Abstract
Succinic semialdehyde dehydrogenase (SSADH) is involved in the final degradation step of the inhibitory neurotransmitter γ-aminobutyric acid by converting succinic semialdehyde to succinic acid in the mitochondrial matrix. SSADH deficiency, a rare autosomal recessive disease, exhibits variable clinical phenotypes, including psychomotor retardation, language delay, behaviour disturbance and convulsions. Here, we present crystal structures of both the oxidized and reduced forms of human SSADH. Interestingly, the structures show that the catalytic loop of the enzyme undergoes large structural changes depending on the redox status of the environment, which is mediated by a reversible disulphide bond formation between a catalytic Cys340 and an adjacent Cys342 residues located on the loop. Subsequent in vivo and in vitro studies reveal that the ‘dynamic catalytic loop' confers a response to reactive oxygen species and changes in redox status, indicating that the redox-switch modulation could be a physiological control mechanism of human SSADH. Structural basis for the substrate specificity of the enzyme and the impact of known missense point mutations associated with the disease pathogenesis are presented as well.
Keywords: GABA, redox switch, ROS, SSADH, SSADH deficiency
Introduction
Succinic semialdehyde dehydrogenase (SSADH) deficiency is a rare autosomal recessive disease caused by inherited deficiency in the final degradation step of the inhibitory neurotransmitter γ-aminobutyric acid (GABA) (Pearl et al, 2003a; Wong et al, 2003). GABA is synthesized from glutamic acid by glutamic acid decarboxylase and converted to succinic semialdehyde (SSA) by GABA transaminase (Porter et al, 1985; Battaglioli et al, 2003; Storici et al, 2004; Fenalti et al, 2007). SSA is converted either to succinic acid by SSADH in the mitochondrial matrix or to γ-hydroxybutyric acid (GHB) by the enzyme SSA reductase (Maitre, 1997; Kelly et al, 2002). The carbon skeleton of GABA therefore enters the tricarboxylic acid cycle in the form of succinic acid. SSADH deficiency disrupts the normal metabolism of GABA, leading to a 30-fold increase of GHB and a 2- to 4-fold increase of GABA in the brains of patients with SSADH deficiency as compared with normal concentrations of these compounds (Gibson et al, 1998). Children with SSADH deficiency exhibit a variable clinical phenotype, including psychomotor retardation, language delay, hypotonia and ataxia. Seizures occur in approximately 50% of patients with SSADH deficiency, and include tonic-clonic, absence and myoclonic seizures, as well as convulsive status epilepticus (Gibson et al, 1998; Pearl et al, 2003b). Recently, Dr Gibson's group has generated a murine SSADH-deficient model (SSADH−/− mice) by deleting the active site of the SSADH gene, to more clearly define the pathophysiology of human SSADH deficiency and to develop preclinical treatment strategies (Hogema et al, 2001a, 2001b). The mutant mice model displays a phenotype that is characterized by ataxia and generalized seizures, and shows increased amount of both GABA and GHB in urine, and homogenates of brain and liver (Gibson et al, 2002; Gupta et al, 2002, 2003). It is also reported that oxidative stress is a pathomechanistic feature contributing to tissue damage in the null mouse model (Latini et al, 2007).So far, more than 50 disease-causing mutations, such as nonsense and missense point mutations, small insertion and small deletion mutations, have been identified from more than 50 SSADH deficiency-suffering families of different geographic origins (Aoshima et al, 2002; Akaboshi et al, 2003; Bekri et al, 2004; Hogema et al, 2001a, 2001b).
Human SSADH is encoded by the ALDH5A1 gene localized on chromosome 6p22, and contains conserved regions of the aldehyde dehydrogenases (ALDHs), suggesting that it is a member of the ALDH superfamily (Trettel et al, 1997). SSADH is known to have a very high substrate specificity compared with other ALDHs, and in the case of Drosophila melanogaster SSADH, the Michaelis constants (KM values) for the substrate SSA and NAD+ were 4.7 and 90.9 μM, respectively (Rothacker and Ilg, 2008). Despite the importance of SSADH in the GABA degradation process, and the involvement of SSADH deficiency in various types of brain dysfunction diseases, there has been a substantial limit, owing to the lack of the structural information of the enzyme, in explaining the detailed catalytic and regulatory mechanisms of the enzyme. In this study, we present that human SSADH is regulated by redox-switch modulation that confers a response to reactive oxygen species (ROS) and changes in redox status. The structural basis for the substrate specificity of the enzyme and the impact of known mutations associated with the disease pathogenesis are presented as well.
Results and discussion
Crystal structure of the oxidized form of human SSADH
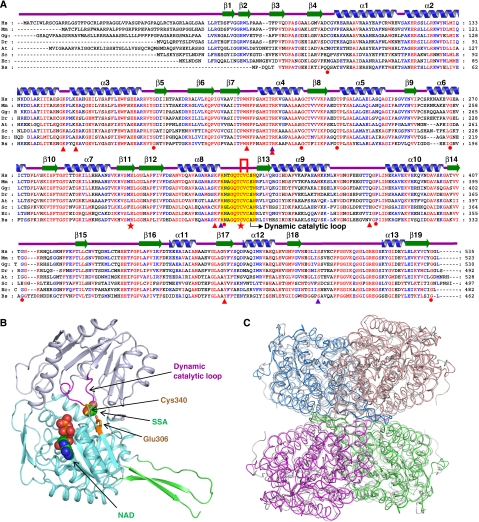
To address the structural basis for the catalytic and regulatory mechanisms of human SSADH, we determined the 2.0-Å crystal structure of the N-terminal signal sequence truncated form of SSADH (amino-acid residues 48–535; Figure 1A). Even though only a monomeric protein is present in the asymmetric unit of our present structure, the tetrameric structure of SSADH can easily be generated by applying the crystallographic F432 symmetry (Figure 1B and C), which is consistent with the size-exclusion chromatography data (data not shown). The overall structure of SSADH shares the general fold of ALDH of classes 1 and 2 (Steinmetz et al, 1997; Moore et al, 1998). The monomeric SSADH comprised three domains: an N-terminal NAD-binding domain (residues 48–173, 196–307 and 509–524), a catalytic domain (residues 308–508) and an oligomerization domain (residues 174–195 and 525–535) (Figure 1B). The NAD-binding and catalytic domains are α/β structure, whereas the oligomerization domain contains three-stranded antiparallel β-sheets.
Figure 1.
Crystal structure of human SSADH. (A) Amino-acid sequence alignment of SSADHs. Secondary structure elements are drawn on the basis of human SSADH structure and shown as a green-coloured arrow (β-sheet) and blue-coloured helix (α-helix) and labelled. The ‘dynamic catalytic loop' is shown as a yellow box and two cysteine residues (Cys340 and Cys342 in human SSADH) are indicated by a red line. Residues involved in enzyme catalysis and SSA binding are indicated by red-coloured stars and violet-coloured triangles. Residues involved in stabilization of the ‘dynamic catalytic loop' are indicated by red-coloured triangles, and those, whose missense point mutations are found in SSASH deficiency patients, are by a red-coloured circle. Hs, Mm, Gg, Dr, At, Sc, Ec and Bs are the representations of Homo sapiens, Mus musculus, Gallus gallus, Danio rerio, Arabidopsis thaliana, Saccharomyces cerevisiae, Escherichia coli and Bacillus subtilis, respectively. (B) Monomeric structure of human SSADH. A monomeric protein is present in the asymmetric unit of our present structure, and shown as ribbon representation in which the substrate-binding domain is coloured light blue, the NAD-binding domain is coloured cyan, and the oligomerization domain is coloured green. The ‘dynamic catalytic loop' is distinguished by magenta colour, and the catalytic residues (Glu306 and Cys30) are presented in stick model in orange colour. SSA and NAD are placed in their binding sites with sphere model. (C) Tetrameric structure of human SSADH. The tetrameric structure is shown as a ribbon diagram showing one dimer in marine and salmon and the other dimer in magenta and green. The tetrameric structure of human SSADH was superimposed with that of human mitochondrial aldehyde dehydrogenase (ALDH2, pdb code 1nzz) that is presented in grey colour. Human SSADH shows the amino-acid sequence identity and the RMS deviation of 30% and 1.27 Å/466 ca, respectively, with the human mitochondrial ALDH2.
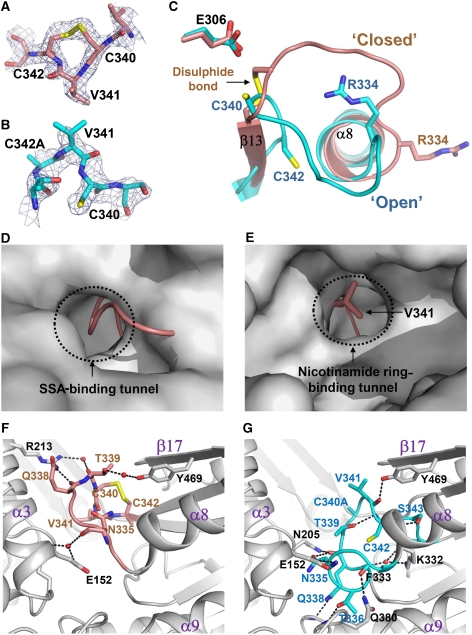
Despite the overall fold of SSADH being similar to those of other ALDHs, a remarkable difference is observed at the active site and the substrate entrance. Interestingly, in our SSADH structure, a disulphide bond formation is found between a catalytic cysteine residue (Cys340) and an adjacent cysteine residue (Cys342) (Figure 2A and C). It seems that, because the protein is crystallized from the condition without a reducing agent, the two cysteine residues are oxidized and form a disulphide bond. Moreover, a catalytic loop (RNTGQTCVCSN, residues ∼334–344) connecting α8 and β13 has a conformation to completely block both the substrate- and cofactor-binding sites (Figure 2D and E). The disulphide-bonded and ‘closed' conformation of the catalytic loop obviously leads the enzyme to an inactive state not only by oxidation of the catalytic residue Cys340 but also by disabled entries of the substrate and the cofactor to the active site. The disulphide-bonded and ‘closed' conformation of the catalytic loop is stabilized by hydrogen bond network between the residues in the loop and the conserved neighbouring residues. Three water molecules are involved in the hydrogen bonding as well (Figure 2F).
Figure 2.
Structural features of the ‘dynamic catalytic loop'. (A, B) A reversible disulphide bond formation. A disulphide bond formation in the oxidized form (A), and a breakage of the disulphide bond in the reduced form (B) of human SSADH are shown as a stick model, and their omit electron densities (blue mesh) are contoured at 3.0σ. (C) Structural changes of the ‘dynamic catalytic loop'. The catalytic loops of the oxidized and reduced forms of the wild-type SSADH (coloured salmon and cyan, respectively) are superimposed. The disulphide bond in the oxidized form, and the two cysteine residues in the reduced form of the SSADH are shown as a stick model and labelled appropriately. The R334 (involved in the substrate binding) and the E306 (functions as a general base) residues of the oxidized and reduced forms of SSADH are presented as stick models. (D, E) Blocking of the SSA-binding tunnel (D) and the nicotinamide ring-binding tunnel (E) by the ‘dynamic catalytic loop' in the oxidized form. The oxidized (ribbon model, coloured salmon) and reduced (surface model, coloured grey) forms of SSADH were superimposed, The SSA and nicotinamide ring-binding tunnels are indicated. The V341 residue that blocks the nicotinamide ring-binding tunnel is shown as a stick model and labelled. (F) Stabilization of the ‘dynamic catalytic loop' in the oxidized ‘closed' form. The ‘dynamic catalytic loop' in the oxidized form is presented in salmon colour, and the residues involved in the hydrogen bonding are as stick model. (G) Stabilization of the ‘dynamic catalytic loop' in the reduced ‘open' form. The ‘dynamic catalytic loop' in the reduced form is presented in cyan colour in the same view with that of in the oxidized form, and the residues involved in the hydrogen bonding are as stick model. For both (F, G), the neighbouring residues involved in the hydrogen bonding with the ‘dynamic catalytic loop' are shown as a stick model in grey colour, and water molecules involved in the stabilization of the loop are presented as red spheres. The hydrogen bond interactions are indicated by black dotted lines.
Crystal structure of the reduced form of human SSADH
The disulphide-bonded and ‘closed' conformation of the catalytic loop observed in the oxidized form of human SSADH raised the possibility of the redox-switch modulation of the enzyme. To investigate the possibility, we crystallized the wild-type (WT) SSADH supplemented with a strong reducing agent tris(2-carboxyethyl)phosphine (TCEP) in the crystallization condition, and determined the crystal structure of the reduced form of the enzyme. As expected, in the reduced form of SSADH, the disulphide bond is found to be broken (Figure 2B). After breakage, the thiol group of Cys340 moves towards the general base Glu306 constituting an active site environment as observed in other ALDHs, and the two cysteine residues involved in disulphide bond in the oxidized form are positioned apart with a distance of 9.24 Å (Figure 2C). The reduction in the enzyme also induces large structural changes in the catalytic loop, switching the loop to the ‘open' conformation, in which the substrate and the cofactor can freely access; hereafter, we call the catalytic loop as a ‘dynamic catalytic loop'. We then determined the crystal structure of the C340A SSADH mutant that crystallized in the condition without a reducing agent to investigate how the artificial breakage of the disulphide bond leads to the conformational changes. In spite of the oxidized condition, the ‘dynamic catalytic loop' of the SSADH mutant shows the ‘open' conformation, and is well superimposed with that of the reduced form (Figure 2C). The result indicates that the breakage of the disulphide bond induces the structural changes of the ‘dynamic catalytic loop', and the ‘open' conformation is structurally preferable to the ‘closed' conformation when the disulphide bond is broken. The ‘open' conformation of the ‘dynamic catalytic loop' is stabilized by hydrogen bond network between the residues in the loop and the conserved neighbouring residues, but with different hydrogen bond interactions from those in the ‘closed' conformation (Figure 2G).
Redox-switch modulation by dynamic catalytic loop
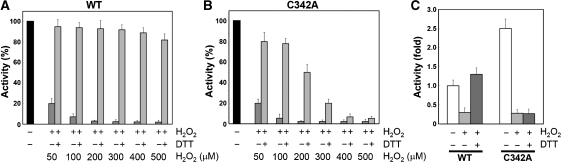
In general, redox-mediated modification of cellular proteins confers a response to ROS and changes in redox status that regulates the initiation of signal-transduction pathways and the induction of gene expression (Choi et al, 2001; Barford, 2004; Ilbert et al, 2007). To verify the redox-switch modulation of human SSADH, first we examined how SSADH responds to redox status using WT and two cysteine-substituted mutants (C340A and C342A). Although no SSADH activity was detected from all three forms without a reducing agent, WT and the C342A mutant showed enzyme activities by the addition of reducing agents such as β-mercaptoethanol (BME), dithiothreitol (DTT) and reduced glutathione (GSH), indicating that SSADH is inactive under oxidation condition and turns into active status when the catalytic residue Cys340 is reduced to thiol (Supplementary Figure S1). Interestingly, the C342A mutant exhibited 1.5 times higher enzyme activity than WT SSADH under the same concentration of a reducing agent, which might be due to the fact that the C342A mutant is constantly maintained in the ‘open' form and provides better access to the oxidized Cys340 for the reducing agents.
To investigate how human SSADH senses ROS, we examined the susceptibility of the enzyme to hydrogen peroxide (H2O2) and activity recovery by a reducing agent, using the recombinant proteins of WT and the C342A mutant. When reduced WT and the C342A mutant were treated with various concentrations of H2O2, both proteins showed almost complete loss of activities in the presence of over 100 μM H2O2 (Figure 3A and B). When the environment was switched to a reduced state by the addition of 10 mM DTT, the activity of WT SSADH was almost completely recovered, whereas the activity recovery of the C342A mutant was dramatically decreased upon the increase of the H2O2 concentration. (Figure 3A and B). The results indicate that WT SSADH is inactivated by oxidative stress through a disulphide bond formation between the catalytic residue Cys340 and an adjacent cysteine residue Cys342, which enables the enzyme to be reactivated upon the environmental switch to reducing condition. However, the C342A mutant might be permanently inactivated through the oxidation of Cys340 to sulfinic acid that cannot be reduced by a reducing agent.
Figure 3.
The ‘dynamic catalytic loop' of human SSADH senses ROS both in vitro and in vivo. (A, B) SSADH senses ROS in vitro. To investigate how SSADH senses ROS through the disulphide bond formation, wild type (WT) and the C342A mutant were treated with various concentrations of H2O2 in vitro and SSADH activities were measured. We then added 10 mM DTT to switch the environment to reduced state, and compared the activity recovery between the two different forms of SSADH. Over 80% of activity was recovered from wild-type SSADH that was treated with even 500 μM H2O2 (A), whereas almost no activities were recovered from the C342A mutant that was treated with more than 400 μM H2O2 (B). (C) SSADH senses ROS in vivo. Signal sequence-containing wild-type (WT) and C342A mutant SSADH were expressed in HEK293 cell. Cells were treated with 500 μM of H2O2 for 1 h to provide oxidative stress, and lysed anaerobically. In addition, SSADH activities were measured with samples with and without H2O2 treatment. To investigate the activity recovery, 10 mM of DTT was added to the H2O2-treated cell lysates, and SSADH activities were measured.
Next, to verify the sensitivity of the enzyme to ROS in a physiological condition, we expressed the signal sequence-containing WT and C342A mutant in HEK293 cell, and examined how the enzyme senses H2O2 in vivo (Figure 3C; Supplementary Figure S2). As expected, both forms of SSADH showed loss of their activities after the H2O2 treatment in cells; however, WT SSADH recovered its activity by the addition of DTT, whereas C342A could not (Figure 3C). The results indicate that human SSADH senses and is protected from oxidative stress through a reversible disulphide bond formation in a physiological condition. Interestingly, WT SSADH exhibited even 1.3 times higher activity by the addition of DTT than before the H2O2 treatment in vivo (Figure 3C), implying that some SSADH proteins exist as inactive forms in a normal physiological redox environment of the mitochondrial matrix that is known to be maintained in a relatively more oxidizing condition than the cytosol (Hu et al, 2008). Even though the in vivo results could not completely represent the true physiological phenomena, the clear tendency of the results drives us to propose that the redox-switch modulation could be a physiological control mechanism of human SSADH, which regulates its activity in a relatively oxidizing environment of the mitochondrial matrix and allows the enzyme to either respond to or be protected from oxidative stress. Considering the role of SSADH in the GABA metabolic pathway, we also propose that the cellular concentration of GABA in human brain could be controlled by the redox status of the mitochondrial matrix and oxidative stress as well. More detailed experiments such as those using the SSADH transgenic mouse might be needed to further investigate the physiological effects of the redox-switch regulation of SSADH in human brain.
SSA binding specificity
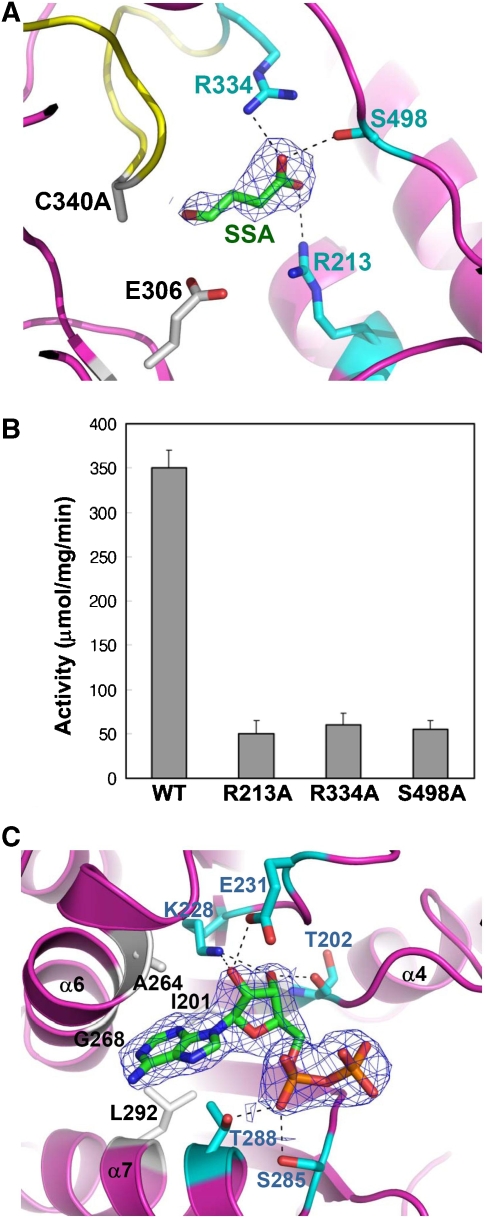
SSADH was known to have very high substrate specificity (Rothacker and Ilg, 2008), and to elucidate the high-fidelity substrate-binding mode of the enzyme, we determined the crystal structure of the C340A SSADH mutant bound with its biological substrate SSA. The aldehyde group of SSA is located near the catalytic nucleophile Cys340 and the general base Glu306, and the carboxyl group is stabilized by forming hydrogen bonds with the hydrophilic side chains of Arg213, Arg334 and Ser498 residues that are completely conserved throughout the organisms from bacteria to mammals (Figure 4A). In particular, the Arg334 residue is positioned far away from the SSA-binding site in the ‘closed' conformation, and upon the structural changes of the ‘dynamic catalytic loop', moves to the position where the side chain of the residue is able to form a hydrogen bond with the carboxyl group of SSA (Figure 2C). The involvement of these residues in the substrate binding is further confirmed by measuring the enzyme activity of the SSADH mutants. Each of the three mutants, R213A, R334A and S498A, exhibits activity of less than 10% than that of the WT SSADH (Figure 4B). Unlike other ALDHs that usually contain a hydrophobic tunnel to accept hydrophobic alkanal and aromatic aldehyde substrates, SSADH seems to use the hydrophilic residues to constitute an optimum environment to accept SSA as a substrate that contains the hydrophilic carboxyl end.
Figure 4.
Substrate- and cofactor-binding properties of human SSADH. (A) Diagram showing the SSA-binding site. To prevent the enzyme reaction and keep the enzyme in ‘open' formation, the C340A mutant was used for the complex crystallization. The C340A mutant is shown as a ribbon model in magenta colour, and its ‘dynamic catalytic loop' is shown in yellow. SSA is shown as a stick model in green colour, and the 2Fo−Fc electron density (blue mesh) of SSA is contoured at 0.8σ. Residues, whose side chains form hydrogen bonds to the carboxyl group of SSA, are shown as stick models in cyan colour, and the hydrogen bond interactions are indicated by black dotted lines. The catalytic residues, E306 and C340 (substituted with alanine in the present structure), are shown as stick models in grey colour. (B) SSADH activity assays. Each of three residues (R213, R334 and S498) involved in the SSA binding was substituted with alanine by site-directed mutagenesis, and SSADH activity assays were performed by measuring the reduction of NAD+ to NADH. (C) SSADH–NAD complex structure. The 2Fo−Fc electron density (blue mesh) of ADP moiety of NAD+ molecule is contoured at 1.0σ. The residues involved in hydrogen bond formations with ADP moiety and in forming hydrophobic cavity are shown as a stick model in cyan and grey colours, respectively. The hydrogen bond interactions are indicated by black dotted lines.
NAD+-binding properties
To elucidate the binding properties of NAD+ to the enzyme, we determined the crystal structure of C340A SSADH mutant bound with NAD+. Although the ADP moiety of the NAD+ molecule was added to the structure using the well-defined electron density, the nicotinamide ring and ribose portions of the cofactor could not be modelled as there was no electron density for them. This phenomenon is also observed in other ALDHs, in which the nicotinamide ring and ribose portions of the cofactor are mobile producing a poor electron density (Di Costanzo et al, 2007). In human SSADH, the ADP moiety of the NAD+ molecule is observed to be stabilized as follows: the adenine base is positioned in a hydrophobic pocket that is constituted by several conserved hydrophobic residues such as Ile201, Ala264, Gly268 and Leu292. For stabilization of the ribose ring, the hydroxyl groups of the ring are hydrogen bonded to the side chains of Lys228 and Glu231, and the backbone CO group of Thr202. Diphosphate oxygen atom AO1 accepts hydrogen bonds from the side chains of Ser285 and Thr288 (Figure 4C).
As described previously, the NAD+ molecule is not able to access to its binding site in the ‘closed' form of the enzyme. Upon the structural changes from the ‘open' to the ‘closed' conformation, the Val341 residue located in the midst of disulphide bond-forming two cysteine residues moves 5.03 Å to the NAD+-binding site, resulting in complete blockage of the tunnel where the nicotinamide ring binds (Figure 2E). Considering the blockage of the substrate-binding site as described previously, the ‘closed' conformation of the ‘dynamic catalytic loop' seems to completely shut down the enzymatic activity in the oxidation state by structurally blocking the entries of both the substrate and cofactor to their corresponding binding sites.
The impact of pathogenic mutations
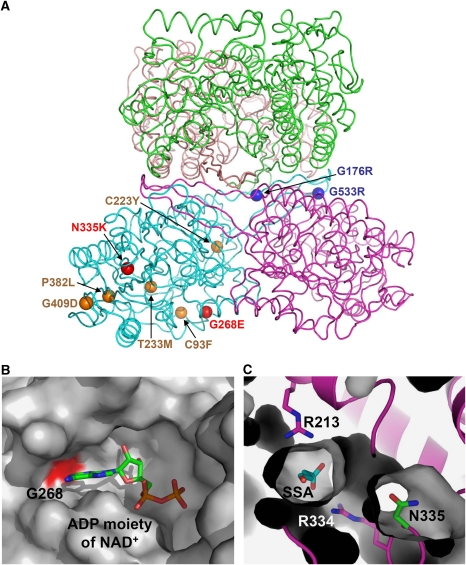
The availability of the three-dimensional structure of the human SSADH and the information on the catalytic and regulatory mechanisms of the enzyme allow us to rationalize the impact of known mutations associated with the disease pathogenesis. Among 18 missense variants causing amino-acid changes identified from the SSADH-deficient patients, 9 of them (C93F, G176R, C223Y, T233M, G268E, N335K, P382L, G409D and G533R) are known to result in an almost complete loss of SSADH activity with less than 5% of the WT when these mutants are reproduced by site-directed mutagenesis and expressed in HEK293 cell (Akaboshi et al, 2003). On the basis of the structural context, the nine missense point mutations can fall, in general terms, into two categories (Figure 5A). The first group of mutations is likely to include mostly buried sites and sites with a distinct structural role. Four such sites are the buried, Cys93, Cys223, Thr233 and Pro382, with the corresponding mutants (C93F, C223Y, T233M and P382L) almost certainly leading to a loss of or reduction in protein stability (Supplementary Figure S3). The residues Gly176 and Gly533 are located in the oligomerization interfaces, and their substitution to bulky basic residue arginine (G176R and G533R mutants, respectively) might disrupt the protein oligomerization and subsequently the protein stability (Supplementary Figure S4). Residue Gly409 is in a conformation disallowed for residues other than glycine; therefore, its mutation (G409R) is potentially destabilizing the protein as well (Supplementary Figure S4). The second group of missense mutations appears to directly affect the enzyme catalysis. The residue Gly268 is involved in constituting the binding pocket of the adenine base of NAD+, and its substitution to glutamate (G268E mutant) might interfere the binding of NAD+ leading to the enzyme inactive (Figure 5B). The residue Asn335 is located on the ‘dynamic catalytic loop', and the corresponding mutant N335K might severely distort the active site environment and/or reduce dynamics of the ‘dynamic catalytic loop' (Figure 5C). Mitochondrial ALDH2, a structurally similar protein with SSADH, is the major enzyme that oxidizes ethanol-derived acetaldehyde, and a nearly inactive form of the enzyme is found in about 40% of the East Asian population (Seitz et al, 2001). This enzyme variant, defined by a glutamate to lysine substitution at residue 487 located within the oligomerization domain, is known to exhibit rigid body rotations of its catalytic- and coenzyme-binding domains relative to the oligomerization domain by the inability of K487 to form important stabilizing hydrogen bonds with arginines 264 and 475 (Steinmetz et al, 1997; Larson et al, 2005). In SSADH, E526 residue (a corresponding residue of ALDH2 K487) is observed to form hydrogen bonds with arginines 302 and 514 (corresponding residues of ALDH2 R264 and R475, respectively).
Figure 5.
Impact of patient mutations on the structure and function of human SSADH. (A) The distributions of nine missense point mutations found in SSADH-deficient patients are shown using a ball representation. Mutations that impact on the enzyme catalysis, oligomerization and protein stability are shown and labelled in red, blue and orange colours, respectively. (B) Impact of G268E mutation. The NAD-binding groove is shown as a surface representation in grey colour, and the Gly268 residue, whose mutation to glutamate might interfere the binding of NAD+, is shown in red colour. The ADP moiety of a NAD+ molecule is shown as a stick model and labelled. (C) Impact of N335K mutation. The SSADH–SSA complex is shown as a ribbon diagram in magenta colour, and SSADH excluding the N335 residue is a surface fill model. SSA, N335 and two arginine residues involved in SSA binding are shown as stick models in cyan, green and magenta colours, respectively. The mutation of N335 to lysine might severely distort the active site environment.
Conclusion
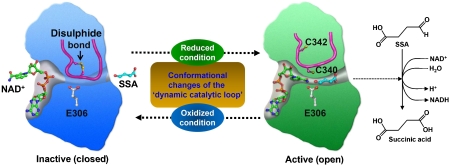
Our results reveal that human SSADH is regulated by redox-switch modulation, which is mediated by the reversible disulphide bond formation and subsequent large structural changes of the ‘dynamic catalytic loop' (Figure 6). We also reveal that human SSADH senses ROS and changes in redox status through the redox-switch modulation. In mammalian brain, redox-switch modulation by sensing ROS and changes in redox status is widely used as well for the regulation of signal transduction and transporter activation; for example, GABAA receptors are known to be regulated by redox modulation using a reversible disulphide bond (Amato et al, 1999; Pan et al, 2000), and the N-methyl-D-aspartate (NMDA) receptor, an important target for the actions of ethanol in brain, is regulated through redox modulation (Trotti et al, 1997; Ikonomidou et al, 2000). It has also been reported that oxidative stress is generated in the SSADH-deficient mice (Latini et al, 2007) and GABA shunt is required to restrict the levels of reactive oxygen intermediates in plants (Bouche et al, 2003; Fait et al, 2005), which implies that changes in redox status and oxidative stress might be ultimate cause and effect in the GABA metabolism. On the basis of the findings presented in the study, we propose that a physiological regulation of human SSADH is mediated by redox-switch modulation through which the enzyme might function as a key enzyme in the regulation of GABA shunt and cellular levels of GABA and GHB concentrations by sensing ROS or changes in redox status.
Figure 6.
Redox-switch-mediated enzyme reaction mechanism of human SSADH. SSADH catalyses the conversion of SSA to succinic acid using NAD+ as a cofactor. In oxidized status, the catalytic residue Cys340 is oxidized forming a disulphide bond with an adjacent cysteine residue (Cys342) and the ‘dynamic catalytic loop' is in a ‘closed' conformation, leading to the enzyme inactive. When the environment is switched to reduced status, the disulphide bond is broken, and the ‘dynamic catalytic loop' undergoes structural changes to an ‘open' form in which both SSA and NAD+ are able to access to. Upon breakage of the disulphide bond, the thiol group of Cys340 moves towards a general base Glu306 to form an active site environment.
Materials and methods
Cloning, expression and purification of human SSADH
The gene coding for N-terminal signal sequence truncated human SSADH (amino-acid residues 48–535) was amplified from the human brain cDNA library by PCR, and the PCR product was then subcloned into pQE30 (Qiagen) with 6His at the N terminus. The resulting expression vector pQE30:hSSADH was transformed into Escherichia coli M15(pREP4) strain and was grown in an LB medium containing 100 μg/ml ampicillin at 37°C to OD600 of 0.6. After induction with 1.0 mM IPTG for a further 20 h at 22°C, the culture was harvested by centrifugation at 5000 g for 15 min at 4°C. Cell pellet was resuspended in ice-cold buffer A (50 mM Tris–HCl, pH 8.0 and 200 mM NaCl) and disrupted by ultrasonication. The cell debris was removed by centrifugation at 11 000 g for 1 h, and lysate was bound to Ni-NTA agarose (Qiagen). After washing with buffer A containing 10 mM imidazole, the bound proteins were eluted with 300 mM imidazole in buffer A. Further purification was carried out by applying the HiTrap Q ion exchange chromatography. The purified protein was concentrated to 20 mg/ml in 50 mM Tris–HCl, pH 8.0 with 200 mM NaCl, and stored at −80°C for crystallization trials. SDS–PAGE analysis of the purified protein showed a single band of ∼53.0 kDa that corresponds to the calculated molecular weight of a human SSADH monomer. The SSADH mutants were similarly prepared as the WT SSADH, after the site-directed mutagenesis method was applied.
Crystallization, data collection and structure determination
The proteins were crystallized by the sitting drop method. The oxidized form of human SSADH and C340A mutants was crystallized in 100 mM HEPES (pH 7.25) and 1.9 M lithium sulphate, and the reduced form was crystallized in the same condition as the oxidized form with 5 mM TCEP. For the crystallization of SSA- and NAD+-bound form of SSADH, C340A mutant crystals were soaked with 30 and 50 mM of SSA and NAD+, respectively, for 30 min. Data were collected at the 6C1(MXII) beamline at the Pohang Accelerator Laboratory using a QUANTUM 210 CCD detector (San Diego, CA, USA). The oxidized form of SSADH crystals diffracted to 2.0-Å resolution, and the crystals of the reduced form, C340A mutant, SSA- and NAD-bound SSADH diffracted to 3.4, 2.3, 2.4 and 2.4 Å, respectively. The data were then indexed, integrated and scaled using the HKL2000 suite (Otwinowski and Minor, 1997). The crystals belonged to the space group F432 and have unit cell dimensions of a=b=c=265.706. With one molecule per asymmetric unit, the crystal volume per unit of protein weight was 3.69 Å3/Da, corresponding to a solvent content of 66.6% (Matthews, 1968). The structure of the oxidized form SSADH was solved by molecular replacement method using Molrep. The crystal structure of E. coli lactaldehyde dehydrogenase (pdb code 2opx), the closest structural homologue with a sequence identity of 36%, was used as a search model. The model building was performed manually using the program WinCoot (Emsley and Cowtan, 2004) and the refinement was performed with CCP4 refmac5 (Murshudov et al, 1997) and CNS (Brünger et al, 1998). The structure of reduced form of SSADH was determined by molecular replacement using Molrep and the refined oxidized form of SSADH model, and the structures of C340A mutant, SSA- and NAD-bound SSADH were by the same method and the refined reduced form of SSADH. Refinement of these structures proceeded as for the oxidized form of SSADH. To verify the formation and the breakage of the disulphide bond in oxidized and reduced conditions, respectively, simulated annealing omit maps were generated by excluding all residues in the ‘dynamic catalytic loop' of human SSADH. The data statistics are summarized in Table I. Five refined models were deposited in the Protein Data Bank.
Table 1.
Data collection and refinement statistics of human SSADH
| Oxidized form | Reduced form | C340A | C340A–SSA | C340A–NAD+ | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | F432 | ||||
| Cell dimensions | |||||
| a=b=c (Å) | 265.706 | 265.910 | 265.895 | 265.400 | 265.925 |
| α=β=γ (deg) | 90.0 | 90.0 | 90.0 | 90.0 | 90 |
| Resolution (Å) | 30.0–2.0 | 30.0–3.4 | 30.0–2.3 | 30.0–2.4 | 30.0–2.4 |
| Rsyma | 9.2 (30.6) | 20.3 (28.0) | 12.1 (29.9) | 10.3 (29.3) | 9.0 (27.4) |
| I/σ(I) | 13.7 (2.4) | 4.8 (2.1) | 12.9 (2.88) | 14.3 (3.7) | 15.3 (2.6) |
| Completeness (%) | 96.6 (92.1) | 95.2 (77.2) | 98.8 (94.5) | 97.8 (88.4) | 97.2 (90.1) |
| Redundancy | 10.2 | 10.0 | 18.6 | 17.7 | 12.8 |
| Refinement | |||||
| Resolution (Å) | 30–2.0 | 30–3.4 | 30–2.3 | 30–2.4 | 30–2.4 |
| No. of reflections | 54 484 | 11 596 | 36 254 | 31 825 | 32 022 |
| Rworkb/Rfree | 22.7/25.0 | 26.1/26.8 | 22.9/26.4 | 22.4/25.3 | 22.6/26.3 |
| No. of atoms | |||||
| Protein | 3627 | 3613 | 3653 | 3649 | 3683 |
| Water | 303 | 5 | 76 | 62 | 48 |
| No. of molecules | |||||
| SSA/NAD+ | 1 | 1 | |||
| SO4/glycerol | 8 | 8/2 | 8/3 | 8/2 | 10/2 |
| r.m.s deviations | |||||
| Bond lengths (Å) | 0.0057 | 0.0077 | 0.0059 | 0.0062 | 0.0066 |
| Bond angles (deg) | 1.2492 | 1.4144 | 1.260 | 1.2501 | 1.2304 |
| B-factor (Å2) | 27.01 | 28.20 | 28.30 | 32.85 | 27.45 |
| pdb code | 2w8n | 2w8o | 2w8p | 2w8q | 2w8r |
| aRsym=∑∣Iobs–Iavg∣/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is average over symmetry equivalents. | |||||
| bRwork=∑∣∣Fo∣–∣Fc∣∣/Ó∣Fo∣, where ∣Fo∣ and ∣Fc∣ are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 5% of the data. | |||||
| The numbers in parentheses are statistics from the highest resolution shell. | |||||
SSADH activity assay in vitro
The spectrophotometric method was used in the assay of SSADH activity. The rate of formation of reduction of NAD+ to NADH (extinction coefficient of 6.22 mM−1/cm) was measured by monitoring the increase in absorbance at 340 nm. The reaction mixture contains 100 mM tetrapotassium pyrophosphate, pH 8.5, containing 0.1 mM EDTA, 50 μM SSA and 3 mM NAD+. The reaction was performed at room temperature for 3 min. One unit of enzyme activity is defined as the amount of enzyme catalysing the production of 1 μmol of NADH per mg protein/min. To examine the enzyme activity in the reduced condition, each of 5 mM of BME, DTT and reduced GSH was added to WT, and two cysteine-substituted mutants (Cys340A and Cys342A). To investigate how human SSADH senses ROS, fully reduced SSADH WT and C340A mutant were treated with various concentrations of H2O2 for 1 h and enzyme–buffer assay mixture was added and the reactions were terminated by adding 5 mM L-methionine. To switch the environment to reduced condition, 10 mM DTT was added to H2O2-treated proteins, and incubated for 10 min. All incubations were performed at room temperature.
SSADH activity assay in vivo
To examine the in vivo enzymatic activity of SSADH, genes coding for signal sequence-containing WT or mutant SSADH were cloned into the pCMV-tag4 expression vector, and HEK293 cells were transfected by 1 μg/ml of the resulting plasmids for 24 h. For checking the expression of each vectors and cell viability, we performed the WB analysis using anti-FLAG Ab, caspase 9 and actin. Enzymatic activity of SSADH was measured as follows: transfected cells were lysed with extract buffer and incubated with assay reagent (50 mM NAD+ and 0.3 mM SSA-containing buffer). After 1 h incubation at 20°C, NADH production was monitored using NADH fluorescence. More detail has been described elsewhere (Ippolito et al, 2005). To test the susceptibility of SSADH to H2O2 in vivo, SSADH WT- and C340A mutant-expressed cells were treated with 500 μM of H2O2 for 1 h, lysed anaerobically and 10 mM of DTT was added. Then SSADH activities were measured with several cell lysate samples, including without H2O2 treatment, and with and without DTT addition after H2O2 treatment.
Supplementary Material
Supplementary Figures S1–S4
Acknowledgments
This study was funded by 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education, Science and Technology (MEST), Republic of Korea.
References
- Akaboshi S, Hogema BM, Novelletto A, Malaspina P, Salomons GS, Maropoulos GD, Jakobs C, Grompe M, Gibson KM (2003) Mutational spectrum of the succinate semialdehyde dehydrogenase (ALDH5A1) gene and functional analysis of 27 novel disease-causing mutations in patients with SSADH deficiency. Hum Mutat 22: 442–450 [DOI] [PubMed] [Google Scholar]
- Amato A, Connolly CN, Moss SJ, Smart TG (1999) Modulation of neuronal and recombinant GABAA receptors by redox reagents. J Physiol 517: 35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshima T, Kajita M, Sekido Y, Ishiguro Y, Tsuge I, Kimura M, Yamaguchi S, Watanabe K, Shimokata K, Niwa T (2002) Mutation analysis in a patient with succinic semialdehyde dehydrogenase deficiency: a compound heterozygote with 103-121del and 1460T>A of the ALDH5A1 gene. Hum Hered 53: 42–44 [DOI] [PubMed] [Google Scholar]
- Barford D (2004) The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol 14: 679–686 [DOI] [PubMed] [Google Scholar]
- Battaglioli G, Liu H, Martin DL (2003) Kinetic differences between the isoforms of glutamate decarboxylase: implications for the regulation of GABA synthesis. J Neurochem 86: 879–887 [DOI] [PubMed] [Google Scholar]
- Bekri S, Fossoud C, Plaza G, Guenne A, Salomons GS, Jakobs C, Van Obberghen E (2004) The molecular basis of succinic semialdehyde dehydrogenase deficiency in one family. Mol Genet Metab 81: 347–351 [DOI] [PubMed] [Google Scholar]
- Bouche N, Fait A, Bouchez D, Moller SG, Fromm H (2003) Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc Natl Acad Sci USA 100: 6843–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103–113 [DOI] [PubMed] [Google Scholar]
- Di Costanzo L, Gomez GA, Christianson DW (2007) Crystal structure of lactaldehyde dehydrogenase from Escherichia coli and inferences regarding substrate and cofactor specificity. J Mol Biol 366: 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fait A, Yellin A, Fromm H (2005) GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Lett 579: 415–420 [DOI] [PubMed] [Google Scholar]
- Fenalti G, Law RH, Buckle AM, Langendorf C, Tuck K, Rosado CJ, Faux NG, Mahmood K, Hampe CS, Banga JP, Wilce M, Schmidberger J, Rossjohn J, El-Kabbani O, Pike RN, Smith AI, Mackay IR, Rowley MJ, Whisstock JC (2007) GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop. Nat Struct Mol Biol 14: 280–286 [DOI] [PubMed] [Google Scholar]
- Gibson KM, Hoffmann GF, Hodson AK, Bottiglieri T, Jakobs C (1998) 4-Hydroxybutyric acid and the clinical phenotype of succinic semialdehyde dehydrogenase deficiency, an inborn error of GABA metabolism. Neuropediatrics 29: 14–22 [DOI] [PubMed] [Google Scholar]
- Gibson KM, Schor DS, Gupta M, Guerand WS, Senephansiri H, Burlingame TG, Bartels H, Hogema BM, Bottiglieri T, Froestl W, Snead OC, Grompe M, Jakobs C (2002) Focal neurometabolic alterations in mice deficient for succinate semialdehyde dehydrogenase. J Neurochem 81: 71–79 [DOI] [PubMed] [Google Scholar]
- Gupta M, Greven R, Jansen EE, Jakobs C, Hogema BM, Froestl W, Snead OC, Bartels H, Grompe M, Gibson KM (2002) Therapeutic intervention in mice deficient for succinic semialdehyde dehydrogenase (gamma-hydroxybutyric aciduria). J Pharmacol Exp Ther 302: 180–187 [DOI] [PubMed] [Google Scholar]
- Gupta M, Hogema BM, Grompe M, Bottiglieri TG, Concas A, Biggio G, Sogliano C, Rigamonti AE, Pearl PL, Snead OC III, Jakobs C, Gibson KM (2003) Murine succinate semialdehyde dehydrogenase deficiency. Ann Neurol 54(Suppl 6): S81–S90 [DOI] [PubMed] [Google Scholar]
- Hogema BM, Akaboshi S, Taylor M, Salomons GS, Jakobs C, Schutgens RB, Wilcken B, Worthington S, Maropoulos G, Grompe M, Gibson KM (2001a) Prenatal diagnosis of succinic semialdehyde dehydrogenase deficiency: increased accuracy employing DNA, enzyme, and metabolite analyses. Mol Genet Metab 72: 218–222 [DOI] [PubMed] [Google Scholar]
- Hogema BM, Gupta M, Senephansiri H, Burlingame TG, Taylor M, Jakobs C, Schutgens RB, Froestl W, Snead OC, Diaz-Arrastia R, Bottiglieri T, Grompe M, Gibson KM (2001b) Pharmacologic rescue of lethal seizures in mice deficient in succinate semialdehyde dehydrogenase. Nat Genet 29: 212–216 [DOI] [PubMed] [Google Scholar]
- Hu J, Dong L, Outten CE (2008) The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem 283: 29126–29134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidou C, Bittigau P, Ishimaru MJ, Wozniak DF, Koch C, Genz K, Price MT, Stefovska V, Hörster F, Tenkova T, Dikranian K, Olney JW (2000) Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science 287: 1056–1060 [DOI] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U (2007) The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol 14: 556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito JE, Xu J, Jain S, Moulder K, Mennerick S, Crowley JR, Townsend RR, Gordon JI (2005) An integrated functional genomics and metabolomics approach for defining poor prognosis in human neuroendocrine cancers. Proc Natl Acad Sci USA 102: 9901–9906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly VP, Sherratt PJ, Crouch DH, Hayes JD (2002) Novel homodimeric and heterodimeric rat g-hydroxybutyrate synthases that associate with the Golgi apparatus define a distinct subclass of aldo-keto reductase 7 proteins. Biochem J 366: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson HN, Weiner H, Hurley TD (2005) Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase ‘Asian' variant. J Biol Chem 280: 30550–30556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini A, Scussiato K, Leipnitz G, Gibson KM, Wajner M (2007) Evidence for oxidative stress in tissues derived from succinate semialdehyde dehydrogenase-deficient mice. J Inherit Metab Dis 30: 800–810 [DOI] [PubMed] [Google Scholar]
- Maitre M (1997) The gamma-hydroxybutyrate signaling system in the brain: organization and functional implications. Prog Neurobiol 51: 337–361 [DOI] [PubMed] [Google Scholar]
- Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33: 491–497 [DOI] [PubMed] [Google Scholar]
- Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN (1998) Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure 6: 1541–1551 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Procession of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pan ZH, Zhang X, Lipton SA (2000) Redox modulation of recombinant human GABAA receptors. Neuroscience 98: 333–338 [DOI] [PubMed] [Google Scholar]
- Pearl PL, Gibson KM, Acosta MT, Vezina LG, Theodore WH, Rogawski MA, Novotny EJ, Gropman A, Conry JA, Berry GT, Tuchman M (2003a) Clinical spectrum of succinic semialdehyde dehydrogenase deficiency. Neurology 60: 1413–1417 [DOI] [PubMed] [Google Scholar]
- Pearl PL, Novotny EJ, Acosta MT, Jakobs C, Gibson KM (2003b) Succinic semialdehyde dehydrogenase deficiency in children and adults. Ann Neurol 54(Suppl 6): S73–S80 [DOI] [PubMed] [Google Scholar]
- Porter TG, Spink DC, Martin SB, Martin DL (1985) Transaminations catalysed by brain glutamate decarboxylase. Biochem J 231: 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothacker B, Ilg T (2008) Functional characterization of a Drosophila melanogaster succinic semialdehyde dehydrogenase and a non-specific aldehyde dehydrogenase. Insect Biochem Mol Biol 38: 354–366 [DOI] [PubMed] [Google Scholar]
- Seitz HK, Matsuzaki S, Yokoyama A, Homann N, Vakevainen S, Wang XD (2001) Alcohol and cancer. Alcohol Clin Exp Res 25: 137S–143S [DOI] [PubMed] [Google Scholar]
- Steinmetz CG, Xie P, Weiner H, Hurley TD (1997) Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 5: 701–711 [DOI] [PubMed] [Google Scholar]
- Storici P, De Biase D, Bossa F, Bruno S, Mozzarelli A, Peneff C, Silverman RB, Schirmer T (2004) Structures of gamma-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5′-phosphate, and [2Fe–2S] cluster-containing enzyme, complexed with gamma-ethynyl-GABA and the antiepilepsy drug vigabatrin. J Biol Chem 279: 363–373 [DOI] [PubMed] [Google Scholar]
- Trettel F, Malaspina P, Jodice C, Novelletto A, Slaughter CA, Caudle DL, Hinson DD, Chambliss KL, Gibson KM (1997) Human succinic semialdehyde dehydrogenase: molecular cloning and chromosomal localization. Adv Exp Med Biol 414: 253–260 [PubMed] [Google Scholar]
- Trotti D, Rizzini BL, Rossi D, Haugeto O, Racagni G, Danbolt NC, Volterra A (1997) Neuronal and glial glutamate transporters possess an SH-based redox regulatory mechanism. Eur J Neurosci 9: 1236–1243 [DOI] [PubMed] [Google Scholar]
- Wong CG, Bottiglieri T, Snead OC (2003) GABA, gamma-hydroxybutyric acid, and neurological disease. Ann Neurol 54 (Suppl 6): S3–S12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S4