Abstract
Germline substitutions in the endothelial cell tyrosine kinase receptor TIE2/TEK cause a rare inherited form of venous anomalies, mucocutaneous venous malformations (VMCM)1-4. We now identified a somatic 2nd hit causing loss-of-function of the receptor in a resected VMCM. We assessed for whether such localized, tissue-specific events play a role in the etiology of the far more common sporadic VM. Eight somatic TIE2 mutations were identified in lesions from 28 out of 57 patients (49.1%), not detected in their blood or in control tissues. The somatic mutations included a frequent L914F change, and a series of double-mutations that occurred in cis, all of which show ligand-independent hyperphosphorylation in vitro. When overexpressed in HUVECs, L914F showed abnormal localization and response to ligand, differing from wild-type and the common inherited R849W mutant, suggesting they may have distinct effects. The presence of the same mutations in multifocal VMs in two patients, suggests a common origin for the abnormal endothelial cells in the distant sites. In conclusion, these data illustrate that a sporadic disease may be explained by somatic changes in a gene causing rare, inherited forms, and pinpoint TIE2 pathways as potential therapeutic targets for VM.
The vascular endothelial cell (EC)-specific receptor tyrosine kinase (RTK) TIE2 plays a crucial role in angiogenesis and cardiovascular development 5-13. Its ligands, the angiopoietins (Ang), induce receptor dimerization and phosphorylation14, with Ang-1 acting as an agonist and Ang-2 as context-dependent antagonist or weak agonist 15,16. In man, heterozygous TIE2 substitutions that induce in vitro ligand-independent hyperphosphorylation cause hereditary mucocutaneous venous malformations (VMCM) 1-4, characterized by multifocal small bluish cutaneous and mucosal lesions, composed of enlarged, tortuous venous channels. The most common is a c.2545c>t change (R849W), which occurs in a total of 10/17 families reported 1,2,4,17. The remaining changes (shown in Fig. 1a) have each been identified in a single family 2,4. We previously postulated a paradominant mode of inheritance to account for the heterogeneous, focal lesions of VMCM 18. Here, we identified a somatic, lesion-associated TIE2 “2nd-hit” alteration in one resected VMCM, from a patient (Sa-I.4, in 1) carrying the germline R849W. A cDNA screen revealed an in-frame deletion of 129-bp, corresponding to a loss of exon 3 and part of exon 4 (amino acids 122-165 of the extracellular Ig2 ligand-binding domain; “Del”, Fig. 1a, b; Supplementary Fig. 1a). Allele-specific PCR showed that the loss, which is part of an intragenic somatic DNA deletion (see Methods), occurs on the wild-type, and not the inherited R849W-mutant allele (Fig. 1b).
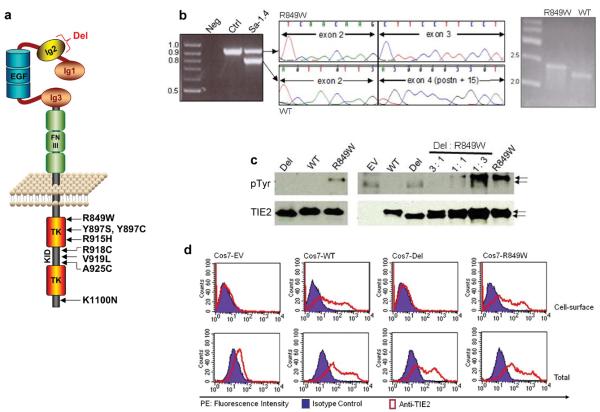
Figure 1. Somatic deletion in Ig2 extracellular domain in a VMCM-tissue.
(a) Schematic representation of TIE2: extracellular region with three immunoglobulin-like loops (Ig1, 2, 3), three epidermal growth factor-like repeats (EGF), and three fibronectin type III-like repeats (FNIII). Intracellular region contains 2 tyrosine kinase domains (TK) separated by kinase insert domain (KID). Locations of missense substitutions previously identified in inherited VMCM are shown. A somatic mutation (“Del”) deletes part of Ig2 in the (wild-type) allele of one VMCM patient (Sa-I.4), carrying the inherited R849W mutation. (b) PCR [exons 1-7] in patient Sa-I.4 shows a 900-bp band (expected: 1029 bp) absent in control (left panel). Sequencing demonstrates a 129-bp deletion, of exon 3 and 14-bp of exon 4 (middle) in 900-bp band. Allele-specific PCR shows deletion on wild-type allele (WT), which runs smaller than expected (by 129 bp); mutant R849W runs at expected size (right panel). (c) Western blot with anti-phosphotyrosine antibody to evaluate TIE2 phosphorylation (pTyr, 140 kD, top). Blots stripped and reprobed with anti-TIE2 for total TIE2 levels (bottom). Lysates from Cos-7 cells expressing WT, R849W, or Del (left panel), or various ratios of R849W: Del (right panel). EV: Empty vector control. 43-amino-acid size difference observed in Del, as compared to WT and R849W (arrows). (d) Flow-cytometry on TIE2 (WT, Del, or R849W) -transfected Cos7 cells, or EV controls. Cells stained for cell-surface (upper panels), or total (intracellular + cell surface; lower panels) TIE2. x-axis: Phycoerythrin (PE) fluorescence intensity with anti-TIE2 antibody and mouse IgG1 isotype control; y-axis: cell-counts.
Unlike inherited intracellular TIE2 mutations, the somatic deletion mutant (“Del”) was not hyperphophorylated in vitro, nor did it exacerbate hyperphosphorylation of R849W in trans (Fig. 1c). The lack of 43 amino acids resulted in loss of cell surface expression of the receptor (Fig. 1d, Supplementary Fig. 1d), as well as its inability to bind Ang1, in contrast to WT and R849W (Supplementary Fig. 1b, c). In retrovirally transfected HUVECs, Del-TIE2 is retained in endoplasmic reticulum (ER), and shows no ability to respond to Ang1 by increased phosphorylation or translocation and clustering (Fig. 2, Supplementary Fig. 1e). It is probable that Del-TIE2 fails to fold into its native form, causing defective protein trafficking and ER-retention 19. Thus, the somatic deletion mutant acts as a null-allele, causing local loss-of-function of wild-type TIE2.
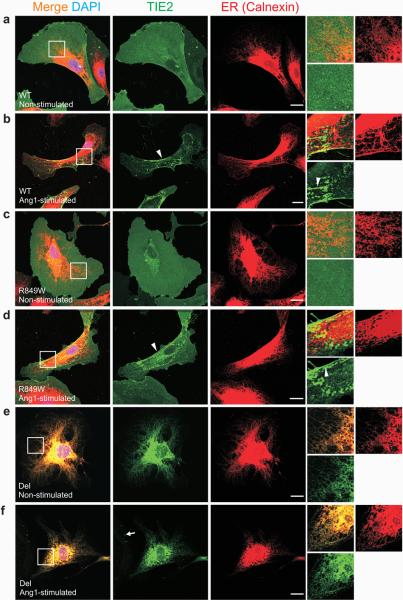
Figure 2. Del-TIE2 is retained in the endoplasmic reticulum (ER).
(a-f) Sparse HUVECs expressing: (a-b) WT, (c-d) R849W, or (e-f) Del mutant-forms of TIE2, stimulated for one hour with Ang1 (b, d and f), or left unstimulated (a, c and e), and stained with antibodies against TIE2 and ER-specific chaperone Calnexin. Magnifications of boxed areas are shown on right. In unstimulated WT and R849W cells, TIE2 is evenly distributed on plasma membranes. Light accumulation of TIE2, with variation between cells, observed in perinuclear area, likely representing newly synthesized or recycled receptors (a and c, middle panels). Ang1-stimulation induced clustering of TIE2-receptors (b and d, arrowheads in insets) and translocation to the retracting cell edges (b and d, arrowheads in middle panels). Del-TIE2 is accumulated in ER in both unstimulated and Ang1-stimulated cells (e-f). Consistent with ER-location, soluble Ang1 failed to induce Del-TIE2 clustering as seen in WT and R849W; only dot-like clusters observed in cell margins, likely representing endogenous TIE2 (arrow). Scale bar 20 μm.
The identification of a 2nd hit in VMCM tissue, led to the hypothesis that somatic changes in TIE2 could also be etiopathogenic for common sporadic VM. We screened blood-DNA from 57 affected individuals, most of whom had extensive, unifocal lesions, typical of sporadic VM20 (Fig. 3a). Histologically, they were characterized by enlarged venous channels with patchy smooth-muscle cell layers (Fig. 3b), and thin, continuous TIE2-positive EC layers (Fig. 3c-e). No TIE2 mutations were detected in blood-DNA. To test for somatic changes, we screened DNAs from 62 VMs from the same 57 patients, and identified 8 missense substitutions in exon 17, in 30 lesions from 28 patients (49.1%; described in Supplementary Table 1). In 24/30 mutation-positive lesions (from 24/28 patients; i.e. 85.7%), a change at codon 914 (CTT to TTT; L914F) was identified, (Supplementary Fig. 2a, Supplementary Table 1, Fig. 4a). In 6 VMs (including 2 lesions each from 2 patients with multifocal VMs; Fig. 3f, g), we detected two mutations in cis, i.e. on the same allele (see Methods; Supplementary Fig. 2b-e, Supplementary Table 1). Each consisted of a Y897 mutation in combination with another change: [Y897H+R915C], [Y897F+R915L], [Y897C+R915C], and [Y897S+S917I] (Fig. 3f, g, Fig 4b; Supplementary Table 1). The necessity for two mutations (897 in the 1st tyrosine kinase [TK] domain, in combination with a residue from the “kinase insert domain [KID]-cluster”; Fig. 4c) on the same allele to confer pathogenicity is uncommon, but was described for lung cancer-associated TK domain epidermal growth factor receptor (EGFR) mutations 21. The 4 patients with double mutations exhibited no clinical differences (Supplementary Table 1), except the occurrence of multiple lesions in 2/4 individuals. Each had a double-mutation in common between two lesions assessed, independent of location, suggesting a common cellular (rather than individual and localized) origin for the multifocal VMs; however, they were not detectable in blood-DNA (Fig. 3f, g). They could be attributed to common EC-progenitor cells circulating to these distinct sites. Late-developing lesions could have their origins in a stem cell niche, where some cells with the capacity for self-renewal and differentiation carry post-zygotic mutations 22, or alternatively, circulating ECs sloughed off from a primary lesion. Resections, however, do not seem to have increased development of additional lesions.
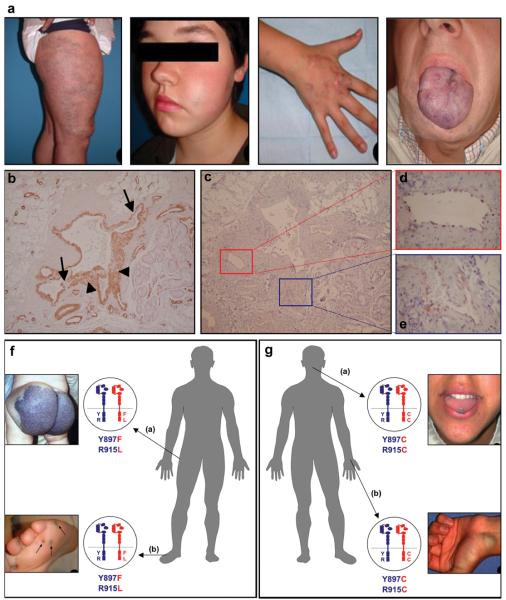
Figure 3. Typical features of sporadic venous malformations.
(a) Lesion on leg, cheek, hand, and tongue of patients #11, 19, 13, and 4, respectively. All have the L914F mutation. (b-e) Immunohistochemical staining of sections of dysmorphic vessels and surrounding areas, from VM of patient # 15, with L914F. (b) Smooth muscle alpha-actin demonstrates variable smooth muscle cell layers, thin (arrow) and thick (arrowhead). (c) TIE2 stains a continuous, thin layer of endothelial cells (ECs), shown at higher magnification (boxed) in (d) and (e). TIE2 expression is not detected outside endothelial cells. (f, g) Double-mutant TIE2 alleles in patients with multifocal VMs. (f) Patient #27: large buttock-VM at birth, appearance of small (<1 cm diameter) lesions (round, hyperkeratotic when on plantar surface) by age 6; [Y897F + R915L] TIE2 cis double-mutations in both buttock and foot lesions. (g) Patient #28: multiple (sub)cutaneous and intramuscular lesions in the face, thorax and abdominal regions, with new lesions appearing by age 18. Tongue and wrist VMs, both with [Y897C + R915C]. Normal allele, blue; mutant, red.
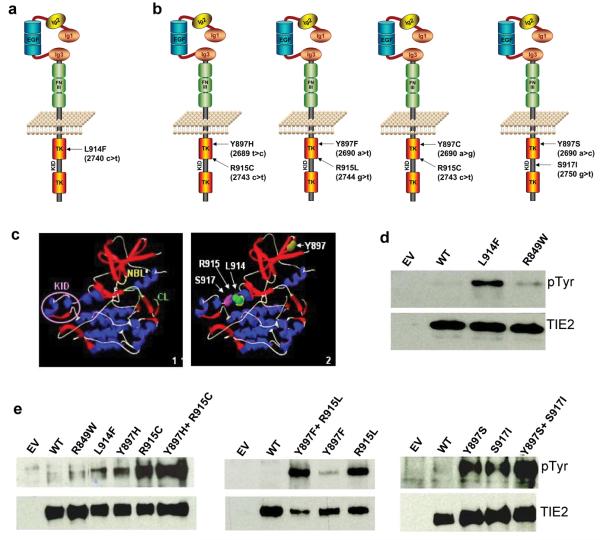
Figure 4. Somatic TIE2 mutations identified in sporadic VM tissues cause ligand-independent hyperphosphorylation.
(a-b) Schematic representations of TIE2 showing the missense substitutions, and corresponding amino acid changes identified in sporadic VMs. Amino acid residues 897, 914 and 915 located in first TK, and 917 in KID. (a) L914F: most frequent change, 24/28 patients. (b) In 4 patients, TIE2 double-mutations occur in cis. [Y897F+R915L] and [Y897C+R915C] in 2 lesions each, from 2 patients with multifocal VMs. (c) Representation of TIE2 intracellular region protein structure (Panel 1, KID = Kinase Insert Domain, NBL = Nucleotide Binding Loop, CL = Catalytic Loop), showing the positions of the somatic mutations (Panel 2). (d,e) Western blot with anti-phosphotyrosine antibody to evaluate TIE2 phosphorylation (pTyr, 140 kD, top). Blots stripped and reprobed with anti-TIE2 antibody to detect total TIE2 levels (bottom). (d) L914F compared to wild-type TIE2 (WT) and common germline mutant R849W. (e) Comparison of double mutants (Y897H+R915C, Y897F+R915L, Y897S+S917I) with WT and constituent single-mutants. EV: empty vector controls.
No exon 17 change was found in 90 control tissues from unrelated individuals, including resected skin, hemangiomas, lymphatic and vascular malformations (p= 9.006e−15; two-tailed Fisher's Exact test), demonstrating that the TIE2 substitutions are not common, non-associated, somatic changes.
We assessed for expression of TIE2 mutants in cDNA from 26/30 mutation-positive tissues. Both wild-type and mutant alleles were detected. To estimate their relative levels, we performed semi-quantitative minisequencing (SNaPshot, Applied Biosystems) on a large subset of DNAs and cDNAs (11 with L914F, i.e. 46%; at least 1 lesion each of the double-mutants; Supplementary Table 1; sample chromatograms: Supplementary Fig. 2, raw data: Supplementary Table 2). The presence of the mutant, relative to wild-type, was consistently enhanced in cDNA as compared to DNA (1.8-18 fold; Supplementary Table 1, 2). In addition, mutant alleles were not detectable by this method in 2 DNAs, but robustly expressed in cDNAs. The most frequent sporadic VM mutation (L914F), which is the only change that occurs alone, has not been identified in the germline in VMCM, suggesting it is strongly deleterious. It may nevertheless require an additional hit for lesion formation: in >60% (7/11) of tissues assessed, the relative level of expressed L914F was over 50% (58-92%) of the total, arguing for overexpression of the mutant, or loss of the wild-type by an unknown mechanism. The double-mutants were also enriched in cDNA, to a lower level (Supplementary Table 1). We compared the relative abundance of pairs of cis-mutations in tissues in order to impute their order of occurrence (see Methods), which would appear to be fairly even, with Y897 mutations appearing first in lesions from two patients, and the other cis mutations (R915C or R915L) occurring first in two others with multifocal lesions. The difference between mutations constituting each pair was small (within 10% of each other; Supplementary Table 1), accounting for the predominance of double-mutants obtained by cloning.
The somatic substitution sites, located within the first TK (Y897, L914, R915) or KID (S917) of TIE2 (Fig. 4a-c) 23, are highly conserved between RTKs across species, and bioinformatic analyses predicted weak-to-strongly damaging effects on protein function (Supplementary Table 3). We found L914F induced higher ligand-independent receptor phosphorylation than the common germline R849W (12.7±5.8 vs. 4.2±1.6 -fold over wild-type, mean±SD; Fig. 4d; Supplementary Fig. 4). Double-mutations had stronger (roughly additive) effects than constituent single mutations, with each Y897 substitution being weaker of the pair (Fig. 4e, Supplementary Fig. 4). There was no correlation between clinical phenotype and level of hyperphosphorylation (Supplementary Table 1).
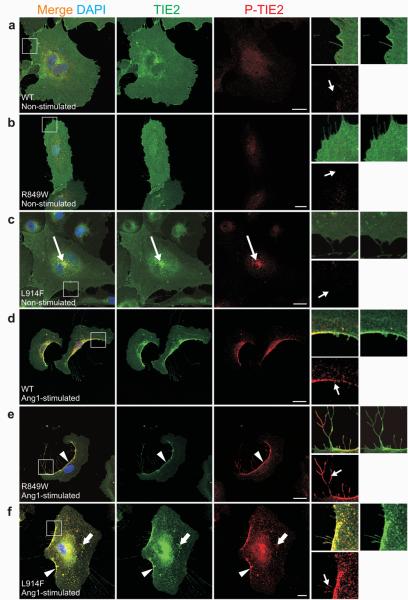
To compare the effects of WT, and the most frequent inherited (R849W) and somatic (L914F) mutants, their sub-cellular localization and activation was studied with and without activating Ang1-ligand (Figs. 5, 6). In the absence of ligand, WT and R849W-TIE2 were distributed evenly on cell-membranes, with occasional and slight increase in perinuclear TIE2 immunoreactivity, and no phosphorylated TIE2 (P-TIE2)-specific staining observed (Fig. 5a, b; Supplementary Fig. 1e). In contrast, L914F showed strong perinuclear accumulation, and ligand-independent activation in a distinct compartment near the nucleus (Fig. 5c). We and others have shown that Ang1-binding induces TIE2 clustering and translocation to cell edges24,25. This was observed in the majority of cells expressing WT and R849W (85.8±3.6% and 84.8±7.6% respectively; Fig. 5d, e), but only in a subset of L914F-HUVECs (12.5±4.1%), in which the protein was largely found in a punctuate pattern dispersed over the plasma membrane (Fig. 5f). In mobile cells, Ang1-activated TIE2 accumulates in the trailing edge and retraction fibers (Fig. 5d). R849W was more activated in retraction fibers than WT in response to Ang1, with L914F showing the presence of fibers similar to those observed with R849W, with intermediate P-TIE2 levels (Fig. 5d-f). Within fibers, WT was typically phosphorylated in the distal ends and TIE2-containing nodules (Supplementary Fig. 5a), whereas R849W showed higher P-TIE2 intensity all along (Supplementary Fig. 5b, d). L914F also showed P-TIE2 immunoreactivity along the fibers, at levels significantly higher than WT, but lower than R849W (Supplementary Fig. 5c, d).
Figure 5. Ang1-induced TIE2 clustering and phosphorylation in HUVECs expressing mutant (R849W and L914F) and wild-type (WT) TIE2.
(a-f) Sparse HUVECs expressing: (a, d) WT, (b, e) R849W, or (c, f) L914F, stimulated for one hour with Ang1 (d-f), or left unstimulated (a-c), and stained with antibodies against total TIE2 (TIE2) and phosphorylated TIE2 (P-TIE2). Boxed areas magnified on right. (a-c) In unstimulated WT and R849W expressing cells, TIE2 distributed evenly on cell membranes with no P-TIE2 specific staining. In contrast, increased L914F immunostaining observed in perinuclear area, where L914F is activated based on P-TIE2 staining (c, long arrows). Without Ang1, neither cell edges nor cellular extensions showed P-TIE2 antibody reactivity (small arrows). (d-f) In response to Ang1-stimulation, TIE2 translocated and phosphorylated in cell rear (arrowheads) and in retraction fibers (small arrows, right). Ang1 promotes WT translocation and activation in cell rear (d). R849W showed enhanced phosphorylation in retraction fibers (e, small arrow) when compared to WT (d, small arrow). Ang1 induced punctuate L914F clustering (thick arrows), and less complete translocation to cell rear (f). Scale bar 20 μm.
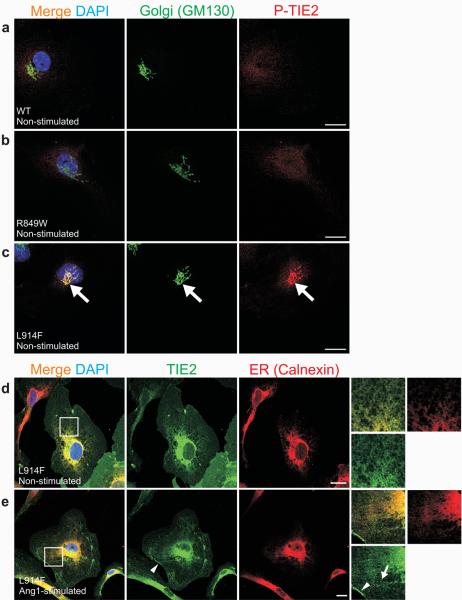
Figure 6. Cellular localization and activation of L914F-TIE2.
(a-e) Sparse HUVECs expressing: (a) WT, (b) R849W or (c-e) L914F forms of TIE2 stimulated for one hour with Ang1 (e), or left unstimulated (a-d), and stained with antibodies against Golgi-specific GM130 and phospho-TIE2 (P-TIE2; a-c), or total TIE2 (TIE2) and ER-specific Calnexin (d-e). (a-c) When compared to WT (a) and R849W (b), L914F showed ligand-independent activation in the Golgi apparatus (c, large arrows). (d-e) In both unstimulated and Ang1-stimulated cells, L914F was found to localize in ER and plasma-membrane. Ang1 induced L914F clustering and translocation, albeit incompletely, to retracting cell edges (e, arrowhead). TIE2 clusters in L914F-expressing cells typically appeared punctuate (e, arrow). Scale bar 20 μm.
To investigate its perinuclear compartmentalization in greater detail, L914F-overexpressing HUVECs were stained with Golgi and ER markers. No P-TIE2 staining was observed in Golgi of WT-transfectants (Fig. 6a), or the majority of R849W expressing cells, (Fig. 6b; weak perinuclear P-TIE2 staining detected in a few cells: 3.2±2% mean±SD, not shown). In contrast, L914F activation was seen in the Golgi in the majority of cells (73.2±7% cells, Fig. 6c). L914F was also retained in ER in both unstimulated (Fig. 6d) and Ang1-stimulated cells (Fig. 6e). Unlike Del-TIE2 however, L914F was not exclusively intracellular, since Ang1 did induce (albeit incomplete) L914F clustering and translocation to cell-cell junctions, cell edges, and retraction fibers, where its activation status was above that of WT (Fig. 5d-f, 6e; Supplementary Fig. 5c, d). ER-retention of L914F may be phosphorylation-dependent (in contrast to the less strongly phosphorylated R849W), as in the case of human disease-associated mutations in other RTKs, including KIT, FLT-3 and FGFR326-28. Thus, the common inherited and sporadic mutations both profoundly affect TIE2 compartmentalization and activation, as well as response to ligand, and may act in overlapping yet distinct ways to potentiate VM -pathogenesis.
In summary, we demonstrate, for the first time, that somatic mutations in TIE2 cause common, sporadic VM, allowing us to generalize findings on a rare, inherited form of a vascular anomaly, to its far more frequent, sporadic counterpart. This may be true for other disorders, and suggests that somatic screens should be performed at least for the numerous non-inherited vascular anomalies of unknown etiology. Finally, the implication of TIE2 in sporadic VMs greatly broadens the potential utility of therapeutic treatments that inhibit angiogenesis in general, and the TIE2 signaling pathway in particular.
Methods
Patient Recruitment
Fifty-seven patients were recruited in collaboration with the Vascular Anomalies Center, Cliniques Universitaires Saint-Luc, Brussels, Belgium. Informed consent was obtained from all participants prior to their enrolment in the study, as approved by the ethical committee of the Medical Faculty at the Université catholique de Louvain, Brussels, Belgium. All patients originated from Belgium.
DNA and RNA samples
Tissues were collected in liquid nitrogen immediately after resection. DNA was extracted from frozen tissue and from peripheral leukocytes using the Puregene DNA extraction kit (Gentra). RNA was extracted from frozen tissue using Tripure (Roche), and cDNA synthesis was performed using iScript cDNA Synthesis Kit (Bio-Rad) according to manufacturer instructions.
Mutational Analysis of a VMCM (Sa-I.4) Tissue
Tissue and lymphoblast RNA was extracted and cDNA synthesized as above. Five overlapping amplicons of ∼1 kb covering the entire TIE2 coding sequence (3.5 kb) were amplified and sized on agarose gels. PCR products were resolved on 1% agarose gels. In order to determine if the somatic deletion and R849W germline mutation were allelic, allele-specific PCR was performed with Reverse primers specific for the wild-type or mutant at the R849W change, located in exon 15, and a common Forward primer located in exon 2. The non-deleted product is of 2276 bp, whereas the allele containing the deletion is of 2147 bp (Fig. 1b). Conditions for the c.2545C>T (R849W) allele-specific PCR have been described previously 1. Upon sequencing an amplicon spanning exons 3 and 4 in both blood and tissue DNA, we observed loss-of-heterozygosity in the tissue for a novel SNP (t/c in blood > t in tissue) in TIE2 intron 3 at position 27159180 bp on chromosome 9, indicating that the identified loss in the ligand-binding domain is part of an intragenic somatic DNA deletion, rather than the result of aberrant splicing due to a single nucleotide change.
Full-length human TIE2 cloning and overexpression
The 3.5 kb human TIE2 cDNA, with 50 bp of 5′ and 60 bp of 3′ UTR, was cloned into the XhoI and NotI sites of pcDNA3.1/Zeo (−) (Invitrogen). To generate mutants, tissue cDNAs were used as templates to amplify regions around the respective mutations (or the deleted region). These were inserted into the full-length TIE2 construct replacing the wild-type fragment, using appropriate unique flanking restriction sites. The wild-type and mutant constructs were sequence-verified and expressed in Cos-7 cells. Their ability to cause ligand-independent hyperphosphorylation of the receptor was tested by immunoblotting with antibodies recognizing phosphotyrosine (PY-99, Santa Cruz) or anti-pTyr1102/1108Tie2 (Calbiochem), followed by stripping and re-probing with antibody for total-TIE2 (C-20, Santa Cruz) (see Supplementary Methods).
Flow cytometry
Cos-7 cells were detached with PBS-EDTA at 24-hrs post-transfection, washed and incubated in FACs buffer (1mM, pH 7.4 sodium phosphate buffer containing 137mM NaCl, 5mm KCl, 0.4mM MsSO4, 0.3mM MgCl2, 5mM glucose, 4mM NaHCO3, 1mM EDTA, and 3% FCS). Cells were stained with PE-conjugated anti-TIE2 mouse IgG1 (R&D Systems) or PE-mouse IgG1 isotype control antibodies (BD Pharmingen), for 30′ on ice at 4°C in the dark, and fixed in 0.6% paraformaldehyde before analysis by flow cytometry (Becton Dickinson). For total (intracellular + cell surface) staining, cells were resuspended in Fixation/Permeabilization solution (eBioscience), incubated for one hour on ice at 4°C in the dark, washed and stained in Permeabilization buffer (eBiosciences) for 30′ on ice at 4°C, with the above antibodies. Cells were washed and resuspended in FACs buffer before flow cytometric analysis.
Immunohistochemistry
Standard protocols were used. Briefly, 5μm sections were deparaffinised using Histosafe (Yvsolab, Beerse, Belgium), rehydrated in propanol, then in water. Endogenous peroxydase was blocked by incubation in 0.3% H2O2 for 30′, and antigens were retrieved using citrate buffer pH 5.7 at 95°C for 95′. Sections were blocked with 10% normal goat serum and 1% BSA, incubated overnight at 4°C with TIE2 antibody (C-20, Santa Cruz at 1:200) or anti-smooth muscle actin (DAKO, 1:200), washed with Tris-HCl 0.05 M, pH 7.4, and visualised with the DAKO EnVision™+/HRP kit following manufacturer recommendations.
Mutation Analysis of the TIE2 Gene in Sporadic VM
The 23 coding-exons of the TIE2 gene, including exon-intron boundaries, were PCR-amplified from genomic DNA. The amplicons were screened by denaturing high-performance liquid chromatography (DHPLC) on the WAVE 3500 HS system (Transgenomic Inc.). For each sample showing an abnormal elution profile, PCR-amplification of that exon was repeated, followed by sequencing on a CEQ2000 fluorescent capillary sequencer (Beckman Coulter) in order to identify the nucleotide change(s). These were confirmed by allele-specific PCR, or by enzymatic digestion on a separately amplified PCR-product (Supplementary Fig. 2). To assess for mutations in exon 17 of the TIE2 in tissue cDNA, direct sequencing of PCR products was carried out for each patient with a TIE2 mutation, using primers located in exons 15 and 18. Where double-mutations occurred, allele-specific PCR using a common forward and specific reverse primer for the more distal mutant residue (915 or 917) was performed on DNA. This product was sequenced in order to assess for whether the mutations occurred in cis (product with mutation at 897) or trans (wild-type at 897; Supplementary Fig. 2). For confirmation, each double-mutant sample (and 2 carrying the L914F mutation), was PCR-amplified for exon 17 on tissue-DNA, using Pfu proofreading polymerase, followed by cloning and sequencing of a minimum of 24 clones each, in order to obtain at least 2 carrying the mutant allele(s).
Semi-quantitative estimation of prevalence of mutant-alleles in DNA and RNA
For a large subset of VMs, the SNaPshot minisequencing assay (Applied Biosystems) was performed, according to manufacturer instructions. Briefly, PCR-amplification of exon 17 was carried out, in duplicate (with Pfu-proofreading, and Taq polymerases) from both tissue DNA and cDNA. Amplicons were cleaned by SAP and ExoI treatment. A Forward or Reverse primer stopping just short of the mutation-site was used in a single-base extension reaction with fluorescently labelled ddNTPs, performed on cleaned amplicons (primer sequences: Supplementary Table 1). These were SAP-treated and run on an ABI3130xl Genetic Analyzer. The peak colors (Supplementary Fig. 3), and the ratio of allele peak-heights obtained (Supplementary Table 1), reflect the composition of the template amplicon, allowing for an estimation of the relative abundance of mutant to wild-type alleles. The absolute peak-heights reflect amplicon-abundance, which do not alter allele ratios. Full-length wild-type or mutant TIE2 plasmid constructs were used as controls; 50:50 mixtures of these provided the heterozygote allele peak-height ratio, to which ratios obtained from tissues were normalized (Supplementary Table 1). The relative levels of pairs of cis-mutations in tissues were compared to each other. Increased presence of one mutation as compared to the other, could be imputed to reflect its precedence: the occurrence of the first in a set of cells is followed by the appearance of the second in a subset of them. SNaPshot data was corroborated by large-scale cloning-and-sequencing of DNA from lesion #28b. We obtained 5 clones with [Y897C+R915C], 6 with R915C alone, and 0 with Y897C alone, out of 80 total. This trend correlates with SNaPshot data (5% for Y897C vs. 11% for R915C), so that by these criteria, R915C may have occurred first.
HUVEC cell culture, TIE2 overexpression, and immunofluorescence
Cell culture and retrovirus based TIE2 overexpression, followed by immunofluorescence staining and confocal microscopy, were performed as previously described in detail25.
Cell statistics
The percentage of cells having phosphorylated TIE2 in the perinuclear area was calculated based on immunofluorescence staining with antibodies detecting phospho-TIE2 (Cell Signaling Technology, Tyr992). The proportion of the cells showing Ang1-induced TIE2 clustering and translocation, was calculated based on the presence of TIE2 clusters, their arrangement in filamentous structures and translocation to retracting cell edges or to cell-cell contacts. At least 320 cells were counted in each group, in three independent experiments run in duplicate.
Supplementary Material
Acknowledgments
We are grateful to all patients for their invaluable contributions. These studies were partially supported by the Interuniversity Attraction Poles initiated by the Belgian Federal Science Policy, network 5/25 and 6/05; Concerted Research Actions (A.R.C.) - Conventions No 02/07/276 and 7/12-005 of the Belgian French Community Ministry; the National Institute of Health, Program Project P01 AR048564-01A1; EU FW6 Integrated project LYMPHANGIOGENOMICS, LSHG-CT-2004-503573; the F.N.R.S. (Fonds national de la recherche scientifique) (to M.V., a “Maître de recherches du F.N.R.S.”); V.W. was supported by a fellowship from F.R.I.A. (Fonds pour la formation à la recherche dans l'industrie et dans l'agriculture), and Patrimoine UCL. L.E. and M.T. were supported by a grant from Academy of Finland (116138). The authors thank Antonella Mendola, Anne Van Egeren, and Jaana Träskelin for their expert technical assistance, Dr. Catherine Godfraind for immunohistochemistry, and Ms Liliana Niculescu for secretarial help.
Footnotes
Accession Numbers
VMCM: MIM 600195; TIE2: MIM 600221
References
- 1.Vikkula M, et al. Vascular dysmorphogenesis caused by an activating mutation in the receptor tyrosine kinase TIE2. Cell. 1996;87:1181–90. doi: 10.1016/s0092-8674(00)81814-0. [DOI] [PubMed] [Google Scholar]
- 2.Calvert JT, et al. Allelic and locus heterogeneity in inherited venous malformations. Hum Mol Genet. 1999;8:1279–89. doi: 10.1093/hmg/8.7.1279. [DOI] [PubMed] [Google Scholar]
- 3.Wouters V, Boon LM, Mulliken JB, Miikka Vikkula. TIE2 and Cutaneomucosal Venous Malformation. In: Epstein C, Erickson RP, Wynshaw-Boris A, editors. Inborn Errors of development. 2nd. Oxford Univeristy Press Inc.; 2007. [Google Scholar]
- 4.Wouters V, Limaye N, Uelbelhoer M, Irrthum A, Boon LM, Mulliken JB, Enjolras O, Baselga E, Berg J, Dompmartin A, Ivarsson SA, Kangesu L, Lacassie Y, Murphy J, Teebi AA, Pennington A, Rieu P, Vikkula M. Hereditary Cutaneomucosal Venous Malformations Are Caused by Variably Activating TIE2 Mutations. submitted.
- 5.Dumont DJ, Yamaguchi TP, Conlon RA, Rossant J, Breitman ML. tek, a novel tyrosine kinase gene located on mouse chromosome 4, is expressed in endothelial cells and their presumptive precursors. Oncogene. 1992;7:1471–80. [PubMed] [Google Scholar]
- 6.Schnurch H, Risau W. Expression of tie-2, a member of a novel family of receptor tyrosine kinases, in the endothelial cell lineage. Development. 1993;119:957–68. doi: 10.1242/dev.119.3.957. [DOI] [PubMed] [Google Scholar]
- 7.Sato TN, Qin Y, Kozak CA, Audus KL. Tie-1 and tie-2 define another class of putative receptor tyrosine kinase genes expressed in early embryonic vascular system. Proc Natl Acad Sci U S A. 1993;90:9355–8. doi: 10.1073/pnas.90.20.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumont DJ, et al. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. Genes Dev. 1994;8:1897–909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 9.Dumont DJ, et al. Vascularization of the mouse embryo: a study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- 10.Sato TN, et al. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–4. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 11.Suri C, et al. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–80. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 12.Partanen J, Dumont DJ. Functions of Tie1 and Tie2 receptor tyrosine kinases in vascular development. Curr Top Microbiol Immunol. 1999;237:159–72. doi: 10.1007/978-3-642-59953-8_8. [DOI] [PubMed] [Google Scholar]
- 13.Jones N, Iljin K, Dumont DJ, Alitalo K. Tie receptors: new modulators of angiogenic and lymphangiogenic responses. Nat Rev Mol Cell Biol. 2001;2:257–67. doi: 10.1038/35067005. [DOI] [PubMed] [Google Scholar]
- 14.Eklund L, Olsen BR. Tie receptors and their angiopoietin ligands are context-dependent regulators of vascular remodeling. Exp Cell Res. 2005 doi: 10.1016/j.yexcr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Maisonpierre PC, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 16.Loughna S, Sato TN. Angiopoietin and Tie signaling pathways in vascular development. Matrix Biol. 2001;20:319–25. doi: 10.1016/s0945-053x(01)00149-4. [DOI] [PubMed] [Google Scholar]
- 17.Nobuhara Y, et al. TIE2 gain-of-function mutation in a patient with pancreatic lymphangioma associated with blue rubber-bleb nevus syndrome: report of a case. Surg Today. 2006;36:283–6. doi: 10.1007/s00595-005-3138-9. [DOI] [PubMed] [Google Scholar]
- 18.Boon LM, et al. Assignment of a locus for dominantly inherited venous malformations to chromosome 9p. Hum Mol Genet. 1994;3:1583–7. doi: 10.1093/hmg/3.9.1583. [DOI] [PubMed] [Google Scholar]
- 19.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 20.Boon LM, Mulliken JB, Enjolras O, Vikkula M. Glomuvenous malformation (glomangioma) and venous malformation: distinct clinicopathologic and genetic entities. Arch Dermatol. 2004;140:971–6. doi: 10.1001/archderm.140.8.971. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, et al. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 2008;27:4336–43. doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- 22.Colmone A, Sipkins DA. Beyond angiogenesis: the role of endothelium in the bone marrow vascular niche. Transl Res. 2008;151:1–9. doi: 10.1016/j.trsl.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Shewchuk LM, et al. Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure. 2000;8:1105–13. doi: 10.1016/s0969-2126(00)00516-5. [DOI] [PubMed] [Google Scholar]
- 24.Fukuhara S, et al. Differential function of Tie2 at cell-cell contacts and cell-substratum contacts regulated by angiopoietin-1. Nat Cell Biol. 2008;10:513–26. doi: 10.1038/ncb1714. [DOI] [PubMed] [Google Scholar]
- 25.Saharinen P, et al. Angiopoietins assemble distinct Tie2 signalling complexes in endothelial cell-cell and cell-matrix contacts. Nat Cell Biol. 2008;10:527–37. doi: 10.1038/ncb1715. [DOI] [PubMed] [Google Scholar]
- 26.Lievens PM, Mutinelli C, Baynes D, Liboi E. The kinase activity of fibroblast growth factor receptor 3 with activation loop mutations affects receptor trafficking and signaling. J Biol Chem. 2004;279:43254–60. doi: 10.1074/jbc.M405247200. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt-Arras DE, et al. Tyrosine phosphorylation regulates maturation of receptor tyrosine kinases. Mol Cell Biol. 2005;25:3690–703. doi: 10.1128/MCB.25.9.3690-3703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabone-Eglinger S, et al. KIT mutations induce intracellular retention and activation of an immature form of the KIT protein in gastrointestinal stromal tumors. Clin Cancer Res. 2008;14:2285–94. doi: 10.1158/1078-0432.CCR-07-4102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.