Abstract
Proton nuclear magnetic resonance relaxation times were measured for the protons of micelles formed by the detergents sodium dodecyl sulfate, dodecyltrimethyl ammonium bromide, and polyethylene glycol sorbitan monolaureate in the presence of ferriprotoporphyrin IX and the antimalarial drugs chloroquine, 7-chloro-4-quinolyl 4-N,N-diethylaminobutyl sulfide, and primaquine. Diffusion coefficients were extracted from pulsed gradient NMR experiments to evaluate the degree of association of these drugs with the detergent micelles. Results indicate that at low or neutral pH when the quinolyl N is protonated, chloroquine does not associate with neutral or cationic detergent micelles. For this reason, chloroquine’s interaction with heme perturbs the partitioning of heme between the aqueous medium and detergent micelles.
Keywords: Malaria, Antimalarials, Chloroquine, Heme, Detergent, Micelle
1. Introduction
Antimalarial drugs such as chloroquine (CQ) and quinine (QN) are known to inhibit the conversion of ferriprotoporphyrin IX (FPIX) into crystalline hemozoin. Although the crystalline structure of hemozoin is known [1], precisely how hemozoin is formed from FPIX, and precisely how CQ, QN and other drugs inhibit this process, are still not fully understood. The measured IC50’s for in vitro hemozoin inhibition by CQ and QN are 24 μM and 65 μM, respectively [2]. It should be noted that these in vitro IC50’s are highly pH dependent and are also influenced by the presence of lipids. In a recent study, it has been shown that CQ and QN differentially perturb the heme monomer – μ-oxo dimer equilibrium [3]. CQ promotes the μ-oxo dimer while QN promotes the monomer. How this difference relates to the drugs’ similar ability to prevent crystallization of hemozoin is puzzling. Promoting the monomer over the μ-oxo dimer can certainly reduce the stacking of heme units and thus retard crystal growth. The stabilization of the μ-oxo dimer, which may easily be the precursor of hemozoin, is more difficult to explain. A compound similar to CQ, in which the 4-amino group has been replaced by a thiol group, 7-chloro-4-quinolyl 4-N,N-diethylaminobutyl sulfide (4S), has recently been shown to associate likewise with the μ-oxo dimer [4]. And like CQ, 4S also favors the μ-oxo dimer in solution. However, as reported recently [4], the ability of 4S to inhibit hemozoin formation is significantly lower than that of CQ, demonstrating that the ability to interact strongly with the μ-oxo dimer does not guarantee inhibition of hemozoin crystallization.
There has been a resurgence of interest in the role of lipids in hemozoin formation. Hemozoin formation has been shown to be catalyzed by pre-formed hemozoin crystals [5], by the presence of histidine-rich proteins [6], or by lipids [7]. Fitch et al. [8] have screened several lipids and detergents for the ability to promote hemozoin formation. Arachidonic, linoleic, oleic and palmitoleic acids, 1-monooleoylglycerol, 2-dioleoylglycerol, SDS, Tween-80, and n-octyl-glucopyranoside have been shown to catalyze hemozoin formation while tri-oleoylglycerol, cholesterol, dioleylphosphatidylethanolamine, stearic acid, and palmitic acid appear not to catalyze. Lecithin (phosphatidyl choline) also promotes hemozoin crystallization [9] and recently, it has been demonstrated that hemozoin readily forms near water/lipid interfaces [10], and that the neutral lipids monopalmitic glycerol and monostearic glycerol can be found associated with hemozoin crystals that have been extracted from malaria parasites [11]. It has also been suggested that physiological formation of hemozoin requires both histidine-rich proteins and lipids in a coordinated two-step process [12]. The histidine rich protein binds as many as 50 heme units at a time, thus bringing the heme monomers close together. And simultaneously, lipids shield the heme from the aqueous environment allowing hydrogen bonds to be formed between nearby propionate side chains. Recently, a protein that is highly conserved across Plasmodium species has been discovered to be very efficient in helping hemozoin crystallize [13]. It has been proposed that this heme detoxification protein binds heme and rapidly initiates the formation of the head-to-tail dimers and delivers them to lipid micelles where stacking of these dimers eventually leads to hemozoin crystals.
This present work uses NMR spectroscopy to help in elucidating the behavior of antimalarial drugs and heme in the presence of detergent micelles. SDS is a negatively-charged surfactant while dodecyltrimethyl ammonium bromide (DTAB) is positively charged. For a neutral species, polyethylene glycol sorbitan monolaureate (Tween-20) is employed. Heme with its unpaired electrons causes major perturbation in the relaxation times of protons in its vicinity. This property is utilized to determine whether heme stays inside detergent micelles. On the other hand, diffusion gradient measurements are used to see whether the antimalarial drugs associate with detergent molecules. CQ is studied and compared against primaquine (PQ) and the 4S analogue of CQ (Both compounds, 4S and PQ, do not effectively inhibit hemozoin formation) to see if there are significant differences between these two compounds and CQ. Magnetic susceptibilities are also measured to monitor if any of the detergent molecules used in this study are perturbing the heme monomer – μ-oxo dimer equilibrium.
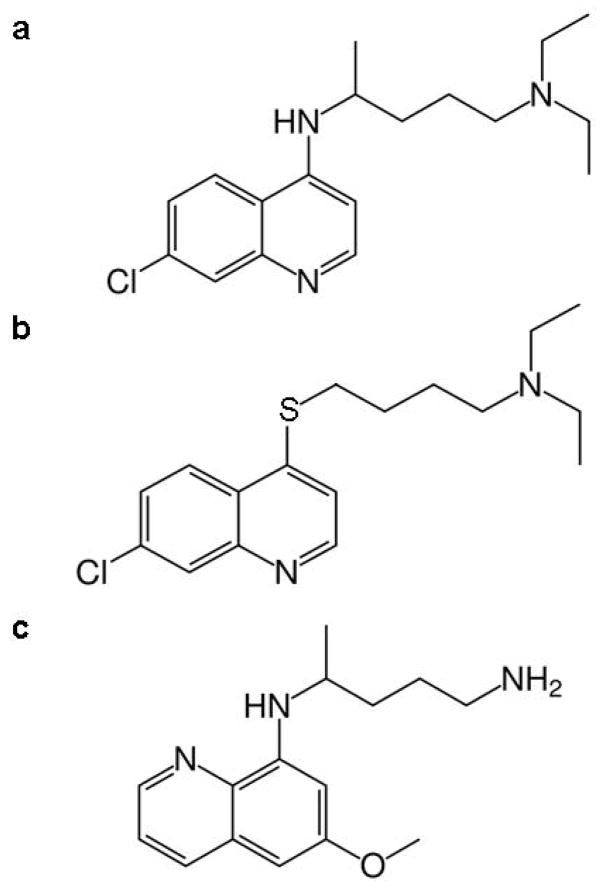
The use of detergent micelles enables concentrations of samples that are suitable for solution NMR spectroscopy. The interaction of the drugs as well as heme with these detergent systems is expected to be similar to how these molecules interact with lipid nanospheres. The structures of the drugs used in this study are shown in Figure 1.
Fig. 1.
Structures of antimalarial compounds used in this study. (a) chloroquine (CQ), (b) 7-chloro-4-quinolyl 4-N,N-diethylaminobutyl sulfide (4S), and (c) primaquine (PQ).
2. Materials and Methods
CQ diphosphate, PQ diphosphate, SDS, DTAB, Tween-20, sodium carbonate, DMSO-d6, and hemin (fluka) were purchased from Sigma-Aldrich (St. Louis, MO). Deuterium oxide was purchased from Cambridge Isotope Labs (Andover, MA). Sodium phosphate monobasic and sodium bicarbonate were purchased from Fisher Chemicals (Fair Lawn, NJ) and sodium phosphate dibasic was from EM Science (Gibbstown, NJ). The chloroquine analogue, 4S, was synthesized as described previously [4].
For samples in aqueous solution, phosphate buffer was used for pH 7 while a carbonate buffer was employed for pH 10. Buffers at these pH values were prepared with a total salt concentration of 200 mM, using D2O as solvent. The actual pH of each sample was measured using a Beckman Φ 11 pH meter. Heme stock solutions were made in 0.1 M NaOH. SDS, DTAB and Tween-20 stock solutions (500 mM) and drug stock solutions (50 mM) were made in D2O. Samples containing 50 mM detergent, 2.5 mM drug and 5 mM heme were prepared by diluting these stock solutions in the appropriate buffer. Samples for measurements of drug and detergent diffusion coefficients contained 5 mM drug and 50 mM detergent and were dissolved in 200 mM of the appropriate buffer. For magnetic susceptibility measurements of heme, samples were prepared by adding the required amount of heme (4 mM) and detergent (50 mM). The concentration used (50 mM) for all the detergents used in this study is well above their critical micelle concentrations.
NMR spectra were recorded on a Varian Unity INOVA 500 MHz spectrometer with a proton frequency of 499.789 MHz using Varian VNMR version 5.1 software. Sample and instrument temperatures were controlled at 298 K. T1 measurements were made using a standard inversion-recovery pulse sequence. Diffusion coefficients were measure via the pulsed gradient experiment [14–15]. Gradient strengths ranged from 0.3–35 Gauss/cm for fast-moving species, and from 0.7–70 Gauss/cm for slow-moving species.
Magnetic susceptibility measurements were made using the Evans method [16], using the equation appropriate for a superconducting magnet;
| (1) |
where χM is the molar susceptibility of the paramagnetic substance in cm3/mol, Δν is the chemical shift difference (in ppm) between a reference proton in the sample and that in a solution lacking the paramagnetic compound, c is the concentration of FPIX in mol/mL and χD is the diamagnetic susceptibility of heme (6.9 × 10−4 cgs units). Measurements were made with the use of 5 mm NMR tubes equipped with 2.5 mm capillary tube inserts. The capillary tube contains the exact same solution (with detergent, drug and buffer) except heme. The corrections for the susceptibility of the solvent and the difference in densities of the solvent and the solution are ignored. The water resonance frequency was used as the reporter for the chemical shift difference. Results using water are found to be identical with those obtained using DMSO or an additional reporter molecule such as tert-butyl alcohol. The molar susceptibility of FPIX is converted to magnetic moment using the following equation:
| (2) |
3. Results and discussion
It has long been known that more than 90% of FPIX becomes encapsulated inside micelles in aqueous detergent solutions [17]. Previous NMR experiments have already demonstrated that heme is embedded inside micelles, with the charged propionate side chains directed toward the charged surface of the micelles [18]. It has been shown previously that measurements of magnetic moments of heme solutions can provide information regarding the relative amounts of FPIX in monomer vs. μ-oxo dimer forms [3]. Our magnetic susceptibility measurements of heme in aqueous solutions in the presence of detergent indicate that both Tween-20 and DTAB do not affect the measured magnetic moment of an aqueous FPIX solution at pH 7. On the other hand, addition of SDS leads to a substantial increase in the magnetic moment (from 2.0 to 3.7 Bohr magneton), suggesting that this anionic surfactant promotes the heme monomer.
Results of NMR diffusion measurements of the drugs in micellar solutions are presented in Table 1. It is known that the principal difference between CQ and 4S lies in the pKa of the quinolyl N so the measurements have been made at two pH values, 7 and 10. At pH 7, CQ is expected to have its quinolyl N protonated while its analogue 4S will still have a neutral quinoline ring. The measured pKa of the quinolyl N in 4S is 4.1 while that of CQ is 8.6 [4]. At pH 10, both CQ and 4S are therefore expected to have an unprotonated quinolyl N. The measured diffusion constants of 4S are very similar to those of the detergent molecules at both pH values, suggesting that 4S associates easily with the micelles. On the other hand, at pH 7, CQ demonstrates significantly larger diffusion coefficients with the cationic detergent DTAB and the neutral detergent Tween-20. At pH 7, CQ appears to associate only with the anionic surfactant SDS. This behavior is in agreement with a previous study on the interaction of PQ and CQ with ionic micelles where it was found that if the drug has two positive charges, that is, the quinolyl N is protonated, interaction with both cationic and neutral zwitterionic surfactants is prevented [19]. Similar to heme, these drugs, if associated with micelles, are likewise found to be embedded inside the micelle with the quinoline ring residing in the hydrophobic interior of the micelle [19]. The decrease in the diffusion constant of CQ at pH 10 is due to its association with the detergent micelles and not self-aggregation because similar fast diffusion constants (≈4 × 10−6 cm2/s) are seen for CQ at pH 10 when detergent molecules are not present.
Table 1.
Measured diffusion coefficientsa (×10−6 cm2/s) for drug and detergent molecules
| Drug (detergent) | pH 7 | pH 10 |
|---|---|---|
| CQ | 3.8 | |
| CQ (SDS) | 0.2 (0.2) | 0.3 (0.4) |
| CQ (Tween-20) | 3.4 (0.7) | 0.9 (0.8) |
| CQ (DTAB) | 3.8 (1.5) | 1.3 (1.5) |
|
| ||
| 4S | 2.4 | |
| 4S (SDS) | 0.2 (0.3) | 0.2 (0.3) |
| 4S (Tween-20) | 0.8 (0.7) | 0.4 (0.4) |
| 4S (DTAB) | 1.2 (1.3) | 0.9 (1.3) |
|
| ||
| PQ | 4.9 | |
| PQ (SDS) | 0.5 (0.6) | 0.5 (0.6) |
| PQ (Tween-20) | 1.7 (0.4) | 1.1 (0.4) |
| PQ (DTAB) | 2.2 (1.5) | 1.1 (1.4) |
Diffusion coefficients for detergent (50 mM) in the presence of drug (5 mM) are enclosed in parenthesis.
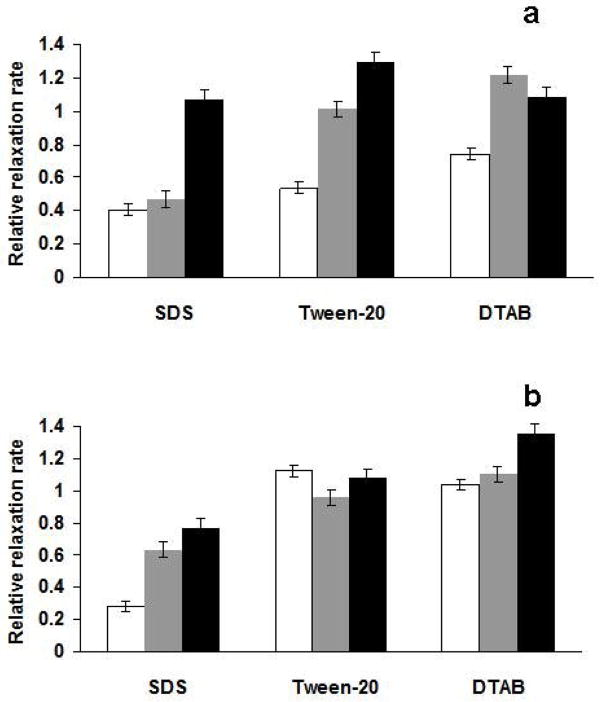
The very short T2 relaxation time of heme protons precludes diffusion measurements by pulsed gradient NMR spectroscopy. However, heme exerts a significant distance-dependent paramagnetic relaxation enhancement on protons of nearby molecules. Thus, measuring T1 relaxation times for the detergent protons can provide a qualitative picture of how heme partitions between the micellar and aqueous portions of the solution. Addition of heme to detergent leads to a significant decrease in the detergent’s proton T1 relaxation times. Tween-20 provides a unique setting since its micelles contain an inner hydrophobic core and a hydrophilic layer formed by its polyethylene glycol (PEG) portion. Protons from these two distinct regions of the micelles are well resolved and their relaxation times can be separately measured. In addition, due to the ester linkage in Tween-20, spin diffusion is limited between the protons of the hydrophobic chain and those of PEG. In the absence of heme, the protons of the terminal –CH3 group (hydrophobic core) of Tween-20 have a T1 of about 1 second while the PEG protons (the largest peak at 3.6 ppm from tetramethylsilane) exhibit a T1 of 0.65 seconds. Upon adding heme, the T1 of the methyl proton is reduced to 0.07 seconds while the relaxation time of the PEG protons is 0.49 seconds, still comparable to the heme-free value, suggesting that heme is indeed located inside the hydrophobic core of the micelle. Thus, in comparing the various drug-heme-detergent solutions, we will simply focus on the proton relaxation rates of the hydrophobic terminal –CH3 group of the detergent molecule. Upon addition of drugs, the measured relaxation rates of the methyl protons will be compared to the values observed when only heme (no drug) is present. In this manner, a reduction in the paramagnetic enhancement of the relaxation rate insinuates that heme is being taken out of the micelle. The relative relaxation rates (measured relaxation rates divided by the rates observed with heme alone) are displayed in Figure 2.
Fig. 2.
(a) The relative relaxation rates (compared to the relaxation rates when 5 mM heme alone is added) of the inner methyl protons of the detergent molecules (50 mM in aqueous solution, at pH 7) in the presence of 2.5 mM CQ (white), 2.5 mM 4S (gray), and 2.5 mM PQ (black). (b) as in (a), but at pH 10.
Increasing amounts of drug (1.25, 2.5 and 5 mM) have been added to the heme (5 mM) – detergent (50 mM) solutions. The effects of adding 2.5 mM are twice as large as those obtained with 1.25 mM, however, results with 5 mM drug are essentially identical with those at 2.5 mM (not shown). This apparent stoichiometry, one drug per 2 FPIX molecule, points to a dimer species interacting with the drug. As mentioned earlier, addition of SDS, unlike with DTAB and Tween-20, leads to an increase in the observed magnetic moment of the heme solution. This ability of SDS to perturb the monomer – μ-oxo dimer equilibrium can be attributed to the substantially lower pH (lower than bulk) near the surface of negatively charged SDS micelles. Upon adding either CQ or 4S, the observed magnetic moment is brought down to the SDS-free value. Both CQ and 4S [3–4] have already been shown to promote the lower magnetic moment μ-oxo dimer. On the other hand, we have observed in this work that PQ does not perturb the heme monomer-μ-oxo dimer equilibrium. This is expected because PQ is known not to interact strongly with heme [20]. Therefore, the differences seen in the relaxation rates of the SDS CH3 protons are mainly due to drug-induced changes in the heme monomer – μ-oxo dimer equilibrium. Based on the diffusion coefficient data CQ, PQ and 4S are all expected to be embedded inside SDS micelles, hence, there should have been no difference in the paramagnetic relaxation enhancement of SDS protons. However, with CQ and 4S, the monomer-promoting effects of SDS on the monomer – μ-oxo dimer equilibrium are canceled, which leads to the lower magnetic moment species and slower relaxation rates in the presence of these two compounds.
The situation with the detergents DTAB and Tween-20 is less complicated. These two detergents do not change the observed magnetic moment of aqueous heme solutions. In Figure 2a, the difference between CQ and 4S becomes apparent. Addition of CQ leads to a decrease in the paramagnetic enhancement of relaxation rates of the detergent methyl protons. The decrease is about 50% in Tween-20 and about 30% in DTAB. This decrease can only be attributed to a removal of heme from the inside of the micelles. The relaxation rates measured when 4S is present are very similar to those observed with PQ, or when no drug is present, suggesting that unlike CQ, 4S is unable to perturb heme’s favorable partitioning into the detergent micelles. The difference between CQ and 4S disappears at higher pH, as shown in Figure 2b. At pH 10, all three drugs do not have a substantial effect on the paramagnetic relaxation enhancement of the detergents’ methyl protons. Therefore, the uniqueness of CQ at pH 7 can be explained by the relatively high pKa of its quinolyl N. With the quinolyl N charged, CQ does not associate with either DTAB or Tween-20, yet it binds strongly to the μ-oxo dimer, thus, enabling CQ to alter heme partitioning between micelles and the aqueous medium. Although 4S likewise interacts strongly with the μ-oxo dimer, its solubility inside DTAB and Tween-20 micelles precludes it from removing the heme from this hydrophobic environment. The above comparisons are valid with the assumption that if heme aggregation inside micelles is occurring, the amount of aggregation is uniform across all samples. The relatively high amount of detergent used in these experiments should minimize heme aggregation.
We conclude that with the exception of the anionic detergent SDS, the heme species that becomes embedded inside the detergent micelles exhibits a low magnetic moment, a property that can only be exhibited by an antiferromagnetically coupled μ-oxo dimer. This suggests that if hemozoin is indeed formed inside a hydrophobic lipid environment, its precursor may be the μ-oxo dimer. This difference between SDS and the neutral and cationic detergents in this study may explain why only neutral lipids are found associated with hemozoin inside the digestive vacuole of malaria parasites [11]. The insolubility of CQ in micelles composed of either cationic or neutral detergent molecules at pH 7, and its strong interaction with the μ-oxo dimer enables CQ to remove heme from the micelle environment. Thus, if indeed hemozoin formation requires a hydrophobic environment that is either afforded by detergent micelles or lipid nanospheres, removal of the μ-oxo dimer from this environment is perhaps another key facet behind CQ’s mechanism of action. This suggestion implies that an effective antimalarial drug that works in this fashion has the ability to form a complex with the μ-oxo dimer and is insoluble in neutral or cationic lipids or micelles. The quinoline ring of CQ with its polarized π–system enables this molecule to interact favorably with the μ-oxo dimer [21]. Its 4-amino group enhances appreciably the basicity of the quinolyl N, giving it a pKa of about 8. Thus, at pH 7 or lower, the quinolyl N is protonated making it insoluble inside neutral or cationic micelles.
Acknowledgments
We thank the NIH (Grant R01AI060792) for financial support. LBC thanks the Achievement Rewards for College Scientists (ARCS) Foundation for financial support. JBK thanks the National Science Foundation Research Experience for Undergraduates program.
Abbreviations
- FPIX
Ferriprotoporphyrin IX
- CQ
Chloroquine
- PQ
Primaquine
- 4S
7-chloro-4-quinolyl 4-N,N-diethylaminobutyl sulfide
- DTAB
Dodecyltrimethyl ammonium bromide
- Tween-20
Polyethylene glycol sorbitan monolaureate
- PEG
polyethylene glycol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pagola S, Stephens PW, Bohle DS, Kosar AD, Madsen SK. Nature. 2000;404:307–310. doi: 10.1038/35005132. [DOI] [PubMed] [Google Scholar]
- 2.Hawley SR, Bray PG, Mungthin M, Atkinson JD, O’Neill PM, Ward SA. Antimicrob Agents Chemother. 1998;42:682–686. doi: 10.1128/aac.42.3.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casabianca LB, An D, Natarajan JK, Alumasa J, Roepe PD, Wolf C, de Dios AC. Inorg Chem. 2008;47:6077–6081. doi: 10.1021/ic800440d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan JK, Alumasa J, Yearick K, Ekoue-Kovi K, Casabianca LB, de Dios AC, Wolf C, Roepe PD. J Med Chem. 2008;51:3466–3479. doi: 10.1021/jm701478a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorn A, Stoffel R, Matile H, Bubendorf A, Ridley RG. Nature. 1995;374:269–271. doi: 10.1038/374269a0. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan DJ, Glizman IY, Goldberg DE. Science. 1996;271:219–222. doi: 10.1126/science.271.5246.219. [DOI] [PubMed] [Google Scholar]
- 7.Bendrat K, Berger BJ, Cerami A. Nature. 1995;378:138–138. doi: 10.1038/378138a0. [DOI] [PubMed] [Google Scholar]
- 8.Fitch CD, Cai GZ, Chem YF, Shoemaker JD. Biochim Biophys Acta. 1999;1454:31–37. doi: 10.1016/s0925-4439(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 9.Trang DTX, Huy NT, Uyewn DT, Sasai M, Hiono TS, Harada S, Kamei K. Anal Biochem. 2006;349:292–296. doi: 10.1016/j.ab.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Egan TJ, Chen JYJ, de Villiers KA, Mabotha TE, Naidoo KJ, Ncokazi KK, Langford SJ, McNaughton D, Pandiancherri S, Wood BR. FEBS Lett. 2006;580:5105–5110. doi: 10.1016/j.febslet.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 11.Pisciotta JM, Coppens I, Tripathi AK, Scholl PF, Shuman J, Bajad S, Shulaev V, Sullivan DJ. Biochem J. 2007;402:197–204. doi: 10.1042/BJ20060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey AV, Babbarwal VK, Okoyeh JN, Joshi RM, Puri SK, Singh RL, Chauhan VS. Biochem Biophys Res Commun. 2003;308:736–743. doi: 10.1016/s0006-291x(03)01465-7. [DOI] [PubMed] [Google Scholar]
- 13.Jani D, Nagarkatti R, Beatty W, Angel R, Slebodnick C, Andersen J, Kumar S, Rathore D. PLoS Pathog. 2008;4:e1000053. doi: 10.1371/journal.ppat.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stejskal EO, Tanner JE. J Chem Phys. 1965;42:288–292. [Google Scholar]
- 15.Tanner JE. J Chem Phys. 1970;52:2523–2526. [Google Scholar]
- 16.Evans DF. J Chem Soc. 1959:2003–2005. [Google Scholar]
- 17.Simplicio J, Schwenzer K. Biochemistry. 1973;12:1923–1929. doi: 10.1021/bi00734a014. [DOI] [PubMed] [Google Scholar]
- 18.Mazumdar S. J Phys Chem. 1990;94:5947–5953. [Google Scholar]
- 19.Perussi JR, Yushmanov VE, Monte SC, Imasato H, Tabak M. Physiol Chem Phys & Med NMR. 1995;27:1–15. [PubMed] [Google Scholar]
- 20.Dorn A, Vippagunta SR, Matile H, Jaquet C, Vennerstrom JL, Ridley RG. Biochem Pharmacol. 1998;55:727–736. doi: 10.1016/s0006-2952(97)00510-8. [DOI] [PubMed] [Google Scholar]
- 21.Casabianca LB, de Dios AC. J Phys Chem A. 2006;110:7787–7792. doi: 10.1021/jp061320t. [DOI] [PubMed] [Google Scholar]