged metaphase delay upon LIC1 depletion likely allows more efficient bleeding of kinetochore BubR1, leading to a lower percentage of metaphase cells with BubR1-positive kinetochores. This assay is discussed further in Supplementary data. Collectively, these data suggest that LIC1 is involved in the selective removal of Mad1, Mad2 and Zw10, but not BubR1, from kinetochores.
LIC1 depletion reduces the ability of dynein to bind Mad2
Removal of SAC proteins such as Mad1 and Mad2 from kinetochores requires cytoplasmic dynein (Howell et al, 2004; Lo et al, 2007). To test whether deficient removal is due to an altered ability of dynein to bind Mad2, we immunoprecipitated the motor (intermediate chain) from synchronised metaphase-arrested cells treated with LIC1 or control siRNAs. Indeed, less Mad2 co-precipitated with dynein in LIC1-depleted metaphase cells (2.8-fold less, Figure 6I) when equivalent levels of dynein were immunoprecipitated, whereas BubR1 binding to dynein was not detectably affected (data not shown). The reduced level of Mad2 in dynein immunoprecipitates suggests that LIC1 is required for dynein-mediated binding of specific SAC cargo proteins like Mad2.
Mad2 is retained on kinetochores with maximal interkinetochore distances in LIC1-depleted cells
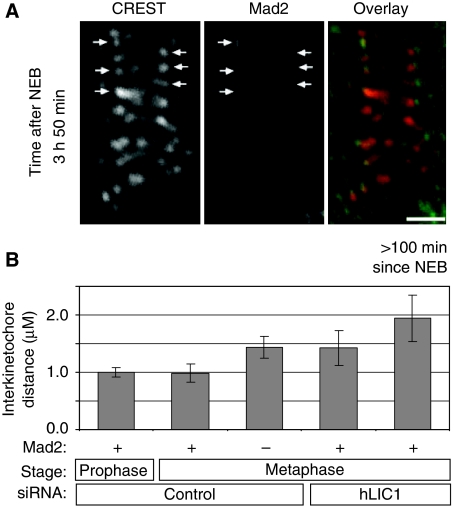
To examine individual LIC1-depleted cells, we used time-lapse imaging to identify cells delayed in metaphase, followed by immunofluorescence staining and imaging of the very same (delayed) cells on gridded cover slips. Most LIC1-depleted cells delayed for >100 min showed Mad2 localisation to several kinetochores (Figure 7A). Closer inspection unexpectedly revealed Mad2-positive kinetochores under tension (Figure 7A and B). As expected, control cell kinetochore pairs positive for Mad2 were under little or no tension in prophase and metaphase (Figure 7B), in agreement with Mad2 moving off kinetochores, as they are attached by microtubules and placed under tension. In contrast, Mad2-positive kinetochores in LIC1-depleted metaphase-arrested cells (>100 min) were widely separated and, thus, under significant tension (Figure 7A and B). In addition, 5/5 LIC1-depleted cells delayed in metaphase >100 min all showed the longest interkinetochore distances that were higher than in control cells. This shows that even when kinetochores are under tension, Mad2 is not effectively removed from these sites.
Figure 7.
Mad2 remains on kinetochores under tension in LIC1-depleted mitotic-delayed HeLa cells. (A) A hLIC1 siRNA-treated cell stained for Mad2 almost 4 h after NEB (arrows—Mad2-positive kinetochores). Note tension (large interkinetochore distance) on Mad2-positive kinetochores. (B) Average distances between sister kinetochore pairs. For metaphase, 16–82 pairs of kinetochores were counted from 7–11 cells per bar from two experiments. Mad2-positive kinetochores in metaphase delayed hLIC1-depleted cells (>100 min past NEB) (n=5 kinetochore pairs from four cells). Bar in A, 2 μM.
Phosphorylation of LIC1 at the S207 Cdk1 site is required for SAC function
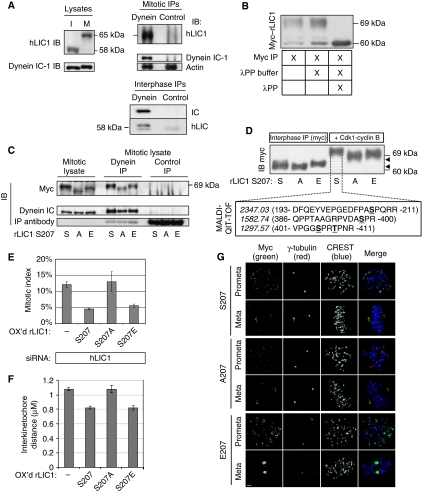
Cdks play a role in the SAC, although few targets of Cdk1 phosphorylation have been identified in this context (D'Angiolella et al, 2003). Here we show that LIC1 undergoes an upward mobility shift on SDS gels in mitotic versus interphase cells (Figure 8A, left), suggesting a mitosis-specific phosphorylation as shown previously for nonhuman forms of LIC1 (Hughes et al, 1995; Dell et al, 2000; Addinall et al, 2001). The mitotic phosphorylated form of LIC1 co-immunoprecipitated with the dynein complex, showing an association with dynein in mitosis (Figure 8A, right); phosphorylation was not essential for LIC1 binding as the nonphosphorylated form bound dynein in interphase cells (Figure 8A, right bottom). The shifted mitotic LIC1 band was reduced to the interphase position after incubation with lambda protein phosphatase, showing that the decreased mobility was due to phosphorylation (Figure 8B). Previous in vitro studies showed that LIC1 was phosphorylated in mitosis by the mitotic kinase Cdk1–cyclin B1 on distinct residues and that serine S197 in chicken and Xenopus (corresponding to S207 in human and rat LIC1) was required for releasing dynein from interphase vesicles during entry into mitosis (Dell et al, 2000; Addinall et al, 2001). To determine whether this phosphorylation site functioned in mitosis and the SAC, we asked whether mutations in the rat LIC1 S207 site affected its ability to restore SAC activity when expressed in LIC1-depleted cells. S207 mutated to alanine (S207A) did not undergo the full mitotic shift seen with the wild-type protein, confirming S207 as a phosphorylation site in this system (Figure 8C). In contrast, the pseudo-phosphorylated form of rLIC1 (S207E) shifted to the wild-type position (Figure 8C).
Figure 8.
Phosphorylation of rLIC1 on S207 is important for resolution of the spindle assembly checkpoint. (A) Endogenous hLIC1 in interphase cells (I) undergoes a gel shift during mitosis (M, left). Both the mitotic form of LIC1 (above) and the interphase form (below) co-immunoprecipitate with dynein IC-1. (B) Myc-tagged rLIC1 also undergoes a mitotic shift and is reduced to the lower form with lambda protein phosphatase treatment (λPP). (C) The mitotic shift of myc–rLIC1 can be partially suppressed by a S207A mutation and restored with a S207E mutation (three left lanes). All myc–rLIC1 mutants co-immunoprecipitate with dynein IC-1 antibodies (centre) but not control IgG (right). S, A, E=S207, S207A, S207E, respectively. (D) Top: Myc–rLIC1 mutants (S, A, E) immunoprecipitated from interphase lysates undergo phosphorylation on incubation with cdk1–cyclin B. Arrows indicate the small difference in upward migration (see text) among the S, A and E mutants. Bottom: Observed masses (italics) and single letter amino-acid sequences (in parentheses with N-terminal and C-terminal residue numbers indicated and phosphorylated residues underlined) from MALDI-QIT-TOF mass spectrometric analysis of the three phosphopeptides obtained from a tryptic digest of the phosphorylated S207 band (indicated by dotted lines). (E) Mitotic index of HeLa cells treated with hLIC1 siRNA and overexpressing myc–rLIC1 or point mutants (for S207, S207A and S207E, n=approximately 1500 cells each from three experiments; for LIC1 knockdown alone, approximately 500 cells were counted from three experiments). OX'd, overexpressed. Mitotic index of control (GFP siRNA treated) Hela cells was approximately 5%. (F) Average interkinetochore distance in LIC1 siRNA treatment (first bar) followed by rescue with myc–rLIC1 constructs. Bars, average interkinetochore distance for 30 kinetochore pairs (five cells) per experiment for each construct, total of three experiments. (G) All three myc–rLIC1 constructs localise to kinetochores (prometaphase) and spindle poles (metaphase). Scale bar=2 μM.
We tested whether cdk1 mediated the phosphorylation at S207 in vitro (Figure 8D). Hela cell interphase lysates overexpressing the three myc–rLIC1 proteins (S207, S207A and S207E) were subjected to immunoprecipitation using a myc antibody, and the purified, bead-bound myc–rLIC1 substrates incubated in vitro with purified cdk1–cyclin B kinase complex. The kinase reactions were quenched and analysed upon SDS–PAGE gels followed by immunoblotting with a myc antibody. The expected phosphorylation-induced gel retardation was observed for all three fusion proteins only upon incubation with Cdk1 (compare Figure 8D with Figure 8B and C). The slightly lesser retardation (approximately 2 kDa) of the phosphodeficient form (S207A) compared with wt S207 and the phosphomimetic S207E proteins is consistent with phosphorylation of the S207 site by cdk1 (also Figure 8C). We employed mass spectrometry to identify the precise sites of phosphorylation on rLIC1. The quenched cdk1 kinase reaction (S207 protein) was resolved on SDS–PAGE gels, and the Coomassie blue stained phosphorylated gel band (approximately 69 kD) subjected to trypsin digestion. The three phosphopeptides enriched from this digest were analysed by Matrix-Assisted-Laser Desorption/Ionization Quadrupole Ion Trap Time-of-Flight (MALDI-QIT-TOF) mass spectrometry. This analysis ascertained that residues S207, S398, S405 and T408 were the exact sites of phosphorylation on rLIC1 (Figure 8D, lower panel) that led to an approximately 9 kDa upward gel shift (Figure 8D, upper panel). These rLIC1 cdk1 sites correspond to the same residue numbers in the highly homologous hLIC1 sequence, and to the analogous S197, S386, S393 and T396, respectively, on Xenopus LIC1, which are known cdk1 consensus sites (Addinall et al, 2001). These results show unequivocally that cdk1 mediates mitotic phosphorylation of LIC1 at four sites, including S207.
We next tested the ability of the LIC1 mutants to suppress the LIC1 depletion-induced phenotype. The mitotic delay and the increased interkinetochore distance induced by LIC1 depletion were indeed suppressed by re-expression of wild-type LIC1 and the S207E mutant, but not by S207A (Figure 8E and F). In contrast, all three forms of rLIC1 (wild type, S207E and S207A) co-immunoprecipitated with dynein (Figure 8C), showing that LIC1 S207 phosphorylation did not regulate the LIC1-dynein interaction. The myc–rLIC1 proteins behaved like endogenous hLIC1. All three myc–rLIC1 constructs localised to kinetochores in prometaphase and to spindle poles in metaphase (Figure 8G) in a manner similar to hLIC1 (Figure 4C and D). On the basis of the results showing dynein/dynactin streaming in living cells from kinetochores in prometaphase to spindle poles in metaphase (Gaglio et al, 1997; Hoffman et al, 2001; Wojcik et al, 2001), we believe our fixed cell data strongly suggest streaming of the LIC1 constructs from kinetochores to spindle poles. In addition, cells overexpressing both S207 and E207 constructs showed mean mitotic timing values comparable to control cells (Supplementary Figure S4), whereas mitosis appeared only marginally delayed in the S207A-transfected cells, perhaps due to a mild dominant negative effect. These results suggest that the S207 and E207 constructs rescue the LIC1 depletion phenotype by genuine functional complementation, and not due to a compromised SAC itself. Taken together, these data show that phosphorylation of rLIC1 at S207 is required for progression through the SAC and suggest that cdk1 regulates this event perhaps by facilitating LIC1 binding to cargoes such as SAC proteins.
Discussion
A novel role for LIC1 in the spindle assembly checkpoint
SAC control by human LIC1 identifies a new role for this dynein subunit. This result has also been shown recently for the solitary LIC in Drosophila (Mische et al, 2008). However, in both Drosophila and C. elegans, the solitary LIC functions in multiple mitotic dynein-associated processes (Yoder and Han, 2001; Mische et al, 2008), which can explain the SAC defect in Drosophila. This is in contrast to our work, which shows that LIC1 functions more specifically in the selective removal of SAC components from kinetochores, but does not appear to participate in other mitotic dynein-associated functions. The disparity between the mammalian and Drosophila SACs (Buffin et al, 2007; Orr et al, 2007) could underlie these mechanistic differences. Alternatively, the additional phenotypes observed after mutation of the solitary LIC in Drosophila could result from loss of functions associated with both human LICs (1 and 2), differences in LIC function between embryonic and somatic cell systems, or incomplete depletion of hLIC1.
Our work suggests that LIC1 is directly involved in the SAC and is not simply a structural component of the kinetochore. The dependence of the LIC1 depletion-induced metaphase delay on Mad1 and Mad2 shows that an active checkpoint is required but does not rule out disruption of kinetochore organisation as a contributing factor. However, localisation of Mad1, Mad2, BubR1 and dynein to kinetochores in nocodazole-treated LIC1-depleted cells shows that LIC1 has no detectable effect on kinetochore structure, and that the checkpoint-signaling platform is intact at the kinetochore. Finally, Mad2 not only localises to but also accumulates at kinetochores in LIC1-depleted cells (Figure 6C and G), which would not be expected with compromised kinetochore structure.
Partial depletion of kinetochore components involved in microtubule-kinetochore attachment (Hec1 and Nuf2, Ndc80 complex components) induces defects in chromosome congression, compromised Mad2 recruitment to kinetochores in nocodazole-treated cells and metaphase arrest (Martin-Lluesma et al, 2002; DeLuca et al, 2003). Importantly, more extensive depletion led to suppression of the checkpoint—consistent with complete loss of the checkpoint signal pathway from the kinetochore (e.g., Mad2) (Meraldi et al, 2004). LIC1 does not appear to fall into this category of kinetochore proteins, as chromosome congression and SAC protein recruitment on nocodazole treatment were normal in LIC1-depleted cells. We conclude that LIC1 is involved in attenuation of the ‘wait anaphase' signal rather than structural organisation of the kinetochore.
Mad2 localisation to kinetochores may be sufficient to produce the ‘wait anaphase' signal
The observation that Mad1 and Mad2, but not BubR1 remain on kinetochores that are under tension is novel. The SAC is normally not active when kinetochore-associated BubR1 is reduced to control levels. This is because Mad2 is typically removed early, upon microtubule-kinetochore attachment, whereas BubR1 is removed later, after sister kinetochores are under tension (Skoufias et al, 2001; Taylor et al, 2001; Shannon et al, 2002). Our results suggest that kinetochore-associated Mad2 may be sufficient to activate or maintain the SAC. This does not rule out the possibility that residual BubR1 on kinetochores or cytoplasmic BubR1 may function in SAC activation possibly through propagation of the signal. This is suggested by data showing that BubR1 depletion allows cells to progress through an active SAC and other data showing that BubR1 is in the cytoplasmic mitotic checkpoint complex (Sudakin et al, 2001; Meraldi et al, 2004). Thus, the LIC1 depletion-induced delay suggests that kinetochore Mad2 may be sufficient to produce the ‘wait anaphase' signal and provides a novel system to study the role of cytoplasmic BubR1 in producing a ‘wait anaphase' signal. However, the role of cytoplasmic BubR1 in cells under checkpoint activation with little kinetochore-associated BubR1 has not been tested. Alternatively, the residual kinetochore BubR1 may also be involved in maintaining the wait anaphase signal.
A model for LIC1 function: adaptor for dynein cargoes at the kinetochore
The role of LIC1 in the SAC represents one of many mitotic functions carried out by cytoplasmic dynein (Gaglio et al, 1997; Hoffman et al, 2001; Wojcik et al, 2001). The role of dynein in the SAC appears to involve movement of SAC proteins such as Mad2, BubR1 and Zw10 from kinetochores to spindle poles (Hoffman et al, 2001; Wojcik et al, 2001). Our data are consistent with a model in which LIC1 serves as an adaptor that links specific cargoes (Mad1/2, Zw10) but not others (BubR1) to the dynein motor for kinetochore removal and silencing the SAC. This could be mediated through direct interactions with specific cargo proteins, or indirectly through contacts with other proteins. Likely candidates include SAC proteins like Mad1 and Mad2 (Hoffman et al, 2001), RZZ complex and its receptor Zwint1 (Karess, 2005), outer kinetochore proteins including components of the KMN network (Cheeseman et al, 2006; Cheeseman and Desai, 2008), newly discovered proteins like Spindly (Griffis et al, 2007) that specifically modulate kinetochore dynein function, or other novel proteins.
The role of the RZZ complex in LIC1 depletion phenotype
The RZZ complex (Rod–Zw10–Zwilch) migrates off the kinetochore in association with Mad2 along spindle microtubules to the spindle poles, in addition to its role in kinetochore localisation of Mad2 (Buffin et al, 2005; Kops et al, 2005). Thus, it is in the right place to be involved in docking, catalysis and kinetochore removal of Mad2 (Buffin et al, 2005; De Antoni et al, 2005; Karess, 2005). Zw10 interacts with the p50 subunit of the dynein/dynactin complex (Starr et al, 1998; Howell et al, 2000, 2001), but this interaction does not appear to be required for RZZ or Mad2 removal from kinetochores. Like LIC1 depletion, dynein disruption inhibits removal of Zw10 and Mad2 from kinetochores. LIC1 depletion does not affect other dynein functions, whereas LIC1 could specifically mediate binding to member(s) of the RZZ or Mad1–Mad2 complexes, or regulate interaction between RZZ or Mad1–Mad2 with the dynein complex.
LIC1 and dynein mediate binding to other cargoes
Mad2 is not the only cargo linked to dynein by LIC1. The centrosome protein pericentrin binds directly to LIC1 (Purohit et al, 1999) and is transported to centrosomes by cytoplasmic dynein (Young et al, 2000). LIC1 also interacts with Rab4A, a GTPase involved in the regulation of intracellular vesicle trafficking (Bielli et al, 2001). Because the interaction of dynein with interphase membranes/vesicles is regulated by mitotic phosphorylation (Addinall et al, 2001), it is possible that mitotic phosphorylation of LIC1 downregulates its affinity for Rab4A and other interphase vesicle proteins to allow targeting of mitotic cargoes. The precise role of LIC1 in the transport of vesicles and proteins in interphase and mitosis is under investigation.
The role of LIC1 in binding dynein cargoes is consistent with similar roles of other dynein and dynactin subunits, which bind NuMA, PCM-1, spectrin and rhodopsin (Zimmerman and Doxsey, 2000; Pfister, 2005). In fact, other dynein subunits may be involved in regulating Mad2 removal from kinetochores as depletion of the Rab6A' GTPase, a potential regulator of dynein/dynactin function through the dynactin complex, induces Mad2 retention at a few kinetochores and activation of the SAC (Miserey-Lenkei et al, 2006). The use of dynein and dynactin subunits to link multiple cargoes to the minus end dynein motor (Pfister et al, 2006) appears to provide a mechanism analogous to amplification of the kinesin gene family, which links cargoes to multiple kinesins (Goldstein, 2001).
LIC1 phosphorylation is required for its role in the spindle assembly checkpoint
Our work extends the observation that mitotic phosphorylation of dynein by cdk1 disengages the motor from interphase membrane cargoes (Addinall et al, 2001), by showing that phosphorylation at the S207 cdk1 site on the LIC1 subunit is required for the SAC. Phosphorylation could promote the interaction of LIC1 with an as yet unidentified cargo. Obvious candidates are Mad1, Mad2 and Zw10 as all remain on kinetochores in LIC1-depleted cells, and perhaps other members of the RZZ complex (Rod, Zwilch). The phosphorylation mutants provide powerful tools to identify LIC1 interacting proteins and to determine the molecular mechanism by which LIC1 functions in the SAC.
Our data show that cdk1 is the mitotic kinase that phosphorylates LIC1 at S207 during mitosis. The 9-kDa gel shift of LIC1 in mitosis (Figure 8B) and the smaller cdk1-dependent shift seen with the phosphodeficient S207A LIC1 protein in vitro (Figure 8C and D) together suggest that LIC1 is phosphorylated at multiple sites by cdk1 in mitosis, of which S207 is one site. Mass spectrometric identification of the phosphorylation sites from an in vitro kinase reaction indeed confirmed four consensus cdk1 sites (including S207) as the specific targets of the kinase (Figure 8D; Supplementary Figure S7). The only caveat in this in vitro assay is the theoretical possibility that another kinase that co-purified in the myc interphase immunoprecipitates could directly phosphorylate LIC1 after activation by cdk1. This is unlikely, however, owing to the fact that the four residues phosphorylated in this assay are consensus cdk1 sites, and also because the gel shift is seen in vivo only during mitosis and not during interphase (Figure 8A). Moreover, cdk1 phosphorylates the analogous S197 in vitro (Dell et al, 2000) and in vivo during mitosis (hLIC1, this study). Additionally, cyclin B accumulates and activates cdk1 specifically in mitosis. Other kinases with similar consensus sites for phosphorylation (e.g., cdk2 and MAPK) are not good candidates for S207 phosphorylation. The cdk2 activators cyclin A and cyclin E are both low in cells arrested in mitosis through the SAC (D'Angiolella et al, 2003). MAPK is also not active under normal conditions or when the SAC is active (Deacon et al, 2003).
It is interesting that inhibition of cdk1 promotes entry into anaphase, in contrast to re-expression of the nonphosphorylatable rLIC1 mutant, which does not promote entry into anaphase in LIC1-depleted cells. One explanation for this difference is the observation that cdk1 appears to have two antagonistic roles in mitosis. Cdk1 activity inhibits anaphase onset, but it also sets up an environment that leads to its own inactivation by cyclin B1 degradation and thus promotes anaphase onset (D'Angiolella et al, 2003). These two roles of cdk1 insure that chromosomes are attached and under enough tension for the SAC to be inactivated before cells transition from metaphase to anaphase.
Materials and methods
Cell culture
Hela (ATCC CCL-2), diploid hTERT-BJ-1 and SAOS cells (ATCC HTB-85) were cultured as described by American Type Cell Collection. hTERT-BJ-1 cells were only used in Supplementary Figure S2.
siRNA treatment
siRNAs (21-nt; Dharmacon Research, Inc.) targeting hLIC1 (GenBank/EMBL/DDBJ accession no. NM_016141;nt 1156–1174), GFP (Gromley et al, 2003), hMad2L1 (NM_002358; nt 503–523), hMad1 (Martin-Lluesma et al, 2002) and BubR1 (NM_001211; nt 1310–1328) were delivered to cells using Dharmafect 1 (Dharmacon Research, Inc.). hLIC1 siRNA-target sequence is specific to hLIC1 and does not completely match rLIC1 or hLIC2.
Constructs and overexpression
The constructs in Figure 8 were generated as described in Supplementary data. Constructs were delivered to cells using Lipofectamine 2000 (Invitrogen).
LIC1 antibody production
A synthetic peptide consisting of the C-terminal 15 amino acids of hLIC1: VFP TTP TSP TEG EAS (Bielli et al, 2001) was used to raise polyclonal antibodies (Q-biogene). Antibody was affinity purified on a CNBr-activated sepharose 4B beads peptide column (Amersham) (Harlow and Lane, 1999).
Time-lapse imaging
HeLa cells were imaged as described previously (Gromley et al, 2003). In Figure 2A, cells while still on the stage were incubated in 0.3 μM Syto13 (Invitrogen) for 5 min and then imaged.
Antibodies, fixation, staining and imaging
Cells were fixed in methanol or formaldehyde (Dictenberg et al, 1998). For staining Mad2, cells were permeabilised and fixed in formaldehyde as described previously (Hoffman et al, 2001). The following antibodies were used: anti-dynein intermediate chain 74.1, anti-cyclin B1 and anti-α-tubulin DM1α culture supernatant (Sigma); anti-hMad1 (De Antoni et al, 2005); anti-LIC1 (described earlier); CREST serum (Brenner et al, 1981); anti-Zw10 (gift from Tim Yen) and anti-BubR1 monoclonal antibody (BD Biosciences). Golgi was directly stained with Helix pomatia agglutinin Alexa 488 (Invitrogen). Images were taken either on a wide field microscope as described previously (Dictenberg et al, 1998) or on a Perkin Elmer Ultraview spinning disk confocal microscope: Zeiss Axiovert 200, 100 × Plan-APOCROMAT NA1.4 Oil, DIC lens and Hamamatsu ORCA-ER camera. The entire fixed cell volume was imaged and displayed as a two-dimensional projection (Meta-Morph; Universal Imaging Corp.). Cyclin B1 levels were measured by integrating the fluorescence intensity of the whole cell. To look at Mad2 and BubR1 localisation to unattached kinetochores, cells were treated with 1-μM nocodazole for 1 h to depolymerise microtubules before fixation.
Image analysis, kinetochore quantification
Kinetochore fluorescence was quantified essentially based on the method described by Hoffman et al (2001), with modifications as detailed in Supplementary data.
Western blotting
Proteins were separated by SDS–PAGE, transferred to PVDF membranes and probed with the following antibodies: anti-dynein intermediate chain 74.1 and anti-actin AC40 (Sigma); anti-dynein heavy chain R-325 and anti-myc 9E10 (Santa Cruz); anti-hMad1 (De Antoni et al, 2005); anti-hMad2 (Covance) and anti-LIC1 (described earlier). Anti-rabbit and anti-mouse HRP antibodies were used along with ECL Plus luminescent reagent (Amersham Biosciences) and then visualised on film or using a 4000-MM Image Station (Kodak).
Immunoprecipitation, phosphatase treatment and cdk1 assays
Lipofectamine 2000 (Invitrogen) was used to transfect cells following the manual protocol. At 24 h, 1 μg/ml NOC was added. At 48 h, mitotic cells were collected by pipetting PBS over the plate several times. Cells were lysed in 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EGTA, 1% Triton X-100, 50 mM NaF, 10 mM β-Glycerophosphate, 5 mM Na4P2O7, 1 mM Na3VO4, 1 μM microcystin. Immunoprecipitations were carried out with myc or 74.1 dynein intermediate chain antibody and Protein G Plus-Agarose beads (Santa Cruz Biotechnology). For phosphatase treatment, beads were treated in one of the three ways: SDS buffer was added; 30-min incubation at 30°C in lambda protein phosphatase buffer and then SDS buffer was added; or 30-min incubation at 30°C in lambda protein phosphatase buffer and lambda protein phosphatase (New England Biolabs, Inc.) and then SDS buffer was added. For detection of Mad2 in dynein immunoprecipitates, see Supplementary data.
In vitro cdk1 activity was assayed by incubating commercially purified cdk1–cyclin B complexes (Invitrogen, at manufacturer recommended concentrations) with myc-immunoprecipitates of interphase lysates coupled to Protein-G PLUS agarose beads (Santa Cruz Biotechnology). The kinase assay was carried out at room temperature with mild shaking for 30 min in kinase assay buffer (25 mM Tris–HCl pH 7.5, 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM sodium orthovanadate, 10 mM magnesium chloride and 200 μM ATP). The reaction was quenched by addition of SDS loading dye and boiling for 10 min, followed by resolution on 8% SDS–PAGE gels before either immunoblotting with a myc antibody (Sigma) or staining with Coomassie brilliant blue dye.
Mass spectrometric analysis
Coomassie blue stained protein bands were digested ‘in gel' with trypsin according to established methods (Lahm and Langen, 2000). Phosphorylated tryptic peptides were enriched for using NuTip TiO2 micropipette tips (Glygen Corporation) using a modified protocol as described in Imanishi et al (2007), followed by further purification using micro-C18 Zip Tip columns (Millipore Corporation). Mass spectrometric analyses were carried out on a Shimadzu Axima QIT (Shimadzu Instruments) MALDI-QIT-TOF mass spectrometer. Tryptic peptides were analysed in positive ion mode in the midmass range 700–3000 Da. Collisionally induced dissociation analyses were carried out on the same instrument using the ion trap for precursor selection and argon as the collision gas. All spectra were processed with Mascot Distiller (Matrix Sciences Ltd.) before database searches, which were carried out in house with the Mascot search engine (Matrix Sciences Ltd.).
Sucrose gradient
Cells were harvested with 10 mM EDTA in PBS and lysed in 50 mM Tris–Cl pH 7.5, 150 mM NaCl, 1% IGEPAL CA-630, 1 mM EDTA, 1 complete protease inhibitor cocktail mini tablet (Roche) per 10 ml of lysis buffer. Lysates were spun at 16 000 RCF at 4°C for 10 min, and the supernatant layered on a 12 ml 5–20% sucrose gradient and centrifuged at 4°C in a Beckman Coulter Optima L-90K ultracentrifuge (35 000 r.p.m., SW41 rotor) for 12.5 h. Sedimentation standards used were thyroglobulin 19.4S, catalase (11.3S), aldolase (7.3S) and BSA (4.4S). Sucrose gradient fractions (500 μl) were collected and analysed by western blots.
Statistical analyses
In most cases, the mean±standard deviation (s.d.) was plotted when three or more experiments were analysed, unless otherwise indicated.
Supplementary Material
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary data
Acknowledgments
We thank John Leszyk for performing the MALDI-QIT-TOF experiment and analysis, and the UMass Diabetes and Endocrinology Research Center for providing support to his lab. We thank Todd Stukenberg, Dannel McCollum and Bill Theurkauf for comments on the work and the following for antibodies: Ted Salmon, Andrea Musacchio, Steve Taylor (all for Mad1/2), Tim Yen (Zw10) and Bill Brinkley (CREST). We thank Paul Furcinitti for introductory microscopy tutorials and microscope management. This work was supported by grants from the National Institutes of Health (GM51994) and the National Cancer Institute (CA82834-01) to SJD and a training grant from the Department of Rheumatology (NIH T32 AR07572) to TLW.
References
- Addinall SG, Mayr PS, Doyle S, Sheehan JK, Woodman PG, Allan VJ (2001) Phosphorylation by cdc2-CyclinB1 kinase releases cytoplasmic dynein from membranes. J Biol Chem 276: 15939–15944 [DOI] [PubMed] [Google Scholar]
- Bielli A, Thornqvist PO, Hendrick AG, Finn R, Fitzgerald K, McCaffrey MW (2001) The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem Biophys Res Commun 281: 1141–1153 [DOI] [PubMed] [Google Scholar]
- Brenner S, Pepper D, Berns MW, Tan E, Brinkley BR (1981) Kinetochore structure, duplication, and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J Cell Biol 91: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E, Emre D, Karess RE (2007) Flies without a spindle checkpoint. Nat Cell Biol 9: 565–572 [DOI] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15: 856–861 [DOI] [PubMed] [Google Scholar]
- Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ (1999) Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol 146: 941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DC, Xu N, Luo KQ (2003) Degradation of cyclin B is required for the onset of anaphase in Mammalian cells. J Biol Chem 278: 37865–37873 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol 143: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 274: 242–246 [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol 1: 82–87 [DOI] [PubMed] [Google Scholar]
- Corthesy-Theulaz I, Pauloin A, Rfeffer SR (1992) Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol 118: 1333–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Mari C, Nocera D, Rametti L, Grieco D (2003) The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev 17: 2520–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, Musacchio A (2005) The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 15: 214–225 [DOI] [PubMed] [Google Scholar]
- Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, Vaughan KT, Gelfand VI (2003) Dynactin is required for bidirectional organelle transport. J Cell Biol 160: 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell KR, Turck CW, Vale RD (2000) Mitotic phosphorylation of the dynein light intermediate chain is mediated by cdc2 kinase. Traffic 1: 38–44 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Howell BJ, Canman JC, Hickey JM, Fang G, Salmon ED (2003) Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol 13: 2103–2109 [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ (1998) Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Dionne MA, Compton DA (1997) Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol 138: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS (2001) Molecular motors: from one motor many tails to one motor many tales. Trends Cell Biol 11: 477–482 [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Chen RH, Murray AW (1998) Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J Cell Biol 141: 1193–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S (2003) A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol 161: 535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D (1999) Using Antibodies, A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol 150: 1233–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED (2004) Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 14: 953–964 [DOI] [PubMed] [Google Scholar]
- Hoyt MA (2001) A new view of the spindle checkpoint. J Cell Biol 154: 909–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517 [DOI] [PubMed] [Google Scholar]
- Hughes SM, Vaughan KT, Herskovits JS, Vallee RB (1995) Molecular analysis of a cytoplasmic dynein light intermediate chain reveals homology to a family of ATPases. J Cell Sci 108: 17–24 [DOI] [PubMed] [Google Scholar]
- Imanishi SY, Kochin V, Ferraris SE, de Thonel A, Pallari HM, Corthals GL, Eriksson JE (2007) Reference-facilitated phosphoproteomics: fast and reliable phosphopeptide validation by microLC-ESI-Q-TOF MS/MS. Mol Cell Proteomics 6: 1380–1391 [DOI] [PubMed] [Google Scholar]
- Karess R (2005) Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol 15: 386–392 [DOI] [PubMed] [Google Scholar]
- King JM, Nicklas RB (2000) Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J Cell Sci 113 (Part 21): 3815–3823 [DOI] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR III, Tagaya M, Cleveland DW (2005) ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 169: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm HW, Langen H (2000) Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis 21: 2105–2114 [DOI] [PubMed] [Google Scholar]
- Lo KW, Kogoy JM, Pfister KK (2007) The DYNLT3 light chain directly links cytoplasmic dynein to a spindle checkpoint protein, Bub3. J Biol Chem 282: 11205–11212 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- Mayer C, Filopei J, Batac J, Alford L, Paluh JL (2006) An extended anaphase signaling pathway for Mad2p includes microtubule organizing center proteins and multiple motor-dependent transitions. Cell Cycle 5: 1456–1463 [DOI] [PubMed] [Google Scholar]
- McIntosh JR (1991) Structural and mechanical control of mitotic progression. Cold Spring Harb Symp Quant Biol 56: 613–619 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK (2004) Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60 [DOI] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW (2000) Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Pines J, Golsteyn R, Standart N, Mackie S, Colman A, Blow J, Ruderman JV, Wu M, Hunt T (1989) The role of cyclin synthesis, modification and destruction in the control of cell division. J Cell Sci Suppl 12: 77–97 [DOI] [PubMed] [Google Scholar]
- Mische S, He Y, Ma L, Li M, Serr M, Hays TS (2008) Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol Biol Cell 19: 4918–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Couedel-Courteille A, Del Nery E, Bardin S, Piel M, Racine V, Sibarita JB, Perez F, Bornens M, Goud B (2006) A role for the Rab6A′ GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J 25: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW (1989) The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature 339: 280–286 [DOI] [PubMed] [Google Scholar]
- Orr B, Bousbaa H, Sunkel CE (2007) Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol Biol Cell 18: 850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK (2005) Dynein cargo gets its groove back. Structure 13: 172–173 [DOI] [PubMed] [Google Scholar]
- Pfister KK, Shah PR, Hummerich H, Russ A, Cotton J, Annuar AA, King SM, Fisher EM (2006) Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet 2: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit A, Tynan SH, Vallee R, Doxsey SJ (1999) Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol 147: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI (2003) Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol 4: 855–864 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G (1995) The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 130: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G (1994) Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol 127: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Cleveland DW (2000) Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell 103: 997–1000 [DOI] [PubMed] [Google Scholar]
- Shannon KB, Canman JC, Salmon ED (2002) Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension. Mol Biol Cell 13: 3706–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL (2001) Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA 98: 4492–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Hays TS, Goldberg ML (1998) ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol 142: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ (2001) Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F (1998) The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol 142: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ (2001) Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci 114: 4385–4395 [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol 141: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T (2001) Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol 3: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL (2007) Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol 17: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JH, Han M (2001) Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol Biol Cell 12: 2921–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ (2000) Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol Biol Cell 11: 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yin S, Jiang MX, Ma W, Hou Y, Liang CG, Yu LZ, Wang WH, Sun QY (2007) Cytoplasmic dynein participates in meiotic checkpoint inactivation in mouse oocytes by transporting cytoplasmic mitotic arrest-deficient (Mad) proteins from kinetochores to spindle poles. Reproduction 133: 685–695 [DOI] [PubMed] [Google Scholar]
- Zhou J, Yao J, Joshi HC (2002) Attachment and tension in the spindle assembly checkpoint. J Cell Sci 115: 3547–3555 [DOI] [PubMed] [Google Scholar]
- Zimmerman W, Doxsey SJ (2000) Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic 1: 927–934 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary data