Abstract
Active site guanines are critical for self-cleavage reactions of several ribozymes, but their precise function(s) in catalysis are unclear. To learn whether protonated or deprotonated forms of guanine predominate in the active site, microscopic pKa values were determined for ionization of 8-azaguanosine substituted for G8 in the active site of a fully functional hairpin ribozyme to determine microscopic pKa values for 8-azaguanine deprotonation from the pH dependence of fluorescence. Microscopic pKa values above 9 for deprotonation of 8-azaguanine in the active site were about 3 units higher than apparent pKa values determined from the pH dependence of self-cleavage kinetics. Thus, the increase in activity with increasing pH does not correlate with deprotonation of G8 and most of G8 is protonated at neutral pH. These results do not exclude a role in proton transfer, but a simple interpretation is that G8 functions in the protonated form, perhaps by donating hydrogen bonds.
Introduction
The hairpin ribozyme has been the prototype of a catalytic RNA that relies exclusively on nucleotide functional groups for catalytic chemistry since a catalytic role for divalent cations was excluded by the ability of cobalt hexammine and monovalent salts to support full activity.1-4 While self-splicing introns and RNase P catalytic RNAs clearly exploit divalent cation cofactors for catalytic chemistry, divalent cations contribute little or nothing to catalysis of self-cleavage by hammerhead, hepatitis delta virus, Varkud satellite, or glmS ribozymes or to RNA catalysis of peptide bond formation in the ribosome.5-7 How RNA functional groups that lack the chemical versatility of amino acid side chains can mediate catalysis is the focus of considerable debate.
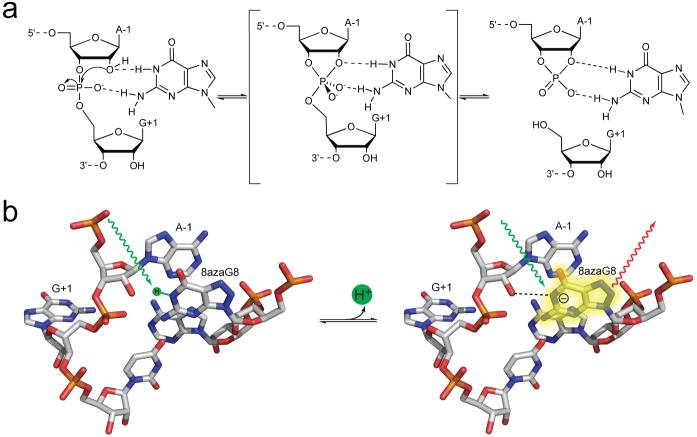
Like other self-cleaving RNAs, hairpin ribozymes catalyze a reversible phosphodiester cleavage reaction in which nucleophilic attack of an adjacent 2′ oxygen on phosphorus and breaking of the 5′ oxygen-phosphorus bond generates 5′-hydroxyl and 2′,3′-cyclic phosphate termini (Fig. 1a).6,7 The reaction proceeds with inversion of configuration8 and crystal structures of ribozyme complexes with transition state mimics show nucleophilic and leaving group oxygens positioned nearly in-line with phosphorus,9,10 consistent with an SN2(P)-type nucleophilic attack mechanism. Two active site purines, G8 and A38, occupy positions similar to the two histidines that mediate catalysis of the same reaction by RNase A, leading to the proposal that hairpin ribozymes use the same concerted general acid base mechanism (Fig. 1).11 N1 of guanine is near the 2′ oxygen that is the nucleophile during cleavage and the leaving group during ligation and the N2 amine lies within hydrogen bonding distance of the pro-RP nonbridging oxygen of the reactive phosphodiester. Conserved guanines occupy virtually identical positions in the active sites of hammerhead and glmS ribozymes,12-14 suggesting that guanines play a common role in RNA self-cleavage. In a general acid base model, the deprotonated form of guanine would act as a general base to accept a proton at the N1 position from the 2′ hydroxyl to activate nucleophilic attack during cleavage and the protonated form would donate a proton from N1 to stabilize the 2' oxyanion leaving group during ligation.
Figure 1.
Structure and function of G8 in the hairpin ribozyme active site. (a) Reversible self-cleavage reaction catalyzed by the hairpin ribozyme. (b) Model of the active site of a hairpin ribozyme-substrate complex based on the X-ray crystal structure of a ribozyme complex with a substrate analog in which cleavage is blocked by a 2' OCH3 modification of A-1 (PDB entry 1M5K).11 To create this image, G8 was replaced with deprotonated and protonated forms of 8azaG8 and the methyl group was removed from A-1 using PyMOL.46
The pH-dependence of hairpin ribozyme activity can be interpreted as support for the concerted general acid-base mechanism.15,16 However, alternative models in which nucleotide protonation states are important have not been excluded, since pH-rate profiles provide limited information about the nature of the ionization events that affect activity,7,17 and certain features of the general acid base mechanism are problematic. Unfavorable electrostatic repulsion between the anionic, deprotonated form of guanine could destabilize a transition state in which five electronegative oxygen atoms associate with phosphorus. Furthermore, only a small fraction of guanine would be in the deprotonated form at neutral pH unless some feature of the active site shifts the pKa in the acidic direction from the value near 10 that is typical for guanine deprotonation in the context of an oligonucleotide.18
Information about the protonation state of the active site guanine would help to distinguish among alternative models for its role in catalysis. Nucleobases in structured RNAs have been shown to ionize with pKa values that differ from the pKa values of nucleotides in solution,19 including a product of HDV ribozyme self-cleavage20 and the isolated A and B domains of hairpin ribozymes,21,22 using NMR spectroscopy. Raman spectroscopy was used to measure pKa values shifted toward the neutral range for an active site cytosine in crystals of an HDV ribozyme.23 Fluorescence assays of 8-azaguanine-substituted RNAs offer an alternative approach for monitoring purine protonation states that can be applied to catalytically active ribozymes under a wide variety of solution conditions. 8-azaguanine (8azaG) is an analog of guanine that displays high fluorescence emission intensity when N1 is deprotonated, and low fluorescence emission intensity when N1 is protonated (Fig. 1b).24 The difference in fluorescent quantum yields between protonated and deprotonated forms allows calculation of ionization equilibria from pH-fluorescence profiles. We showed previously that 8-azaguanosine-5′-triphosphate (8azaGTP) is a good substrate for T7 RNA polymerase transcription, allowing site-specific 8-azaguanine incorporation into RNA transcripts.25 8-azaguanine differs from guanine only in having nitrogen in place of carbon at the 8 position. It retains the Watson-Crick hydrogen bonding face of guanine, so substitution of 8-azaguanine for guanine has little effect on RNA secondary structure stability.25
In the present study, we introduced a single 8-azaguanine at position 8 in the active site of a fully functional hairpin ribozyme, determined microscopic pKa values for 8azaG8 deprotonation, and compared microscopic pKa values for 8azaG8 deprotonation with apparent pKa values determined from the pH dependence of self-cleavage kinetics. Under a variety of conditions, apparent pKa values determined from kinetics were about 3 units lower than the microscopic pKa values above 9 obtained for deprotonation of 8-azaguanine in the active site. Thus, most of G8 is protonated at neutral pH and the increase in self-cleavage activity with increasing pH does not correlate with G8 deprotonation. While these results do not exclude a role for G8 in proton transfer, a simple interpretation is that G8 contributes to catalysis as the protonated species, perhaps by donating hydrogen bonds to provide electrostatic stabilization of the transition state and facilitate positioning and orientation.
RESULTS
A hairpin ribozyme with 8azaG in place of G8 is fully functional
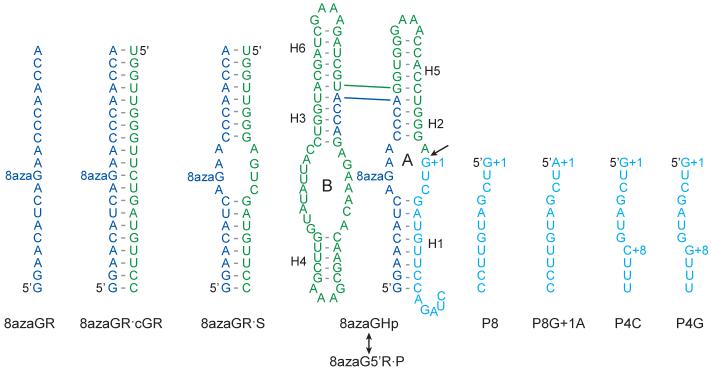
We previously measured the pH dependence of 8-azaguanine fluorescence in single-stranded RNA (8azaGR), in a perfectly paired duplex in which 8azaGR was annealed to a complementary oligonucleotide (8azaGR·cGR), and in a bulged duplex (8azaGR·S) that is analogous to domain A of the hairpin ribozyme (Fig. 2).25 In the current study, we designed a hairpin ribozyme variant (8azaGHp) that allows incorporation of a single 8-azaguanine at the G8 position in the active site by ligating the 8azaGR oligonucleotide to a second transcript prepared with unmodified guanosines to form a complete ribozyme. Self-cleavage of 8azaGHp generates 5' and 3' cleavage products that associate through eight base pairs in the intermolecular H1 helix. The ligated form of this ribozyme predominates at equilibrium under standard conditions with 10 mM MgCl2 at pH 7.5 and 25°C because the equilibrium dissociation constant for this 5'R·P complex with 8 base pairs in H1 is in the subpicomolar range and the internal equilibrium between cleavage and ligation of bound products favors ligation by about 30-fold.26
Figure 2.
RNAs and complexes used for fluorescence experiments. An RNA transcript with a single 8-azaguanine (8azaGR) was annealed to a perfectly complementary oligonucleotide, cGR, or to a partially complementary oligonucleotide, S, to form a bulged duplex analogous to the loop A domain of a hairpin ribozyme as described previously.25 8azaGR was ligated to a second transcript to create a full hairpin ribozyme (8azaGHp) that contains A and B domains and base-paired helices H1 through H6. Cleavage occurs at the site indicated by the arrow to create a stable complex with a 5' product (8azaG5'R) and a 3' product (P) that anneal through eight base pairs in H1. RNAs that correspond to a functional 3' cleavage product (P8) and a cleavage product analog with an inactivating G+1A mutation (P8G+1A) were prepared through chemical synthesis. Unstable ribozyme complexes in which H1 has 4 base pairs and a flanking cytosine or guanine (P4C or P4G) were used for functional assays to ensure that measurements of cleavage rate constants were not complicated by religation of bound products.
To incorporate a unique 8azaG at position 8, the G:C base pairs that normally flank loop A had to be replaced by C:G base pairs. These mutations reduce activity of minimal hairpin ribozymes,27,28 so it was important to learn how they affect activity in the four-way junction form of the ribozyme used in the current study. We used an established ligation-chase assay26 to measure cleavage activity for ribozymes with the same mutations but with guanine or 8-azaguanine at position 8. The ribozymes used for self-cleavage assays have the same sequence as 8azaGHp except that the H1 helix has just four base pairs instead of eight (Fig. 2). Rapid dissociation of the 5'R·P complex with a short H1 helix ensured that observed cleavage rates reflected the catalytic step and not assembly or dissociation steps in the reaction pathway. Under standard conditions with 10 mM MgCl2 at pH 7.5, the unmodified ribozyme displayed a cleavage rate constant of 0.22 min-1, similar to the cleavage rate constant of 0.6 min-1 measured previously for other hairpin ribozyme sequences with the same configuration (Table 1).26 We obtained virtually identical cleavage rate constants with two different 3' product RNAs that have cytosine or guanine at the +8 position and bind to 5' ribozyme RNAs with affinities that differ by threefold. Since the two product RNAs would exhibit a corresponding 3-fold difference in product dissociation kinetics, the fact that they display the same cleavage rate constant confirmed that product dissociation is much faster than ligation and observed cleavage rates specifically monitor the cleavage step without complications from re-ligation of bound products. 8azaGHp exhibited a cleavage rate constant of 0.65 min-1, confirming that the 8azaG8 modification had no significant effect on activity.
Table 1.
pH dependence of cleavage kinetics
| 8azaGHp |
Hp |
|||
|---|---|---|---|---|
| pKa,app | kmax (min-1) | pKa,app | kmax (min-1) | |
| 6.8 ± 0.1 | ||||
| 10 mM MgCl2 | 0.65 ± 0.04 | 7.0 ± 0.2 | 0.22 ± 0.02 | |
| 9.9 ± 0.1 | ||||
| 6.8 ± 0.1 | ||||
| 1 mM MgCl2 | 0.98 ± 0.06 | 6.7 ± 0.1 | 0.22 ± 0.02 | |
| 10.3 ± 0.2 | ||||
| 6.3 ± 0.1 | ||||
| 1 M LiCl | 1.9 ± 0.1 | 6.2 ± 0.2 | 1.04 ± 0.06 | |
| 10.1 ± 0.1 | ||||
| 6.6 ± 0.1 | ||||
| 1 M NaCl | 1.9 ± 0.1 | 6.1 ± 0.2 | 0.67 ± 0.05 | |
| 10.4 ± 0.1 | ||||
The fraction of ligated 8azaGHp observed at equilibrium provides information about the fraction of ribozyme RNA that is properly folded and retains the 2',3' cyclic phosphate terminus required for ligation. Since the internal equilibrium between cleavage and ligation of bound products favors ligation by more than 30-fold, perfectly assembled ribozymes will display 30-fold more ligated ribozyme than cleaved 5'R·P complex at equilibrium, that is, flig,∞=kligation/(kligation +kcleave).26 The fraction of ligated ribozyme ranged from 55% to 80% under the conditions used for fluorescence experiments, similar to other ribozymes with four-way helical junctions,26 indicating that most of the 8azaGHp RNA folded into a functional ribozyme.
Some fluorescence quenching occurs when 8-azaguanine is incorporated into RNA and quenching increases when 8azaGR is located in a base-paired helix.25 Fluorescence titration experiments confirmed that complexes contained stoichiometric amounts of complementary RNAs, as expected (Supplementary Fig. 1 online). Fluorescence emission intensity decreased linearly as increasing amounts of complementary RNAs were combined with 8azaGR or 8azaG5'R RNAs and maximum fluorescence quenching occurred with nearly equimolar amounts of complementary strands, as expected. The degree of quenching varied with RNA structure and with salt conditions. Two to three-fold greater fluorescence emission intensity was observed in 10 mM MgCl2 in the context of single-stranded 8azaGR or the 8azaG5'R cleavage product than in solutions without added salt. Consistent with previous results, the most efficient quenching occurred upon formation of a perfectly paired duplex, 8azaGR·cGR, for which fluorescent emission was virtually undetectable in the presence or the absence of 10 mM MgCl2 even at a concentration as high as 20 μM. An analog of the ribozyme cleavage product complex with an inactivating mutation (8azaG5'R·P8G+1A) and a functional ribozyme complex both displayed similar fluorescence emission intensity in the absence of MgCl2 but somewhat lower emission intensity was observed for the functional complex in 10 mM MgCl2.
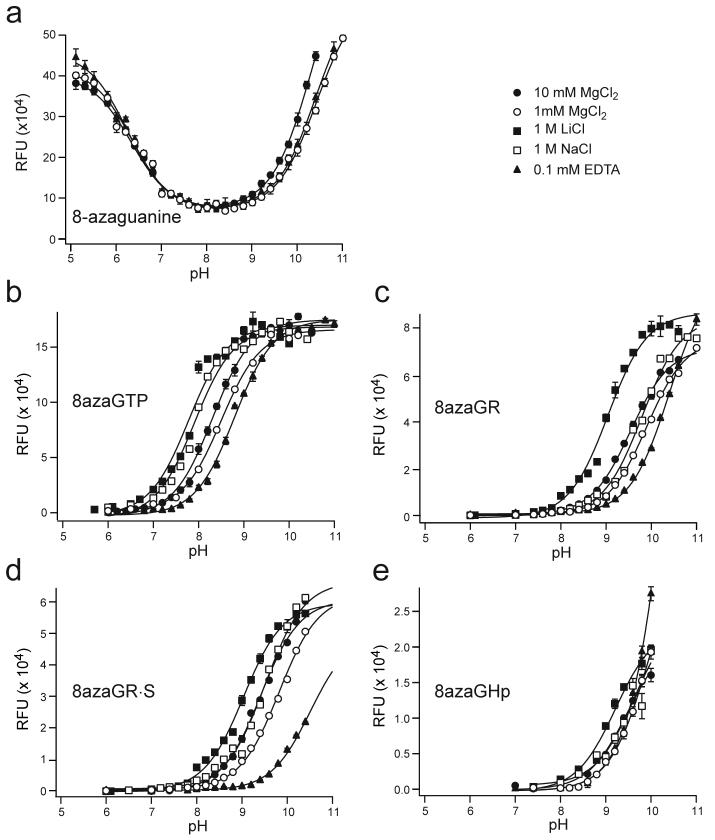
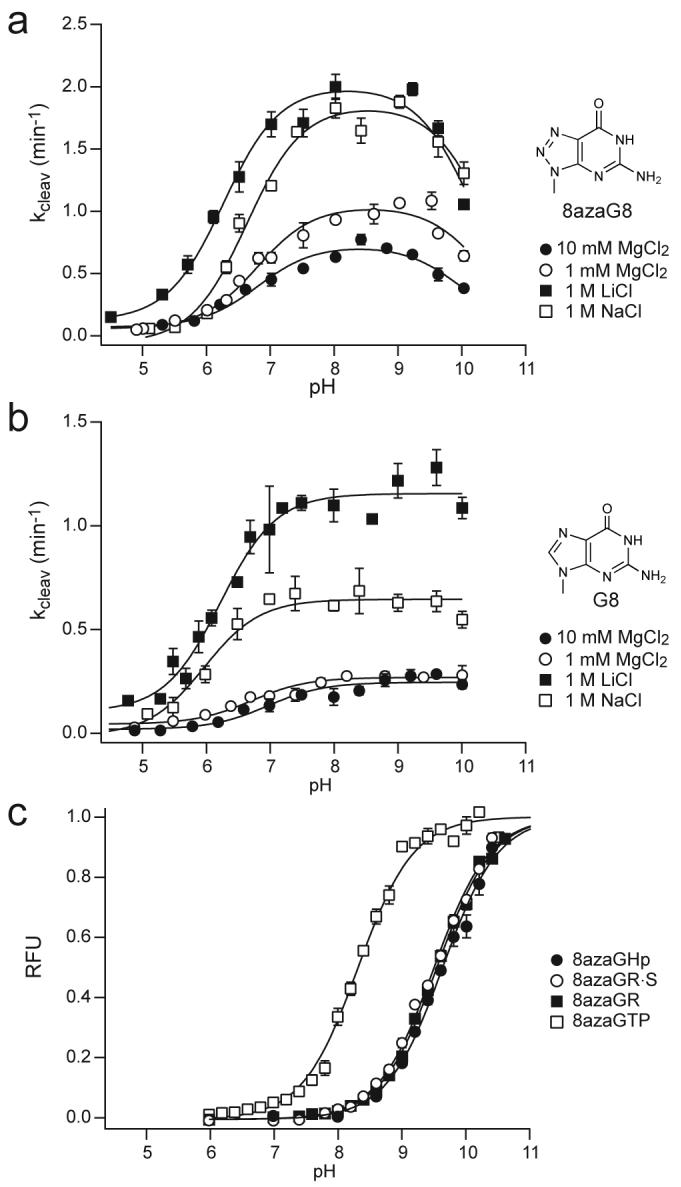
Self-cleavage rate constants increased by about 10-fold with increasing pH between pH 5 and pH 8 under standard conditions with 10 mM MgCl2 for ribozymes with G8 or 8azaG8, similar to previously characterized ribozymes (Fig. 3a,b).29,30 Activity began to decline at pH 9.5 for the 8azaG variant (Fig. 3a) while the unmodified ribozyme remained fully active at higher pH (Fig. 3b). The pH-rate profile for 8azaGHp self-cleavage fit an equation that includes two variables for ionizable groups to give apparent pKa values of 6.8 and 9.9 (Table 1). The pH-rate profile for self-cleavage of the unmodified ribozyme fit an equation that includes a single ionizable group to give an apparent pKa value of 7. These apparent pKa values are similar to values between 6.1 and 6.5 reported previously for other hairpin ribozymes with four-way helical junctions.29,30 Taken together, the results of these functional studies suggest that substitution of 8-azaguanine for G8 had little effect on ribozyme activity, apart from a modest decline in activity at high pH.
Figure 3.
pH dependence of ribozyme cleavage kinetics and 8-azaguanine fluorescence emission intensity under various salt conditions. (a) Ribozyme with 8-azaguanine at position 8. Lines represent the fits to kobs=kmax/(1+10(pKa1-pH)+10(pH-pKa2)+10(pKa1-pKa2)+kmin and apparent pKa values are reported in Table 1. (b) Ribozyme with guanine at position 8. Lines represent the fits to kobs=kmax/(1+10(pKa-pH))+kmin and apparent pKa values are reported in Table 1. (c) pH dependence of fluorescence emission intensity for 8azaG RNAs in 10 mM MgCl2. Lines represent fits of pH-fluorescence profiles to equation (1) and pKa values are reported in Table 2. Error bars are s.d. of two or more measurements.
8azaG ionization equilibria
Since fluorescence emission intensity depends on whether N1 is protonated, microscopic pKa values for 8azaG deprotonation can be determined from pH-fluorescence profiles. The fluorescence properties of 8-azaguanine, 8-azaguanosine, 8azaGTP, and the 8azaGR oligonucleotide in single-stranded form and in perfectly paired and bulged duplexes were characterized previously under low salt conditions.24,25 These previous studies showed that 8azaG deprotonation is significantly less favorable in the context of an oligonucleotide or duplex RNA than in the free nucleobase or in 8azaGTP. Under low salt conditions for example, the pKa value for deprotonation of 8azaG in duplex RNA is higher than the pKa for deprotonation of 8azaGTP by more than 2 units.25
In the present study, we determined microscopic pKa values for 8azaG deprotonation in 10 mM MgCl2, the salt condition typically used for analysis of hairpin ribozyme activity (Fig. 3c and Table 2). In 10 mM MgCl2, the pKa values for 8azaG deprotonation in the context of single-stranded and duplex RNAs shifted in the acidic direction by about 1 unit relative to values determined under low salt conditions. Strikingly, the microscopic pKa values for 8azaG deprotonation in the ribozyme active site were not significantly different from the values determined in a single-stranded oligonucleotide (8azaGR), or in the catalytically inactive complex analogous to domain A (8azaGR·S), which all displayed microscopic pKa values near 9.5 in 10 mM MgCl2. The microscopic pKa value of 9.5 for deprotonation of 8azaG in the active site is significantly more alkaline than the apparent pKa value of 6.8 calculated from the pH dependence of 8azaGHp cleavage kinetics, with a ΔpKa value of +2.7 under equivalent conditions (Tables 1 and 2).
Table 2.
Microscopic pKa values for 8azaG ionization determined from pH-fluorescence profiles
| 8-azaguanine | 8azaGTP | 8azaGR | 8azaGR·S | 8azaGHp | |
|---|---|---|---|---|---|
| 6.24 ± 0.04 | |||||
| 0.1 mM EDTA | 8.79 ± 0.02 | 10.40 ± 0.02 | 10.51 ± 0.04 | 10.35 ± 0.08 | |
| 10.40 ± 0.01 | |||||
| 6.33 ± 0.04 | |||||
| 10 mM MgCl2 | 8.31 ± 0.03 | 9.54 ± 0.02 | 9.49 ± 0.03 | 9.48 ± 0.06 | |
| 10.30 ± 0.04 | |||||
| 6.33 ± 0.06 | |||||
| 1 mM MgCl2 | 8.50 ± 0.03 | 9.90 ± 0.02 | 9.83 ± 0.03 | 9.95±0.04 | |
| 10.40 ± 0.02 | |||||
| 5.88 ± 0.05 | |||||
| 1 M LiCl | 7.82 ± 0.03 | 9.16 ± 0.06 | 9.04 ± 0.04 | 9.22 ± 0.09 | |
| 9.58 ± 0.05 | |||||
| 6.10 ± 0.07 | |||||
| 1 M NaCl | 7.91 ± 0.03 | 10.01 ± 0.07 | 9.51 ± 0.04 | 9.60 ± 0.2 | |
| 10.23 ± 0.04 |
Salt effects on 8azaG ionization and cleavage activity
Ribozymes with G8 or 8azaG8 exhibited virtually the same cleavage activity and the same apparent pKa values in 1 mM MgCl2 as in 10 mM MgCl2 (Fig. 3a,b and Table 1). These ribozymes also remained fully functional in monovalent salts, as shown previously for minimal hairpin ribozymes that assemble in the context of a two-way helical junction.1-4,31 In fact, cleavage rate constants were 3- to 5-fold higher in reactions with 1 M LiCl or 1 M NaCl than in 10 mM MgCl2. Apparent pKa values in molar concentrations of LiCl and NaCl shifted slightly in the acidic direction, with ΔpKa,app values all less than 1 unit.
Salt had larger effects on microscopic pKa values for 8azaG deprotonation in 8azaGTP and in single-stranded and base-paired RNAs (Fig. 4, Supplementary Fig. 2 online and Table 2). Microscopic pKa values shifted in the acidic direction as salt concentrations increased and pKa values were about 1 unit higher in the absence of any added salt than in 10 mM MgCl2. Maximum pKa shifts occurred in the molar range with monovalent salts and the microscopic pKa value for deprotonation of 8azaGTP approached neutral pH in high concentrations of LiCl (Supplementary Fig. 2 online). Ionization equilibria for the 8-azaguanine nucleobase alone displayed significantly less salt dependence. Under all salt conditions tested, however, microscopic pKa values remained above 9 for 8azaG deprotonation in the hairpin ribozyme active site, almost 3 units higher than the apparent pKa values determined from cleavage kinetics.
Figure 4.
pH dependence of 8azaG fluorescence emission intensity under various salt conditions. (a) Lines represent fits to equation (2). (b-e) Lines represent fits to equation (1). pKa values are reported in Table 2. Error bars are s.d. of two or more measurements.
DISCUSSION
The hairpin ribozyme mechanism has been the subject of considerable interest as an example of catalytic chemistry mediated exclusively by RNA functional groups. Cleavage and ligation rate constants increase with pH for the four-way junction form of the ribozyme, and pH-rate profiles can be fit to an equation that includes a variable for a single ionizable group, giving an apparent pKa value near 6.5.29,30 One explanation for the increase in activity with increasing pH is that it reflects accumulation of the deprotonated form of G8 needed for general base catalysis, assuming that active site interactions shift the pKa relative to the pKa values above 9 for deprotonation of guanine nucleotides in solution. Substitutions of G8 with nucleobases that have different pKa values for protonation at the N1 position change the pH dependence of cleavage activity.16 Furthermore, hairpin ribozymes that are inactivated by an abasic substitution of G8 can be rescued by exogenous nucleobase analogs provided in solution and the pH dependence of rescue varies with the pKa for protonation of the exogenous nucleobase.29,32 The small changes in pH-rate profiles caused by an 8-azaguanine substitution are consistent with these earlier results. These studies do not distinguish whether G8 contributes to catalysis as a general acid base catalyst or through some other mechanism that requires hydrogen bonding from the N1 position, such as electrostatic stabilization or orientation of reactive groups in the transition state. Interpretation of pH-rate profiles in terms of the functional protonation state of an active site nucleobase is further complicated by the fact that identical pH dependencies can arise from alternative mechanisms that involve different combinations of general acid and base catalysts and hydronium or hydroxide ions.15,33
A hairpin ribozyme with an 8azaG8 substitution in the active site exhibits virtually the same catalytic activity as an unmodified ribozyme. Thus, 8-azaguanine reports on the ionization state of a close guanine analog in a functional active site. Strikingly, the microscopic pKa values determined for 8azaG ionization in the active site were not significantly different from its ionization equilibria in a single-stranded oligonucleotide or in catalytically inactive complexes, which all displayed microscopic pKa values near 9.5 in 10 mM MgCl2. Emission intensity reflects context-dependent quenching as well as the N1 protonation state so it is not possible to determine whether some 8azaG remains protonated and displays no fluorescence emission throughout the accessible pH range. However, we detected no evidence of a shift toward a different pKa for the ribozyme relative to the inactive structures under any salt conditions.
Consistent with previous studies of minimal hairpin ribozymes,1-4,31 the four-way junction form of the ribozyme retained full catalytic activity in reactions with monovalent Li+ and Na+ salts instead of Mg2+, confirming that Mg2+ plays no special role in hairpin ribozyme function (Fig. 3a,b and Table 1). Salt conditions had smaller effects on apparent pKa values determined from cleavage kinetics than on microscopic pKa values determined from fluorescence. The highest pKa values for 8azaG deprotonation in 8azaGTP and in RNA were measured in buffers without added salts and pKa values became more acidic with increasing salt concentrations. In contrast, pKa values measured for the 8-azaguanine nucleobase did not decrease much with increasing salts. The acidic shift of pKa values in 8azaGTP and in RNA likely reflects the ability of cations to counteract electrostatic repulsion between anionic guanine and negatively charged phosphates. The observation that millimolar concentrations of Mg2+ facilitate deprotonation of 8azaG while molar concentrations of monovalent salts are required to produce the same effect is consistent with evidence that divalent ions form a macrochelate with N7 and phosphate group(s) of guanosine and its derivatives.34,35
In contrast to the discrepancies between the pH dependence of hairpin ribozyme activity and G8 deprotonation, microscopic pKa values measured spectroscopically for an active site cytosine in an HDV ribozyme agree well with apparent pKa values calculated from the pH dependence of cleavage kinetics.23 36 The pKa value near 6 for cytosine in the HDV ribozyme active site is less acidic than the pKa near 4 for protonation of free cytosine under low salt conditions, an effect that was attributed to its proximity to negatively charged phosphates.36,37 Anticooperative coupling between apparent pKa values for cytosine protonation and Mg2+ a also was observed for the active site cytosine in the HDV ribozyme and attributed to electrostatic repulsion between Mg2+ and protonated cytosine.36,37
Under standard conditions with 10 mM MgCl2, 8-azaguanine displays a pKa value of 9.5 in the ribozyme active site and pKa values for 8-azaguanine deprotonation in the ribozyme remained above 9.2 under all salt conditions tested. Since the pKa value of 9.2 for deprotonation of guanosine35 is more alkaline than the pKa value of 8.05 for deprotonation of 8-azaguanosine24 by about 1.1 units, a pKa value near 10.6 can be estimated for deprotonation of an unmodified guanine in the ribozyme active site. Consequently, less than 0.1% of G8 would be in the deprotonated form that could accept a proton from the nucleophilic 2' hydroxyl. If water is present in the active site at a comparable effective concentration, consistent with computational and X-ray crystallographic studies, general base catalysis by G8 might be no more effective than specific base catalysis in activating the 2' oxygen nucleophile.10,38-42 The results herein provide no support for the idea that G8 acts as a general base, but they do not conclusively exclude such a role. An effect of G8 deprotonation on activity could be obscured by a requirement for protonated or deprotonated forms of other functional groups that make larger contributions to activity. A discrepancy between microscopic and apparent pKa values also could occur if apparent pKa values reflect a change in the rate-determining step rather than deprotonation of a single functional group. Nonetheless, the simplest interpretation of these results is that the functional form of G8 is the predominant neutral species. G8 that is protonated at the N1 position could donate hydrogen bonds from N1 to the 2' oxygen, and from the exocyclic N2 amine to the pro-RP nonbridging oxygen, to stabilize the in-line attack geometry (Fig. 1a). Indeed, a model that requires protonated G8 is more consistent with the difference in pH-rate profiles between an unmodified and 8azaG-substituted ribozymes (Fig. 3a,b) if the loss of activity observed in 8azaGHP reactions at pH 9.5 results from 8azaG8 deprotonation.
The large discrepancies between microscopic pKa values for 8azaG8 deprotonation determined through fluorescence and apparent pKa values determined from the pH dependence of cleavage kinetics clearly establish that deprotonation of G8 is not responsible for the pH-dependent transition in catalytic activity. This result confirms previous evidence that elimination of G8 through an abasic substitution does not affect the pH-dependent transition in activity near pH 6.5 that is observed in the unmodified ribozyme.29 Current efforts to understand the molecular basis of the pH dependent transition in hairpin ribozyme activity focus on A38, a second active site purine whose protonation state is important for activity.30 It also will be important to learn whether conserved guanosines in the active sites of glmS and hammerhead self-cleaving RNAs exist predominantly in the protonated form.
MATERIALS AND METHODS
Preparation of RNAs
RNAs were prepared by bacteriophage T7 RNA polymerase transcription of linearized plasmid templates or partially duplex synthetic oligonucleotide templates or by chemical synthesis (Fig. 2) (Dharmacon), as described in detail in Supplemental Methods.26,43 Self-cleaving ribozyme variants assemble in the context of a four-way helical junction and have three or eight base pairs in the intermolecular H1 helix that forms between the 5' and 3' cleavage product RNAs. With eight base pairs in H1, 5' and 3' self-cleavage products form a stable complex useful for fluorescence experiments (8azaG5'R·P, Fig. 2). With just four base pairs in H1, self-cleavage products dissociate rapidly to prevent re-ligation facilitate cleavage rate measurements.
Self-cleavage kinetics
Self-cleavage rates were determined using ligation-chase assays, as described previously,26 in order to avoid potential complications that could result from slow assembly of functional complexes. Briefly, 1 nM of 32P-labeled 5′R and varying concentrations of 3′ product RNA (P4C or P4G, Fig. 2) were combined at a concentration approximately equal to the apparent equilibrium dissociation constant for product binding and allowed to undergo ligation to yield a final fraction of ligated ribozyme of at least 20%. Ligation reactions were then diluted 20-and 40-fold to initiate cleavage at 25°C in 50 mM buffer at various pH (MES pH 4.8-7, HEPES pH 7-8, TAPS pH 8-9, CHES pH 9-10), 0.1 mM EDTA, and the indicated salts. Comparison of rates observed after two different dilutions confirmed that the dilution was sufficient to ensure that rapid and irreversible product dissociation prevented re-ligation of bound products. Aliquots were quenched after various times, fractionated on denaturing gels and bands corresponding to ligated ribozyme (Hp) and 5′ cleavage product RNAs (5'R) were quantified by radioanalytic imaging. Cleavage rate constants were calculated by fitting to the equation FPt = FPt=∞ × (1 - ekcleavt) + FPt=0 where FP = [5'R]/([Hp]+[5'R]). Reported values represent the mean of two or more independent measurements and reported errors are standard deviations. Reported results were obtained from ligation-chase experiments using P4C except that P4G was used in reactions with 1 mM MgCl2 because the binding affinity of P4C was too low to obtain a sufficient amount of ligated ribozyme.
Fluorescence measurements
Fluorescence was measured using a SpectraMax M2e plate reader (Molecular Devices). 2 μM 8azaGTP, 3 μM 8azaGR, or 5 μM 8azaGHP were prepared in 50 mM buffer (MES pH 6-7, HEPES pH 7-8, TAPS pH 8-9, CHES pH 9-10, CAPS 10-11), 0.1 mM EDTA, and 0-40 mM MgCl2, 0-2 M NaCl, or 0-2 M LiCl. Samples were heated to 85°C for 1 min and slowly cooled to 25°C for at least 30 min before measurements. 8 μL of sample were added to 384-well black microplates (Greiner). Samples were excited at 290 nm and emission spectra were recorded between 330 and 450 nm. Previous work using a different fluorescence plate reader identified the maximum emission wavelength as 378 nm.25 Under similar conditions, however, we observed a maximum emission wavelength at 365 nm. To rule out the possibility that the discrepancy reflected an error in the calibration of our plate reader, we confirmed that the observed maximum fluorescence emission wavelengths of fluorescein, 2-aminopurine, and 2,6-diaminopurine agreed with literature values.44,45
pKa values were determined by fitting the integrated fluorescence intensities between 330 and 450 nm to equation (1):
| (1) |
where FB and FBH are the fluorescence of deprotonated and protonated 8azaG respectively and Ka is the acid dissociation constant for protonation of 8azaG in 8azaGTP and in RNAs. or equation (2):
| (2) |
for protonation of 8-azaguanine.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Fedor lab for help with manuscript preparation. This work was supported by NIH Grant GM046422 and by a postdoctoral fellowship provided by the Skaggs Institute for Chemical Biology to L. Liu.
REFERENCES
- 1.Hampel A, Cowan JA. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- 2.Nesbitt S, Hegg LA, Fedor MJ. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 3.Young KJ, Gill F, Grasby JA. Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 5.Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol. Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008 doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 7.Fedor M. Comparative enzymology and structural biology of RNA self-cleavage. Annu. Rev. Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 8.van Tol H, Buzayan JM, Feldstein PA, Eckstein F, Bruening G. Two autolytic processing reactions of a satellite RNA proceed with inversion of configuration. Nucleic Acids Res. 1990;18:1971–1975. doi: 10.1093/nar/18.8.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupert PB, Massey AP, Sigurdsson ST, Ferré-D'Amaré AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 10.Torelli AT, Krucinska J, Wedekind JE. A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupert PB, Ferré-D'Amaré AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 12.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein DJ, Ferré-D'Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 14.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 2007;14:95–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bevilacqua PC. Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 16.Pinard R, et al. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 18.Knobloch B, Sigel H, Okruszek A, Sigel RK. Acid-base properties of the nucleic-acid model 2'-deoxyguanylyl(5'-->3')-2'-deoxy-5'-guanylate, d(pGpG)3-, and of related guanine derivatives. Org. Biomol. Chem. 2006;4:1085–1090. doi: 10.1039/b517904a. [DOI] [PubMed] [Google Scholar]
- 19.Puglisi JD, Wyatt JR, Tinoco I., Jr. Solution conformation of an RNA hairpin loop. Biochemistry. 1990;29:4215–26. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- 20.Lupták A, Ferré-D'Amaré AR, Zhou K, Zilm KW, Doudna JA. Direct pKa measurement of the active-site cytosine in a genomic hepatitis delta virus ribozyme. J. Am. Chem. Soc. 2001;123:8447–8452. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- 21.Cai Z, Tinoco I., Jr. Solution structure of Loop A from the hairpin ribozyme from Tobacco Ringspot Virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 22.Ravindranathan S, Butcher S, Feigon J. Adenine protonation in domain B of the hairpin ribozyme. Biochemistry. 2000;39:16026–16032. doi: 10.1021/bi001976r. [DOI] [PubMed] [Google Scholar]
- 23.Gong B, et al. Direct measurement of a pKa near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J. Am. Chem. Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 24.Wierzchowski J, Wielgus-Kutrowska B, Shugar D. Fluorescence emission properties of 8-azapurines and their nucleosides, and application to the kinetics of the reverse synthetic reaction of purine nucleoside phosphorylase. Biochim. Biophys. Acta. 1996;1290:9–17. doi: 10.1016/0304-4165(95)00181-6. [DOI] [PubMed] [Google Scholar]
- 25.Da Costa CP, Fedor MJ, Scott LG. 8-Azaguanine reporter of purine ionization states in structured RNAs. J. Am. Chem. Soc. 2007;129:3426–3432. doi: 10.1021/ja067699e. [DOI] [PubMed] [Google Scholar]
- 26.Fedor MJ. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 27.Joseph S, Berzal-Herranz A, Chowrira BM, Burke JM. Substrate selection rules for the hairpin ribozyme determined by in vitro selection, mutation, and analysis of mismatched substrates. Genes Dev. 1993;7:130–138. doi: 10.1101/gad.7.1.130. [DOI] [PubMed] [Google Scholar]
- 28.Anderson P, et al. Mutagenesis of the hairpin ribozyme. Nucleic Acids Res. 1994;22:1096–1100. doi: 10.1093/nar/22.6.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuzmin YI, Da Costa CP, Fedor MJ. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmin YI, DaCosta CP, Cottrell J, Fedor MJ. Contribution of an active site adenosine to hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Nesbitt SM, Erlacher HA, Fedor MJ. The internal equilibrium of the hairpin ribozyme: temperature, ion and pH effects. J. Mol. Biol. 1999;286:1009–1024. doi: 10.1006/jmbi.1999.2543. [DOI] [PubMed] [Google Scholar]
- 32.Lebruska LL, Kuzmine II, Fedor MJ. Rescue of an abasic hairpin ribozyme by cationic nucleobases. Evidence for a novel mechanism of RNA catalysis. Chem. Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- 33.Jencks WP. Catalysis in Chemistry and Enzymology. Dover Publications, Inc.; New York: 1969. Chapter 3. General Acid-Base Catalysis; pp. 163–242. [Google Scholar]
- 34.Da Costa CP, Sigel H. Acid-base and metal ion binding properties of guanylyl(3'-->5')guanosine (GpG-) and 2'-deoxyguanylyl(3'-->5')-2'-deoxyguanosine [d(GpG)-] in aqueous solution. Inorg. Chem. 2003;42:3475–34782. doi: 10.1021/ic020672l. [DOI] [PubMed] [Google Scholar]
- 35.Sigel H, Griesser R. Nucleoside 5 '-triphosphates: self-association, acid-base, and metal ion-binding properties in solution. Chem. Soc. Rev. 2005;34:875–900. doi: 10.1039/b505986k. [DOI] [PubMed] [Google Scholar]
- 36.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 37.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 38.Park H, Lee S. Role of solvent dynamics in stabilizing the transition state of RNA hydrolysis by hairpin ribozyme. J. Chem. Theory Comp. 2006;2:858–862. doi: 10.1021/ct0503015. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes MM, Reblova K, Sponer J, Walter NG. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc. Natl. Acad. Sci. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter NG. Ribozyme catalysis revisited: is water involved? Mol. Cell. 2007;28:923–929. doi: 10.1016/j.molcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nam K, Gao J, York DM. Electrostatic interactions in the hairpin ribozyme account for the majority of the rate acceleration without chemical participation by nucleobases. RNA. 2008;14:1501–1507. doi: 10.1261/rna.863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 44.Johnson I. Fluorescent probes for living cells. Histochem. J. 1998;30:123–140. doi: 10.1023/a:1003287101868. [DOI] [PubMed] [Google Scholar]
- 45.Virta P, et al. Synthesis, characterisation and theoretical calculations of 2,6-diaminopurine etheno derivatives. Org. Biomol. Chem. 2005;3:2924–2929. doi: 10.1039/b505508c. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.