Abstract
Telomere lengths are maintained in many cancer cells by the ribonucleoprotein enzyme telomerase but can be further elongated by increasing telomerase activity through the overexpression of telomerase components. We report here that increased telomerase activity results in increased telomere length that eventually reaches a plateau, accompanied by the generation of telomere length heterogeneity and the accumulation of extrachromosomal telomeric repeat DNA, principally in the form of telomeric circles (t-circles). Telomeric DNA was observed in promyelocytic leukemia bodies, but no intertelomeric copying or telomere exchange events were identified, and there was no increase in telomere dysfunction-induced foci. These data indicate that human cells possess a mechanism to negatively regulate telomere length by trimming telomeric DNA from the chromosome ends, most likely by t-loop resolution to form t-circles. Additionally, these results indicate that some phenotypic characteristics attributed to alternative lengthening of telomeres (ALT) result from increased mean telomere length, rather than from the ALT mechanism itself.
Keywords: alternative lengthening of telomeres, extrachromosomal telomeric repeat DNA, t-circle, telomerase, telomere
Introduction
In human cells, telomeric DNA is composed of double-stranded (ds) arrays of a (TTAGGG)n repeat unit, terminating in a single-stranded (ss) G-rich 3′ overhang (Moyzis et al, 1988; de Lange et al, 1990; Makarov et al, 1997; Wright et al, 1997). Telomeres have been shown to exist in a stable secondary structure called a telomere-loop (t-loop), which is formed by strand invasion of the 3′ overhang into internal telomeric repeats on the same chromosome end and the consequent formation of a displacement loop at the site of sequestration (Griffith et al, 1999).
The shelterin complex, which comprises six telomere-specific proteins (TRF1, TRF2, POT1, TIN2, TPP1 and Rap1), associates with the telomeric repeat DNA to form a protective structure that prevents the chromosome terminus from being inappropriately processed by DNA repair pathways (de Lange, 2005). It has been proposed that shelterin is responsible for the structural stability of the t-loop and acts dynamically with a number of interacting partners, which are involved in DNA processing, to shape the telomere and ensure telomere integrity (de Lange, 2005; Palm and de Lange, 2008). Nevertheless, the details of t-loop formation and their dynamics throughout the cell cycle remain elusive.
To counteract telomere shortening, which occurs as a result of end-replication limitations, the majority of immortal cells as well as cells of the germline use the ribonucleoprotein enzyme telomerase, the catalytic core of which comprises the reverse transcriptase hTERT and the RNA component hTR (Feng et al, 1995; Nakamura et al, 1997). Levels of hTR and hTERT are both limiting for telomerase activity and telomere length in immortalised cells, and telomerase-positive cancer cell lines typically have a very small number of telomerase molecules and stably short telomeres (Cristofari and Lingner, 2006; Cao et al, 2008). In such situations, telomerase is preferentially recruited to the shorter telomeres (Bianchi and Shore, 2008).
Telomerase-positive immortal cells maintain a balance between telomere attrition and telomere elongation by negative feedback of telomere length on telomerase activity, mediated in cis at each individual chromosome end by the binding of shelterin proteins, especially TRF1, along the telomeric tract (Smogorzewska et al, 2000; Smogorzewska and de Lange, 2004; Bianchi and Shore, 2008). This feedback mechanism is likely to involve monitoring of the number of telomere-binding proteins and resultant changes in telomere accessibility by telomerase. Telomeres can be lengthened by increasing telomerase activity through exogenous expression of hTR and hTERT, resulting in an increase in telomere-bound shelterin proteins in proportion to the increased telomere length (Cristofari and Lingner, 2006).
In this study, we examined the long-term effects of exogenous hTR expression in telomerase-positive human cancer cell lines and the effects of elongation on telomere function and stability. The results indicate the existence of a mechanism in human cells to negatively regulate telomere length by trimming of telomeric DNA from the chromosome ends.
Results
Increased telomerase activity resulted in telomere elongation and telomere length heterogeneity in telomerase-positive cells
HT1080 and HeLa telomerase-positive cells were transduced with pBABEpuro containing hTR driven by the U3 promoter, or with an empty vector control. Cells were selected and passaged as stable mass cultures for 300 population doublings (pds) from the point at which they were transduced. After transduction, hTR expression increased approximately 20-fold in both HeLa and HT1080 cells, compared with vector controls, and was maintained at this level throughout the 300 pds (Figure 1A). hTERT expression remained unchanged after hTR overexpression in both cell lines (data not shown).
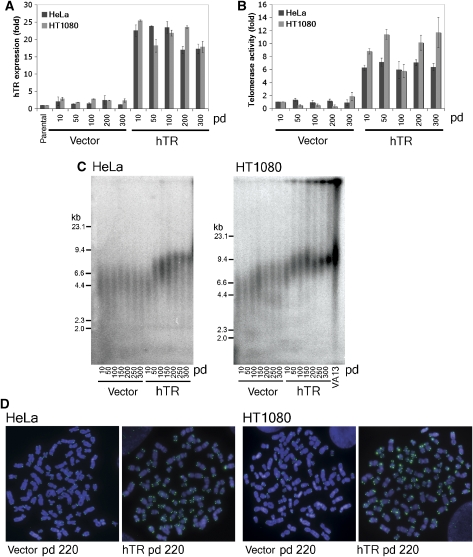
Figure 1.
hTR overexpression increases telomerase activity, telomere length and telomere length heterogeneity in telomerase-positive cells. (A) Expression of hTR as fold change relative to parental control cells at increasing pd of HeLa and HT1080 cells after transduction with pBABEpuro (vector) or pBABEpuroU3hTR (hTR), measured by quantitative RT–PCR. Error bars represent standard deviation between three separate experiments. (B) Telomerase activity measured by IP-RQ-TRAP and expressed as fold change relative to vector sample at pd 10. Error bars represent standard deviation between three separate experiments. (C) TRF analysis of genomic DNA digested with HinfI and RsaI from HeLa and HT1080 vector and hTR cells at increasing pds. Digested DNA from the ALT cell line VA13 was included as an example of telomere length heterogeneity. (D) Telomere-FISH of HeLa and HT1080 vector and hTR cells using a PNA probe against the G-rich telomeric DNA strand. Chromosomes were counterstained with DAPI.
Measurement of telomerase activity by conventional TRAP is sometimes unreliable due to the presence of telomerase inhibitors in cell extracts. To avoid this potential complication, we developed a modified real-time quantitative (RQ)-TRAP assay containing a telomerase purification step. Telomerase was immunoaffinity purified (IP) from cell lysate using an hTERT antibody, which has been verified previously for this application (Cohen and Reddel, 2008). After purification, telomerase-mediated extension of a nontelomeric primer was measured by quantitative PCR (IP-RQ-TRAP) (Wege et al, 2003; Cohen and Reddel, 2008). Linearity of the IP-RQ-TRAP technique was shown (Supplementary Figure S1). Telomerase activity increased approximately 6-fold in HeLa hTR cells and approximately 10-fold in HT1080 hTR cells (Figure 1B).
Terminal restriction fragments (TRFs) were generated by digestion of genomic DNA with HinfI and RsaI restriction endonucleases, separated by pulsed-field gel electrophoresis (PFGE), and then hybridised in-gel to a telomere-specific probe. Mean TRF lengths in HeLa vector control cells remained constant throughout the course of experimentation, at approximately 4.5 kb, whereas mean TRF lengths in HT1080 vector cells grew slightly at later pds from approximately 6.5–7.5 kb (Figure 1C).
HeLa hTR cells showed increasing telomere lengths over 150 pds, after which TRFs stabilised at approximately 8 kb (Figure 1C). Telomere lengths of HT1080 hTR cells strikingly continued to elongate with increasing pd, producing an increasingly heterogeneous TRF length profile. This was compared with the TRF profile of the cell line WI-38 VA13/2RA (referred to here as VA13) (Figure 1C). VA13 cells use the recombination-based alternative lengthening of telomeres (ALT) mechanism of telomere length maintenance, which is characterised by marked telomere length heterogeneity (Bryan et al, 1995). TRF analysis of ALT cells frequently shows residual telomeric signal in the wells of the gel. The nature of this DNA and whether it originates from ALT-associated promyelocytic leukemia (PML) bodies (APBs), is simply too large to be resolved, or whether it represents highly branched t-complex DNA (Nabetani and Ishikawa, 2009) is not known. At later pds, HT1080 hTR cells increasingly phenocopied the ALT TRF profile, including the retention of telomeric DNA in the wells, yet notably lacked the smallest sized telomeric fragments (Figure 1C).
Fluorescence in situ hybridisation (FISH) analysis of metaphase spreads using a telomeric peptide nucleic acid (PNA) probe confirmed telomere length heterogeneity (Figure 1D). Telomere FISH from vector transduced control cells showed homogeneous telomeric signals indicative of short and stable telomere lengths, whereas both HeLa hTR and HT1080 hTR cells showed an overall increase in telomeric signal, accompanied by marked signal heterogeneity between different telomeres within a single metaphase. This was particularly apparent in HT1080 hTR cells at later pd (Figure 1D).
Telomere lengthening in telomerase-positive cells resulted in the formation and accumulation of extrachromosomal telomeric repeat DNA
To determine whether extrachromosomal telomeric repeat (ECTR) DNA accounted for some of the increased telomeric content of HeLa hTR and HT1080 hTR cells, 2D gel electrophoresis was used to detect extrachromosomal t-circles. For optimal size resolution, TRFs were separated by 2D PFGE and hybridised to a telomere-specific probe. Bacteriophage lambda DNA, that was digested with HindIII endonuclease and then circularised by DNA ligase was loaded as ds circular DNA markers.
The prominent arc represents ds linear telomeric DNA of high molecular weight, which increases in size and signal intensity in HeLa and HT1080 hTR cells at pd 235, consistent with elongated telomeres (Figure 2A). An arc of open-form ds DNA circles containing telomeric sequence was present in the HeLa and HT1080 hTR cells, but not evident in HT1080 vector controls (Figure 2A). A faint arc of circular DNA could, however, be seen after longer exposure of the HeLa vector gel. The circular molecules were detected at all molecular weights within the range of detection (∼2–9 kb) and are consistent with the t-circles observed previously in ALT cells (Cesare and Griffith, 2004). A lower arc that has been identified previously as consisting of ss telomeric DNA (Cesare et al, 2008; Raices et al, 2008; Nabetani and Ishikawa, 2009) migrated beneath the linear ds DNA in HeLa hTR cells and was not present in the HT1080 hTR cells (Figure 2A).
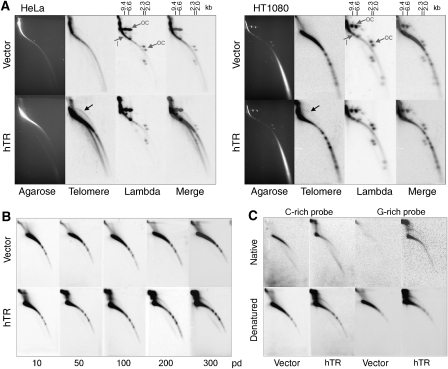
Figure 2.
Telomere lengthening results in t-circle accumulation and an increase in ss G-rich and t-complex telomeric DNA. (A) 2D PFGE of TRFs from HeLa and HT1080 vector and hTR cells at pd 235, digested with HinfI and RsaI. Gels were Southern blotted and hybridised with a C-rich telomeric DNA probe. The black arrows indicate circular telomeric DNA. Circularised HindIII-digested bacteriophage lambda DNA was electrophoresed simultaneously as a marker for ds circular DNA. Open circular (oc) spots are indicated, as is the remaining linear (l) telomere-derived signal. (B) 2D gel electrophoresis of TRFs from HT1080 vector and hTR cells at increasing pd. Telomeric DNA was detected in-gel with a γ-32P-labelled C-rich probe. (C) 2D gel electrophoresis of TRFs from HT1080 vector and hTR cells at pd 300. Hybridisation was carried out in-gel with a γ-32P-labelled C-rich or G-rich probe under native and denaturing conditions, to detect ss and ds telomeric DNA, respectively.
To determine at what stage the t-circles became abundant and to confirm that the circles are the result of progressive telomere elongation, standard 2D gels of HT1080 vector and hTR cells at increasing pds were hybridised to a telomeric probe (Figure 2B). Several prominent lower molecular weight spots, specific to the HT1080 cells were identified. These spots did not change in size and intensity and are therefore most likely to represent interstitial bands of telomeric sequence. By using these spots as a marker of exposure, the gels shown in Figure 2B and C appear to be suitably comparable.
T-circles were detected at early pds in the hTR-expressing cells after longer exposure, and increased substantially in intensity over successive pds (Figure 2B). In addition, an arc of telomeric repeat DNA migrating beneath the linear material was detected at later pds. Very faint arcs of circular telomeric DNA could also be observed after extended exposure in the HT1080 vector cells at later pds (Figure 2B).
HT1080 hTR cells show strong telomeric signal near the wells, consistent with t-complex DNA, which comprises highly branched telomeric repeats with large amounts of internal ss portions (Nabetani and Ishikawa, 2009). This type of telomeric DNA was recently described as a characteristic feature of ALT cells and was proposed to arise from Holliday junctions in the DNA (Nabetani and Ishikawa, 2009). Circular telomeric DNA appears to be present at low levels in control HT1080 and HeLa cells but, most strikingly, we show that ECTR DNA including t-complex DNA accumulates with increasing telomere lengths in the cells with overexpressed telomerase.
To determine more accurately the nature of the ECTR DNA, 2D gels were used to analyse DNA isolated from HT1080 vector and hTR cells at pd 300 and probed in-gel under native or denaturing conditions using a C-rich or G-rich telomeric probe (Figure 2C). Hybridisation under native conditions showed that the arc migrating beneath the linear ds telomeric DNA in the HT1080 hTR cells was composed of ss G-rich telomeric sequence, and that a smaller, but detectable amount was also present in the vector control cells (Figure 2C). Somewhat surprisingly, an increase in C-rich linear telomeric signal was seen under native conditions in hTR expressing cells, indicating the existence of either ss C-rich overhangs at the chromosome termini, or gapped ds linear TRFs, which have been identified previously in ALT cells (Nabetani and Ishikawa, 2009). T-complex DNA was detected in the hTR expressing cells under both native and denaturing conditions, consistent with this type of telomeric DNA containing portions of ss telomeric repeats (Nabetani and Ishikawa, 2009). T-circles were clearly visible with both strand-specific probes under denaturing conditions in HT1080 hTR cells, but not vector cells (Figure 2C). This shows that the t-circles are predominantly ds, although some ss C-rich t-circles can also be detected in HT1080 hTR cells, but are likely to represent ss regions of gapped circles.
Promyelocytic leukemia bodies were associated with telomeric DNA in telomerase-positive cells with extended telomeres
PML bodies are discrete nuclear foci that are present in the majority of mammalian cells and have been implicated in numerous cellular processes (Bernardi and Pandolfi, 2007). A subset of PML bodies known as APBs, which are thought to be specific to ALT cells, are distinguished from other PML bodies by containing telomeric DNA, telomeric-binding proteins and proteins involved in recombination (Yeager et al, 1999).
HeLa and HT1080 vector and hTR interphase cells were immunostained using an antibody against PML protein and then hybridised with a telomeric PNA probe. True colocalisations were confirmed by deconvolution of z-stacked images, followed by 3D imaging (Figure 3A and B). PML protein typically exists in a spherical or doughnut-shaped body (Jiang and Ringertz, 1997) and colocalisations were only counted when the telomeric DNA localised, throughout the z-stacks, to the centre of the PML body (Figure 3B).
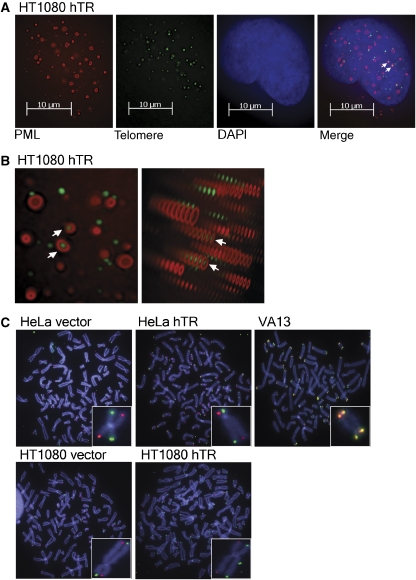
Figure 3.
Elongated telomeres are not substrates for telomeric exchange events. (A) Colocalisation of PML protein and FITC-conjugated telomeric PNA probe in interphase HT1080 hTR cells at pd 200 by deconvolution microscopy of z-stacked images as a 2D-image and (B) as a 3D-image. Arrows indicate colocalisations. (C) CO-FISH analysis of HeLa and HT1080 vector and hTR cells, compared with VA13 (ALT+) positive control, using a Texas Red PNA probe to the C-rich strand and an Alexa 488 PNA probe to the G-rich strand. Chromosomes were counterstained with DAPI. Insets show a representative chromosome with no telomere exchange event in the HeLa and HT1080 cells and with telomere exchange events detected at both telomeres in the VA13 cells.
APB-like foci were identified in approximately 3% of both HeLa and HT1080 cells with elongated telomeres at varying pds (Figure 3A and B), whereas no colocalisations were detected in any vector controls (data not shown). PML protein also colocalised with TRF2 and the components of the Mre11/Rad50/Nbs1 (MRN) homologous recombination complex in both HeLa and HT1080 hTR cells, but not in vector controls (Supplementary Figure S2). In view of this unexpected result, DNA fingerprinting of HeLa and HT1080 vector and hTR cells was used to confirm that there had been no cell line cross-contamination with ALT cells (data not shown).
The frequency of telomere exchange events was unaltered in telomerase-positive cells with elongated telomeres
Long, heterogeneous telomeres, the presence of ECTR DNA and the colocalisation of telomeric DNA with PML protein partly phenocopied the telomeres of cells that use the ALT mechanism, so we investigated whether the hTR-transduced HeLa and HT1080 cells had other features of ALT, including elevated telomeric exchange events, usually referred to as telomere sister chromatid exchanges (t-SCEs) (Bailey et al, 2004; Bechter et al, 2004; Londono-Vallejo et al, 2004; Muntoni and Reddel, 2005). Chromosome-orientation-FISH (CO-FISH) was carried out on metaphase spreads from BrdU:BrdC treated cells using a Texas Red-conjugated PNA probe against the C-rich strand and an Alexa 488-conjugated PNA probe against the G-rich strand, to detect the parental telomeric DNA strands of the two sister telomeres at each end of metaphase chromosomes (Londono-Vallejo et al, 2004). The ALT cell line VA13 was included in the analysis as a positive control.
As described previously, the basal rate of t-SCE events in telomerase-positive cells was very low (Londono-Vallejo et al, 2004), in contrast to the high frequency of exchange events detected in VA13 cells. The rate of t-SCEs did not increase in either the HeLa hTR or HT1080 hTR cells compared with vector controls (Table I; Figure 3C). We therefore conclude that the extended telomeres of HT1080 and HeLa hTR cells are not subjected to postreplicative telomere exchange events.
Table 1.
T-SCE analysis in HeLa and HT1080 vector- and hTR-expressing cells at pd 300 and VA13 cells as positive control
| Cell line | No. of chromosome ends analysed | No. of T-SCE | Frequency of T-SCE per chromosome end (%) |
|---|---|---|---|
| HeLa vector | 4830 | 1 | 0.02 |
| HeLa hTR | 4646 | 0 | 0 |
| HT1080 vector | 4680 | 1 | 0.02 |
| HT1080 hTR | 4794 | 0 | 0 |
| VA13 | 3438 | 778 | 22.6 |
Telomere–telomere copying was not detected in telomerase-positive cells with extended telomeres
Integration of a telomere-targeting plasmid tag has been used previously to examine intertelomere copy templating in ALT cells (Dunham et al, 2000). This technique was used in this study to investigate whether homologous recombination-mediated telomere replication events also occurred in telomerase-positive cell lines overexpressing hTR and showing some of the phenotypic characteristics of ALT.
A neomycin resistance gene (neoR) cassette flanked on either side by 800 bp of telomeric repeats was used as a telomeric DNA tag by targeting into the telomeres of HeLa and HT1080 vector and hTR cells at pd 230. Integration of the DNA tag was detected in clonal cell populations by FISH at early pds (pd +25). Two HT1080 vector clones (clones 13 and 20) and three HT1080 hTR clones (clones 3, 15 and 16), all of which contained a single telomeric integration site (Figure 4A), were picked and cultured separately. HT1080 hTR clone 15 also contained an interstitial tag integration site. No telomeric integration of the tag was obtained in any HeLa vector clones analysed. Two HeLa hTR clones (clones 2 and 4) with telomeric integration of the tag were chosen for analysis (Figure 4A). Clone 2 had a single telomere integration site, and clone 4 contained three telomeric integration sites (Table II). The variation in telomere tag signal intensities within a single metaphase spread, in clones that have more than one tagged telomere, indicates that at least some telomeres contain more than one copy of the tag; this is consistent with the possibility that the tag has integrated as an array.
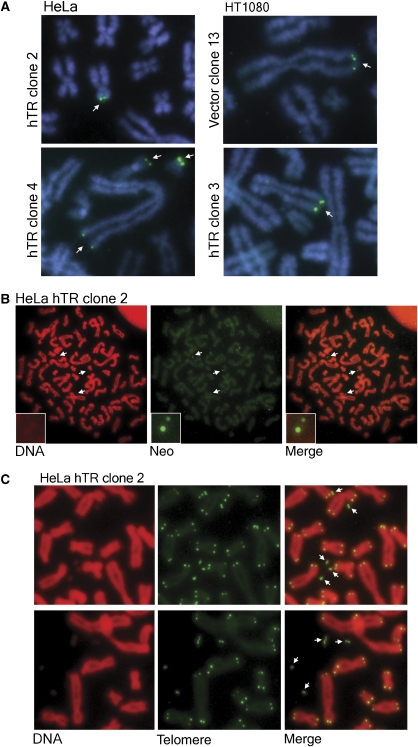
Figure 4.
Telomere targeting results in loss of telomeric repeats, but no intertelomere copying. (A) Telomere tagged clones of HeLa and HT1080 vector and hTR cells, indicating the position of tag integration at the earliest timepoint (pd +25). The neoR tag was detected by FISH using the pSXneo plasmid as a probe. Chromosomes were counterstained with DAPI. (B) Tagged DNA was progressively excised from the telomere in HeLa hTR clone 2, resulting in tagged extrachromosomal fragments, where the tag FISH signal colocalised with the propidium iodide DNA stain (indicated by arrows). Insets show an enlarged colocalisation, indicated by the top arrow. (C) Telomere signal was detected as extrachromosomal fragments using an FITC-telomeric PNA probe in HeLa hTR clone 2. Chromosomes were counterstained with propidium iodide. Arrows indicate extrachromosomal telomere fragments.
Table 2.
Telomere-tagged chromosomes in HeLa and HT1080 clones at early population doubling following tag integration (pd +25) and at late population doubling (pd +60)
| Cell line and clone | Early pd (pd +25) | Late pd (pd +60) |
|---|---|---|
| HeLa | ||
| hTR clone 2 | 12pter | None |
| hTR clone 4 | 2qter, 19pter, 21pter | 2qter, 19pter, 21pter |
| HT1080 | ||
| Vector clone 13 | 1qter | 1qter |
| Vector clone 20 | 11q+ter | 11q+ter |
| hTR clone 3 | 9qter | 9qter |
| hTR clone 15 | 6p or 3pdelinter, 21pter | 6p or 3pdelinter, 21pter |
| hTR clone 16 | Bpter | Bpter |
All clones were passaged for 60 pds after integration of the tag. The number of tagged telomeres did not increase in any of the vector or hTR clones, as observed previously in telomerase-expressing cell lines (Dunham et al, 2000); that is, no telomeric copying events involving a tagged chromosome end were detected (Table II). Therefore, despite the increase in chromosomal telomeric and ECTR DNA, there is no evidence in these telomerase-positive cells of the recombination-mediated telomere replication events observed in ALT cells.
In HeLa hTR clone 2, the telomere tag was entirely lost after 60 pds. Loss of the telomere tag in this clone and not in any of the other analysed clones is likely to reflect a more distal position of tag insertion in the telomere repeat array, resulting in the tag being readily subjected to loss by telomeric deletion events.
This clone provided us with an informative system for investigating the partial loss of telomeric DNA from a specific chromosome end. NeoR-FISH was carried out at pd +28, a timepoint after verification of telomeric tag integration and, intriguingly, deletion of the tag from the telomere could be visualised in metaphase spreads (Figure 4B). We observed neoR-FISH signals separate from chromosomes, that colocalised with propidium iodide or DAPI stained DNA fragments, although this was only evident when the signal intensity of the red or blue channel, respectively, was increased substantially, indicating that the fragment was very small. Several deleted fragments could be seen in a single metaphase (Figure 4B). We hypothesise that this resulted from the tag integrating as an array and one or more copies being lost in progressive rounds of telomere truncation.
Telomeric deletion events were confirmed in HeLa hTR clone 2 pd +28 by telomere-FISH (Figure 4C). The deleted telomere fragments were identified as doublets, irregularly shaped fragments, and also as single signals within the cells. No telomere fragments were identified in vector controls. Importantly, no telomere signal-free ends were identified.
Telomerase-positive cells with elongated telomeres showed no increase in telomere dysfunction-induced foci
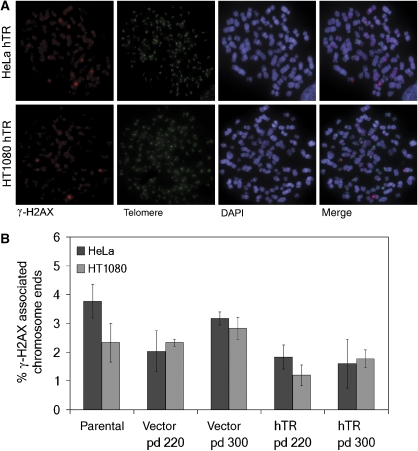
Telomere uncapping results in telomere dysfunction, shown by the formation of telomeric foci containing DNA damage response factors (telomere dysfunction-induced foci, TIFs) and the consequent activation of a DNA damage response (de Lange, 2002; Takai et al, 2003). We quantitated the number of TIFs, as visualised by the presence of the DNA double strand break marker phosphorylated histone H2AX (γ-H2AX), at telomere ends in HeLa and HT1080 vector and hTR cells at pd 220 and pd 300 (Figure 5A).
Figure 5.
No increase in TIFs was detected in cells undergoing telomeric deletion events. (A) Cytospun HeLa and HT1080 hTR chromosomes were immunostained for γ-H2AX in conjunction with FITC-telomeric PNA probe. Chromosomes were counterstained with DAPI. (B) Quantitation of TIF data from HeLa and HT1080 parental cells compared with vector and hTR cells at pd 220 and pd 300, expressed as percentage γ-H2AX associated chromosome ends. Approximately 3000 chromosome ends were counted per experiment. Error bars represent standard deviation between three separate experiments.
The proportion of γ-H2AX associated chromosome ends in HeLa or HT1080 hTR cells undergoing telomere-trimming events and t-circle formation did not increase compared with vector controls (Figure 5B). On the contrary, a slight decrease in the number of TIFs was identified in HeLa and HT1080 hTR cells compared with vector cells (Figure 5B), probably due to a decrease in the number of very short telomeres as the result of increased telomerase activity.
Discussion
In this study, we used the generation of artificially elongated telomeres by overexpression of hTR and the consequent increase in telomerase activity in telomerase-positive cell lines, to reveal a mechanism by which human cells negatively regulate telomere length by trimming telomeric repeats from the chromosome ends (Figure 6). This process is distinct from the gradual telomere attrition seen with each round of cell replication, and from the cis-acting telomere length control mechanism operative in telomerase-positive cells that appears to depend on modulation of telomere accessibility to telomerase (Bianchi and Shore, 2008).
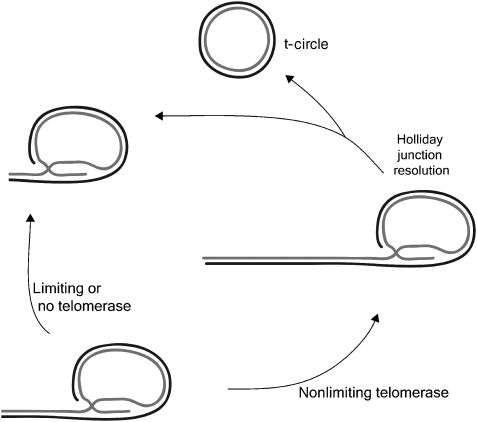
Figure 6.
Schematic of proposed telomere length control by deletion events. In situations of nonlimiting telomerase, telomeres extend beyond a threshold length, after which resolution of the t-loop Holliday Junction intermediate produces a truncated telomere and a t-circle. In situations of limiting or no telomerase, telomere lengths are below the threshold required to trigger these controlled telomere-trimming events.
Cells with 6- to 10-fold increased telomerase activity developed elongated telomeres that became heterogeneous in length but lacked very short telomeres or signal-free ends. This was accompanied by an accumulation of t-circles, ss G-rich ECTR DNA and t-complex DNA in both HeLa and HT1080 cells. Telomere extension by telomerase is thought to occur during S phase (Jady et al, 2006; Tomlinson et al, 2006), after which cells traverse a checkpoint at G2/M before progressing through mitosis. Although we cannot rule out an indirect effect of the over-expressed telomerase, the data presented here are most consistent with telomere length being subject to checkpoint control. We speculate that when telomeres become over-elongated, there is an increased probability that they will be trimmed by resolution of a t-loop intermediate to form t-circles, and therefore that t-loops are likely to be present after DNA synthesis and through G2/M.
We were also able to visualise the partial removal of telomeric sequences in metaphase cells. The identification of some doublet deletion products at this phase of the cell cycle indicates that the truncation events can occur during the G2 or M phase of the cell cycle, after DNA synthesis, but before sister-chromatid separation. Deleted products from the two sister-chromatids may remain associated with cohesins, which are involved in linking sister-chromatids together during cell division, thus appearing as doublets.
Importantly, no increase in the number of TIFs was seen in the cells displaying telomere-trimming events. T-circles do not have exposed DNA ends and would not be expected to trigger a DNA damage response. What is very interesting, however, is that the telomere truncation events do not result in TIFs at the linear chromosome ends, implying that there is no loss of telomere capping function or the formation of critically short telomeres, either of which would be detected as telomere dysfunction. The trimming events, therefore, appear to be a well-controlled process.
Telomere truncation events have been reported previously for unicellular eukaryotes. It has long been known that during continuous culture of Tetrahymena thermophila, macronuclear telomeric sequences in vegetatively dividing cells are added and removed, a process thought to be regulated by cellular signals (Larson et al, 1987). In addition, it has been known for some time that mutant Saccharomyces cerevisiae strains with elongated telomeres undergo a process referred to as telomere rapid deletion (TRD) (Li and Lustig, 1996; Lustig, 2003). Consistent with the conclusion that resolution of t-loops into t-circles is involved in TRD, both t-loop intermediates and t-circles have been identified in yeast mutant strains with long telomeres (Groff-Vindman et al, 2005; Cesare et al, 2008).
In human cells, expression of the TRF2 mutant, TRF2ΔB, which lacks the basic domain, induces catastrophic deletions of telomeric DNA, creating t-loop sized t-circles, accompanied by a DNA damage response and induction of senescence (Wang et al, 2004). This process is dependent on the MRN complex and XRCC3, which has been implicated in resolvase activity. An increase in the number of ECTR DNA fragments and signal-free ends has also been observed in cells from patients with ataxia-telangiectasia, following oxidative stress (Tchirkov and Lansdorp, 2003). Another context in which telomeric homologous recombination and t-circles occur is in cells that use the ALT mechanism (Cesare and Griffith, 2004). Notably, telomere length is much less uniformly controlled in these contexts and, like cells exposed to TRF2ΔB, ALT+ cells frequently have chromosomes with telomere signal-free ends (Perrem et al, 2001; Wang et al, 2004).
The cells used in this study are tumour-derived, and it is conceivable that their telomeres may behave atypically. Nevertheless, the deletion events described here in telomerase-positive cells with elongated telomeres appear to be well regulated. The deletion events appear to occur at a low frequency in the control telomerase-positive cells, as indicated by the low level of t-circles; trimming of over-elongated telomeres is presumably required only infrequently in cells that usually maintain telomeres at a somewhat homogeneously short length. We suggest that this mechanism is also likely to operate in normal human cells with longer telomeres, albeit at a lower frequency. It has been suggested previously that sudden telomere shortening events are responsible for the stochastic onset of senescence in cultures of normal human cells (Rubelj and Vondracek, 1999).
No telomeric exchange or intertelomere copying events were detected in any of the telomerase-positive cells with elongated telomeres. Therefore, telomere elongation in telomerase-overexpressing cells is not sufficient to activate the telomeric recombination events characteristic of ALT (Dunham et al, 2000). Similarly, yeast TRD events involve intrachromosomal telomeric deletions and are distinct from the interchromosomal subtelomeric and telomeric repeat exchange events that occur in telomerase-null yeast type I and type II survivors, respectively (Lustig, 2003).
Nevertheless, a number of similarities do exist between telomerase-positive cells with elongated telomeres and ALT cells: telomere length heterogeneity, the presence of ECTR DNA in the form of t-circles, ss G-rich DNA and t-complex DNA, and the formation of APB-like colocalisations between PML protein and both telomeric repeat DNA and telomere-associated proteins. As in ALT cells, these PML bodies also contained the MRN homologous recombination complex. The lack of any intertelomeric recombination events, however, shows that the ALT mechanism is not activated and that these previously described ‘markers' of ALT are more likely to be consequential to the over-elongated telomeres that are characteristic of that mechanism.
In conclusion, this study has revealed a process in human cells in which telomere-trimming events involving t-circle formation contribute to telomere length regulation in the presence of increased telomerase activity. This mechanism appears to occur at low frequency in telomerase-positive control cells and is exacerbated in the situation of increased telomerase activity and extended telomere lengthening. We propose that telomere length is controlled by the previously described factors, namely telomere attrition resulting from the end-replication problem, telomerase activity levels, and factors acting at the telomere to control access of telomerase to its substrate, together with trimming of over-lengthened telomeres.
Materials and methods
Retroviral infection and cell culture
Retroviruses were prepared by transient transfection of the viral packaging cell line PA317 with pBABEpuro empty vector control or the hTR expression vector pBABEpuroU3hTR. Supernatant was collected and filtered 48 and 72 h after transfection. HT1080 and HeLa cells were transduced with viral supernatant containing 4 μg/ml polybrene twice overnight. After a recovery of 24 h, 0.25 μg/ml puromycin was added to the growth medium (DMEM containing 10% foetal bovine serum), at which time pd was arbitrarily set to 0. Uninfected cells were included as a selection control. After 2 weeks, selection was removed and mass cultures of HeLa and HT1080 cells were passaged at a 1:16 split ratio for 300 pds.
Telomere targeting by transfection
Cells were transfected with KpnI linearised telomere targeting plasmid, Tel (Dunham et al, 2000), using siPORT transfection reagent (Ambion, Applied Biosystems). Forty-eight hours after transfection, cells were plated at low density and selected in 500 μg/ml G418. Clones were isolated and assayed at early (pd +25) and late (pd +60) pd for telomere integration of plasmid sequence by neoR-FISH.
Quantitative RT–PCR
Total RNA was extracted using the RNeasy miniprep kit (Qiagen), digested with DNaseI (Invitrogen) and quantified by spectrophotometry. cDNA was synthesised from 1 μg total RNA by random priming using SuperScript III reverse transcriptase (Invitrogen) and diluted to 100 μl. A measure of 2 μl of cDNA was used in 25 μl qRT–PCR reactions for hTR and GAPDH containing 0.4 μM forward (F) and reverse (R) primers (hTRF: CTAACCCTAACTGAGAAGGGCGTA, hTRR: GGCGAACGGGCCAGCAGCTGACATT, GAPDHF: ACCCACTCCTCCACCTTTG, GAPDHR: CTCTTGTGCTCTTGCTGGG) with SYBR green PCR mastermix (Applied Biosystems). RT minus controls were included. PCR conditions were 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s, 60°C for 60 s, followed by melt curve analysis. Matched PCR efficiencies for the two primer sets were confirmed by standard curve comparison and analysis was carried out by the comparative cycle threshold (Ct) method using GAPDH as the reference gene and expressed as fold change to the parental cell line.
IP-RQ-TRAP
Telomerase was immunoaffinity purified from whole cell lysate from 106 cells in a total volume of 200 μl, as described previously (Cohen and Reddel, 2008). A measure of 2 μl of purified telomerase solution was used in a 25 μl qPCR reaction using 0.1 μg telomerase primer TS, 0.05 μg reverse primer ACX (Kim and Wu, 1997; Wege et al, 2003) and SYBR green PCR mastermix (Applied Biosystems). Telomerase extension was carried out at 25°C for 20 min, followed by PCR amplification of telomerase extension products using 95°C for 10 min and 35 cycles of 95°C for 10 s, 60°C for 60 s. Triplicate serial dilutions of hTR overexpressing HeLa and HT1080 cells were used to draw a standard curve of the form y=mx+c, where y is the Ct value and x is log10(protein quantity). Telomerase activity was expressed as a ratio to the vector sample at pd 10.
TRF analysis
Telomeric restriction fragments were prepared by digestion with HinfI and RsaI, as described previously (Cesare and Griffith, 2004). The fragments were separated by 1D or 2D PFGE, or standard 2D gel electrophoresis and hybridised in-gel using a telomere-specific oligonucleotide probe (Cesare and Griffith, 2004; Cesare et al, 2008). For hybridisation under native conditions, agarose gels were dried at 50°C without denaturation and hybridised in-gel to γ-32P-ATP-labelled (CCCTAA)4 or (TTAGGG)4 telomeric probes (Grudic et al, 2007). After imaging, the gels were denatured, neutralised and rehybridised to the same probe overnight and subsequently washed and exposed to a PhosphorImage screen overnight.
Telomere FISH and CO-FISH
Chromosome preparations from colcemid-arrested cells were obtained according to standard cytogenetic methods. FISH was carried out with 0.3 μg/ml FITC- or Cy3-(CCCTAA)3 PNA probe (Applied Biosystems) for 2 h at 37°C (Perrem et al, 2001). DNA was counterstained with 0.6 μg/ml DAPI (Sigma) and 120 μg/ml propidium iodide (Sigma). Images of metaphase spreads were captured on a Carl Zeiss Axio-Imager M1 using Metafer 4 software (Metsystems, Germany) and analysed with Isis software.
For CO-FISH, cells were grown in 10 μM BrdU:BrdC (3:1) for 18 h, with 0.1 μg/ml colcemid added for the final 2 h. Chromosome preparations were obtained according to standard cytogenetic methods. Slides were stained in 0.5 μg/ml Hoechst 33258 (Sigma) in 2 × SSC for 15 min at room temperature (rt), overlaid with 2 × SSC and exposed to 365 nm UV light (Stratalinker 1800) for 30 min. Slides were rinsed with distilled water and allowed to dry. No exonuclease III treatment was included. Instead, chromosomes were denatured in 70% formamide, 2 × SSC at 70°C for 2 min, and the fragmented DNA of the newly synthesised strand was removed by rinsing twice in water. Chromosomes were hybridised sequentially with Alexa 488-(CCCTAA)3 and Texas Red-(TTAGGG)3 PNA probes (Panagene, Korea), as described previously (Perrem et al, 2001). DNA was counterstained with DAPI. Metaphase spreads were captured on a Carl Zeiss Axio-Imager M1 using Metafer 4 software and analysed using Isis software. Individual chromosome ends were scored as positive for a telomere-exchange event if a signal doublet could be detected in both strand-specific fluorochromes on both sister chromatids of the respective chromosome.
NeoR-FISH
The plasmid backbone pSXneo was labelled by biotin nick translation (Roche) according to the manufacturer's instructions. FISH was carried out as described previously (Dunham et al, 2000), with 10 ng/μl of pSXneo probe followed by detection with FITC-conjugated avidin. Individual chromosome ends were scored as positive for the neoR signal if a signal doublet could be detected on both sister chromatids of the respective chromosome.
Immunofluorescence
Cells were grown in 2-well chamber slides for 2 days, fixed in ice-cold methanol twice for 10 min and incubated with 1:500 rabbit anti-PML (Chemicon) in 0.2% fish gelatin (Sigma) in PBS at 37°C for 1 h. Slides were washed in PBS and incubated with 1:1000 goat anti-rabbit Alexa 594 at 37°C for 30 min. Slides were washed, fixed in 4% formaldehyde and subjected to telomere FISH, as described above. Alternatively, fixed cells were incubated with anti-PML antibody and anti-TRF2 (Calbiochem), anti-Mre11 (BD Biosciences), anti-Rad50 (BD Biosciences) or anti-ppNbs1 (Novus Biologicals), followed by the appropriate Alexa 594 or Alexa 488 secondary antibody. Imaging was carried out on ten 250-nm sections using a Carl Zeiss Axio-Imager M1. Deconvolution was carried out by the inverse filter method using Axiovision software.
TIFs were visualised in metaphase spreads prepared by cytocentrifugation. Briefly, cells were collected by trypsinisation and centrifugation, resuspended in 0.2% KCl, 0.2% tri-sodium citrate at a concentration of 5.5 × 104 cells/ml for 5–10 min, then centrifuged onto glass slides at 2000 rpm for 10 min with medium acceleration in a Shandon Cytospin 4 (Thermo Scientific) cytocentrifuge. Slides were fixed in 1 × PBS, 4% formaldehyde at rt for 10 min and permeabilised for 10 min with KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris pH 7.5, 0.1% Triton-X 100). Slides were blocked in antibody dilution buffer (ABDIL: 20 mM Tris pH 7.5, 2% BSA, 0.2% fish gelatine, 150 mM NaCl, 0.1% Triton, 0.1% sodium azide) with 100 μg/ml RNase A at 37°C for 15 min. Slides were then incubated with 1:1000 mouse anti-γ-H2AX (Upstate clone JBW301) in ABDIL at rt for 1 h, washed in 1 × PBS, 0.1% Tween-20, incubated with 1:750 goat anti-mouse Alexa 594 at rt for 30 min in ABDIL and washed in 1 × PBS, 0.1% Tween. Slides were fixed in 4% formaldehyde at rt for 10 min, dehydrated with ethanol and subjected to telomere FISH, as described previously.
Supplementary Material
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figure Legends
Acknowledgments
We thank Tracy Bryan for critical review of the manuscript. We gratefully acknowledge the following grant support: Promina Research Fellowship (HAP), USA National Science Foundation International Research Fellowship (0602009) and Cure Cancer Australia (AJC), University of Sydney Summer Research Scholarship Program (RLJ) and Cancer Council New South Wales Program Grant and National Health and Medical Research Council (NHMRC) Senior Principal Research Fellowship (RRR).
References
- Bailey SM, Brenneman MA, Goodwin EH (2004) Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res 32: 3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter OE, Zou Y, Walker W, Wright WE, Shay JW (2004) Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res 64: 3444–3451 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D (2008) How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell 31: 153–165 [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR (1995) Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14: 4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Huschtscha LI, Nouwens AS, Pickett HA, Neumann AA, Chang AC, Toouli CD, Bryan TM, Reddel RR (2008) Amplification of telomerase reverse transcriptase gene in human mammary epithelial cells with limiting telomerase RNA expression levels. Cancer Res 68: 3115–3123 [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD (2004) Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous T-loops. Mol Cell Biol 24: 9948–9957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD (2008) Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol 28: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Reddel RR (2008) A sensitive direct human telomerase activity assay. Nat Methods 5: 355–360 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J (2006) Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2002) Protection of mammalian telomeres. Oncogene 21: 532–540 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE (1990) Structure and variability of human chromosome ends. Mol Cell Biol 10: 518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR (2000) Telomere maintenance by recombination in human cells. Nat Genet 26: 447–450 [DOI] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu JH, Le SY, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B (1995) The RNA component of human telomerase. Science 269: 1236–1241 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Groff-Vindman C, Cesare AJ, Natarajan S, Griffith JD, McEachern MJ (2005) Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol Cell Biol 25: 4406–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudic A, Jul-Larsen A, Haring SJ, Wold MS, Lonning PE, Bjerkvig R, Boe SO (2007) Replication protein A prevents accumulation of single-stranded telomeric DNA in cells that use alternative lengthening of telomeres. Nucleic Acids Res 35: 7267–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T (2006) Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WQ, Ringertz N (1997) Altered distribution of the promyelocytic leukemia-associated protein is associated with cellular senescence. Cell Growth Differ 8: 513–522 [PubMed] [Google Scholar]
- Kim NW, Wu F (1997) Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res 25: 2595–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DD, Spangler EA, Blackburn EH (1987) Dynamics of telomere length variation in Tetrahymena thermophila. Cell 50: 477–483 [DOI] [PubMed] [Google Scholar]
- Li B, Lustig AJ (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR (2004) Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res 64: 2324–2327 [DOI] [PubMed] [Google Scholar]
- Lustig AJ (2003) Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Cancer 4: 916–923 [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666 [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA 85: 6622–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni A, Reddel RR (2005) The first molecular details of ALT in human tumor cells. Hum Mol Genet 14 (Suppl 2): R191–R196 [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F (2009) Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol 29: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR (2001) Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol 21: 3862–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J (2008) C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Rubelj I, Vondracek Z (1999) Stochastic mechanism of cellular aging—abrupt telomere shortening as a model for stochastic nature of cellular aging. J Theor Biol 197: 425–438 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Tchirkov A, Lansdorp PM (2003) Role of oxidative stress in telomere shortening in cultured fibroblasts from normal individuals and patients with ataxia-telangiectasia. Hum Mol Genet 12: 227–232 [DOI] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP (2006) Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wege H, Chui MS, Le HT, Tran JM, Zern MA (2003) SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res 31: E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev 11: 2801–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR (1999) Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59: 4175–4179 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figure Legends