Abstract
We report the discovery of a new prodrug, 6-chloro-9-nitro-5-oxo-5Hbenzo[a]phenoxazine (CNOB). This prodrug is efficiently activated by ChrR6, the highly active prodrug activating bacterial enzyme we have previously developed. The CNOB/ChrR6 therapy was effective in killing several cancer cell lines in vitro. It also efficiently treated tumors in mice with up to 40% complete remission. 9-amino-6-chloro-5H-benzo[a]phenoxazine-5-one (MCHB) was the only product of CNOB reduction by ChrR6. MCHB binds DNA; at non-lethal concentration, it causes cell accumulation in the S-phase; and at lethal dose it induces cell surface annexinV and caspase 3 and 9 activities. Further, MCHB co-localizes with mitochondria and disrupts their electrochemical potential. Thus, killing by CNOB involves MCHB, which likely induces apoptosis through the mitochondrial pathway. An attractive feature of the CNOB/ChrR6 regime is that its toxic product, MCHB, is fluorescent. This feature proved helpful in in vitro studies, as simple fluorescence measurements provided information on the kinetics of CNOB activation within the cells; MCHB killing mechanism; its generally efficient bystander effect in cells and cell spheroids; and its biodistribution. The emission wavelength of MCHB also permitted its visualization in live animals, allowing noninvasive qualitative imaging of MCHB in mice and the tumor microenvironment. This feature may simplify exploration of barriers to the penetration of MCHB in tumors and their amelioration.
Keywords: Prodrug, CNOB, MCHB, Nitroreductase, ChrR, ChrR6, Cancer, Imaging, GDEPT
Introduction
Prodrugs are non-toxic in their native state but are converted to toxic products by appropriate enzymes [1–7]. Their effectiveness in treating cancer depends on enzymes that are either highly expressed in malignant cells, or on foreign activating enzymes that are targeted to tumors. Mitomycin C (MMC) is an example of the former condition: it is effective because the concentration of the enzyme that activates it, the mammalian NQO1 (also called DT diaphorase), increases in cancer cells [8], making them more vulnerable than normal cells to its action. In contrast, the effectiveness of 5-aziridinyl-2,4-dinitrobenzamide (CB 1954), which is currently in clinical trial for cancer treatment [9], depends on the delivery specifically to tumors of the gene encoding the Escherichia coli nitroreductase enzyme, NTR (encoded by nfsB/nfnB genes) [5]. The latter approach is termed gene-delivered enzyme prodrug therapy (GDEPT); other examples of DEPT therapy are directed delivery of enzymes, catalytic antibodies and antibody subunits [10, 11]
We previously reported on an improved Escherichia coli nitroreductase, ChrR6 (also called Y6) with much greater activity for generating lethal agents from MMC and CB1954 [7]. We show here that ChrR6 is highly effective also in activating a new reductive prodrug that we have discovered, namely, 6-chloro-9-nitro-5-oxo-5H-benzo[a]phenoxazine (CNOB). We were led to a study of this Molecular Probes compound initially because its reduced product, 9-amino-6-chloro-5H-benzo[a]phenoxazine-5-one (MCHB; Fig. S1), is fluorescent at an emission wavelength that can be visualized noninvasively; and because CNOB resembles CB1954 in having a nitro substituted benzene ring. This raised the possibility that we could indirectly visualize CB1954 reach within animals and tumors by the quenching it might produce of MCHB fluorescence. Indeed, we found that CB1954 competitively inhibited CNOB reduction by our nitroreductase enzymes ChrR (also called YieF) and ChrR6 [7]. Subsequent studies, however, showed that CNOB is an effective prodrug in its own right that should permit direct noninvasive visualization of its activation. We show that reduction of CNOB by ChrR6 generates MCHB, which is the agent responsible for cell killing, and present evidence on the probable mechanism of this effect. MCHB fluorescence (excitation 575 nm; emission 625 nm) greatly facilitated examination of the kinetics of CNOB reduction by ChrR6-generating cancer cells and its bystander effect in vitro, as well as qualitative visualization of its generation in living mice. CNOB/ChrR6 proved to be an effective treatment of implanted tumors in mice, especially if the distribution of the enzyme within the tumor is improved. The fact that MCHB can be visualized in living mice is likely to facilitate improving its production specifically in tumors and increasing its reach within them.
Materials and Methods
Bacterial strains, plasmids, and cell lines
Salmonella typhimurium strain SL7838 has been described previously [7, 12]. This strain contains deletions in the aroA and sopE genes, which make it non-virulent and more specific for targeting tumors. The bacterium was transformed by electroporation [13] with expression plasmids (Table S1) encoding the bacterial Lux operon (pCGSL1), GFP (pFVP25.1), and ChrR6 (pET28a+); ChrR6 [7]) is an improved version of the Escherichia coli ChrR wild type nitroreductase with markedly increased capacity for the activation of reductive prodrugs CB 1954 and MMC [7]. Pure ChrR and ChrR6 were generated as described previously [7]. JC (murine mammary cancer) cells were obtained from Cancer Research UK; 4T1 (murine mammary cancer), MCF-7 (human mammary cancer), HeLa (human cervical cancer) HCT 116 (human colorectal cancer), and 293T human embryonic kidney cancer cells were obtained from ATCC (Manassas, VA). Cells were grown as adherent cultures in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin (0.5 U and 0.5 µg/ml, respectively; Invitrogen, Carlsbad, CA). CNOB/ChrR6 regime was equally effective against these cell lines; the studies presented here focus on three of them: JC (in vitro and in vivo killing and visualization), 4T1 (in vivo killing, visualization, and MCHB killing mechanism); and HCT 116 (MCHB killing mechanism). Results presented are representative of different cell lines.
In vitro viability assays
The effect of converted prodrug on cell lines was measured as described before [7]. Prodrug reduction mixtures containing CNOB (Molecular Probes-Invitrogen, Carlsbad, CA) and pure ChrR6, 1 mM NADPH, and DMEM added to a final volume of 0.5 ml were allowed to carry out drug reduction at 37°C for 30 minutes before addition to cancer cells. The extent of drug activation was inferred from the loss of cell viability after 30 minutes. Cell viability was determined by MTS assay according to the manufacturer’s instructions (CellTiter 96®AqueousOne, Promega, Madison, WI). A490 was measured in an ASYS UVM 340 plate reader (Asys Hitech, Cambridge, UK). In control experiments, CNOB was added to cells without ChrR6 and viability was determined by the MTS assay as before.
ChrR6 was also delivered to cancer cells, as specified from a GDEPT system involving Salmonella typhimurium strain SL7838-chrR6, with strain SL7838 being used as control. The strains also expressed GFP which permitted visualization of cell infection. Different multiplicities of infection [MOI; bacterial colony forming units (CFU)/cancer cell] were added to cells in black walled 96 well plates along with 15 µM CNOB. After one hour incubation, fluorescence (GFP: excitation 488 nm, emission 525 nm; MCHB: excitation 575 nm, emission 625 nm) was measured using a plate reader with appropriate filters (SpectraMax, Molecular Devices/MDA Analytic Technologies, Sunnyvale, CA). Cell viability was determined as described above.
Kinetic constants and killing mechanism
To determine enzyme kinetics, 100 µl of 0.1 M Tris-HCl buffer (50 mM; pH 7), 20 µg ml−1 pure ChrR6 and 100 µM NADPH were mixed. The concentration of CNOB was varied from 0.5–2.5 µM and the progress of the reaction was followed by measuring fluorescence at intervals of 1 minute for 10 minutes. Kinetic parameters were estimated using Excel plots of linear regression of reciprocals of Vmax and Km. Standard curves relating fluorescence and MCHB (ChemBridge, San Diego, CA) concentrations were generated and used to determine its levels. CNOB reduction product was determined by high performance liquid chromatography (HPLC), using the following conditions: flow rate, 3 ml/min; detection wavelength, 280 nm (for CNOB), 500 nm (for MCHB); mobile phase, RP C18, 5 µm; column-packing, Phenomenex Luna 5μ C18(2); dimensions: 250×10 mm; injection volumn: 100 µL. Gradients of acetonitrile, containing 0.1% TFA (“A”) and of water containing 0.1%TFA (‘B’) used were as follows: 0, 3, 28, and 30 min, A: 10% B: 90%; 25 and 28 min, A: 100% , B:0%.
The redox balance method to determine the proportion of electrons utilized in CNOB reduction and reactive oxygen species (ROS) generation was determined by quantifying the NADPH consumed and H2O2 produced during ChrR6-catalysed CNOB reduction. The former was done spectrophotometrically; the latter fluorometrically, using the Amplex Red kit (Molecular Probes/Invitrogen), as before [14, 15].
For DNA binding assays, 1 µg of pUC19 plasmid DNA was mixed with 1 µM MCHB, or Tris buffer (control) for 10 minutes. The DNA was then purified by phenol:chloroform extraction, ethanol precipitated and re-dissolved in Tris-EDTA buffer. The differently treated DNA samples were run on 1% agaraose in parallel; bands were then cutout from the gel and MCHB fluorescence was read in a plate reader (SpectraMax, Molecular Devices).
To conduct cell cycle assays, non-confluent cells were treated with 0.1 µM MCHB for 24 hours before collection and staining with 7-amino-actinomycin D (7-AAD). Flow cytometry was used to remove doublets and gates representing G1, S and G2/M phase cells were set. Annexin-PE staining was used to examine apoptosis 3 hours after MCHB (1 µM) addition to cells, using flow cytometry.
For assaying caspase activity, MCHB was added to cancer cells (1 µM) for 1h, and caspase activities assayed using Apoalert Caspase assay (Clontech, Mountain View, CA). MCHB-mitochondrial co-localization was determined using MitoTracker™ green FM dye (Molecular Probes). Cells were incubated for 1 hour with 1 µM MCHB; 200 nM of MitoTracker™ green FM was then added, followed by an additional 1 hour incubation. Confocal microscopy (TCS SP2 Leica Microsystems) was used to visualize the co-localization of MCHB and the mitochondria. Mitochondrial membrane potential was assayed using the JC Mitochondrial Assay (Molecular Probes) 3 hours after addition of MCHB (1 µM).
In vitro visualization of CNOB reduction in live cell
Cells were grown as adherent cultures and mixed with SL7838 expressing both GFP and ChrR6 at specified MOI values (Results). One hour later, the cells were washed to remove extracellular bacteria and CNOB was added (15 µM) along with gentamycin (20 µg ml−1) to suppress bacterial growth. The conversion of CNOB to fluorescent product (MCHB) and location of the GFP-expressing bacteria were followed using a confocal microscope (TCS SP2, Leica). Spheroids were formed by growing JC cells on non-tissue culture treated plates, transferred to chamber well slides (Lab-Tek, Nunc/Thermo Fisher, Rochester, NY), and treated with bacteria, CNOB and gentamycin as before. Prodrug conversion and bacterial GFP were imaged using a 2-photon microscope (Carl Zeiss Inc., Thornwood, NY).
In vivo qualitative visualization of MCHB and efficacy of CNOB/ChrR6 tumor treatment
Immunocompetent BALB/c mice were subcutaneously implanted with 4T1 cells (1×105) expressing luciferase. Tumors were allowed to form for 14 days, at which point they were approximately 100 mm3 in size. Animals were then intratumorally injected with 1×105 CFU of SL7838 expressing the Lux operon and ChrR6. 1, 3 and 5 days later, they were injected intravenously with 0.1 mg of CNOB (in 100 µl; 3.3 mg/kg) each day (ca. 10 mg total CNOB/kg), or PBS (n = 8 mice/group). (CNOB was initially dissolved in DMSO and then diluted 1:10 in PBS immediately prior to injection.) MCHB production was qualitatively imaged in living animals using a Maestro system (CRI Inc., Woburn, MA) with dsRed filter sets and spectral unmixing. Imaging of bacterial Lux activity was performed using an IVIS100 system (Xenogen/Caliper, Alameda, CA); imaging of firefly luciferase (Luc)-expressing 4T1 cells was performed 5 minutes after intraperitoneal injection of luciferin (150 µl of 30 mg ml−1). The signal generated after luciferin addition (Luc expression) includes the signal due to the bacterial Lux expression; however, since the former was more than 50 fold greater, the latter was negligible.
Generating 4T1 cells transfected to express ChrR6
In one experimental regime the efficacy of CNOB/ChrR6 treatment was determined by initiating tumors with bioluminescent 4T1 cells with constitutive capacity to produce ChrR6. These cells were generated by transposon-mediated gene transfer of human codon optimized chrR6 gene (encoding “HChrR6” enzyme; GeneScript Corp. NJ USA); and firefly luciferase (luc) in the transposon vector pKT2/BsdR=EGFP-fLuc, (pKT2/BGL for short) [16]. Briefly HchrR6 gene was cloned into the Sleeping Beauty transposon, pKT2/UXbG (Table S1), using HindIII/ApaI restriction sites creating "pKT2/hU-HchrR6-SN". Cells were grown to 90–95% confluency in DMEM without antibiotics in a 6 well plate. 0.8 µg transposase vector (pUb-SB11) and 7.2 µg transposon DNA (pKT2/hU-HchrR-SN and pKT2/BGL) were added to 0.5 ml OptiMem (Invitrogen). In a second vial 20 µl of Lipofectamine 2000 (Invitrogen) was added to 0.5 ml of OptiMem, and incubated at room temperature for 5 minutes.
The medium was aspirated and cells were washed once with PBS. The above solutions were combined (total 1 ml/well), added to each well and incubated for 18–24 hours. The transfection solution was then aspirated and replaced by regular complete DMEM. Transfection efficiency was monitored by adding 2–5 µl of luciferin (30 mg/ml) per well and imaged immediately using the IVIS50 system; while the untranfected cells expressed no Luc luminescence, the transfected ones showed high expression. Cells were incubated for an additional 48 hours, selected with blasticydin and geneticin (Invitrogen) (5 and 2 µg/ml, respectively; these concentrations were predetermined as the minimal killing dose for 4T1 cells). To ensure homogeneity of HchrR6 expression, cells expressing luciferase were diluted to about 30 cells per 10 ml DMEM, supplemented with the selection antibiotics and 100 µl aliquots were dispensed into a 96 well plate. This dilution generates a ~30% probability of a well receiving a cell, ensuring that colonies developing in a well originated from a single cell. Tumors from these cells were generated as before.
Tumor burden was measured by caliper measurement at specified times after CNOB treatment. Whole blood counts, and the chemistry panel measurements were performed by the Stanford Veterinary Service Center. All animal studies were performed according to approved institutional IACUC and biosafety committee protocols.
Statistical Analyses
Student’s T test was performed for all statistical analyses, except for the Kaplan-Meier survival curves, where Logrank tests were used. p-values are indicated (significance was assigned at p-values less than 0.05).
Results
CNOB reduction kinetics and killing mechanism
As for MMC and CB 1954 [7, 17], pure ChrR6 showed improved kinetics for CNOB reduction with an almost 20- and 10-fold increased Vmax and Kcat/Km, respectively, over the parent ChrR enzyme (Table S2), and was much more active in CNOB-mediated killing of cancer cells than ChrR (not shown). CNOB/ChrR6 treatment was as effective in killing cancer cells as treatment with CB1954/ChrR6: exposure to either regime (drug concentration, 15 µM) for one hour generated >80% killing of murine (4T1, JC) and human (MCF-7) breast cancer as well as those of human colorectal (HCT 116) kidney (293T), cervical (HeLa), and other cancer cells (listed in Materials and Methods). Control experiments with CNOB alone were conducted as described in Materials and Methods; these generated no cell killing. (All experiments were conducted in triplicate; p<0.05.)
HPLC was used to characterize the products of CNOB reduction by ChrR6. The only compound besides CNOB found in a reaction mixture containing pure ChrR6, NADPH and CNOB was MCHB (not shown). Generation of MCHB from CNOB requires a two-electron reduction, but the nitro group present in CNOB has a predilection for accepting one-electron, which would result in the formation of CNOB nitro-radical anions (Fig S1). These are short lived and cannot be detected by HPLC analysis. As these anions have a redox potential lower than the oxygen couple, they redox cycle. In this process the one-electron-reduced CNOB anion would be rapidly oxidized back to CNOB, giving its electron to dioxygen, resulting in ROS generation. Due to its repetitive nature, this process would produce large quantities of ROS. To detect redox cycling occurs during CNOB reduction by ChrR6, we used our redox balance method [14, 15]. This entailed quantification of the reductant (NADPH) electrons partitioned between MCHB and ROS generation. Little ROS production was found with >80% of NADPH electrons being utilized in CNOB reduction. Thus, there was minimal or no single-electron reduction and redox cycling of the prodrug. (Note that some ROS can result from disproportionation of MCHB [18]). The results strongly suggest that MCHB is the sole product of CNOB reduction by ChrR6.
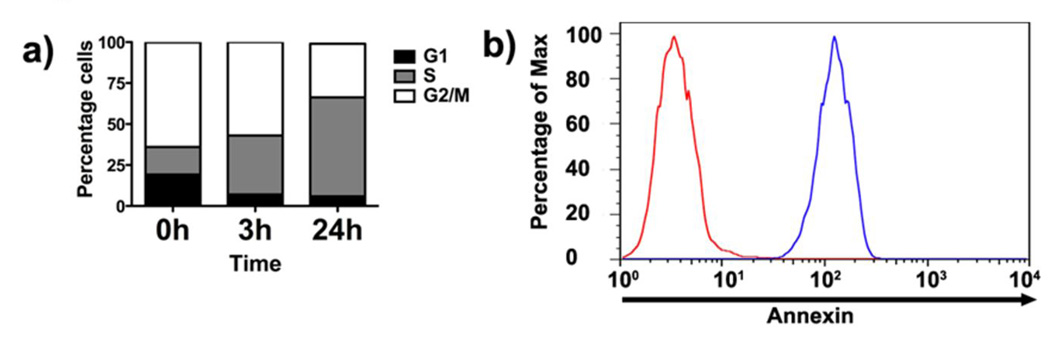
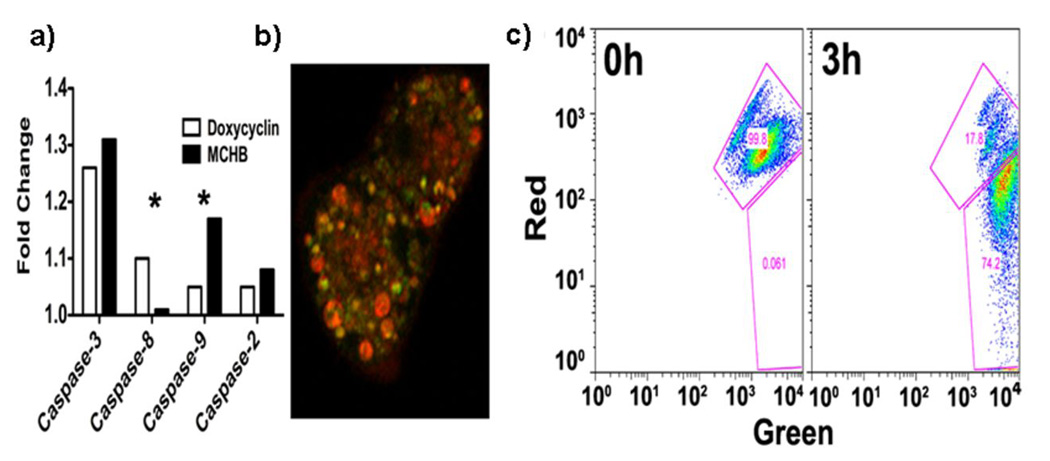
What might be the mechanism of MCHB killing of cancer cells? In addressing this question, we were guided by the example of CB 1954, because CNOB and CB 1954 are both nitroprodrugs and, as stated above, kill mammalian cells with equal efficiency when activated by ChrR6. The reduction product of CB 1954 kills cells through DNA intercalation [19]. The CNOB reduction product, MCHB, did indeed bind DNA (Fig S2) and, as is typical of DNA intercalating agents, caused HCT 116 cell accumulation in the S-phase at non-lethal concentration (0.1 µM (Fig 1a). At lethal dose (1.0 µM), MCHB was apoptotic as indicated by rapid increases in cell-surface annexinV (Fig 1b) and caspase-3 (Fig 2a) activity. Microscopic evidence pointed to focal localization of MCHB mainly within the cytoplasm (Fig 3c and S3), raising the possibility that it was taken up by mitochondria and bound to mitochondrial DNA. When the latter were stained with MitoTracker™ green FM, co-localization of MCHB and mitochondria was seen (Fig 2b). MCHB also caused rapid disruption of mitochondrial membrane potential, as indicated by cell population shift toward the “green” axis (Fig 2c). Furthermore, MCHB increased caspase-9 activity (Fig 2a), indicative of apoptosis initiated through the intrinsic mitochondrial pathway and cytochrome c release. The results suggest that MCHB kills the cells by intercalating with mitochondrial DNA and causing apoptosis involving the mitochondrial pathway. Treatment with doxycycline served as control for this experiment. This drug induces apoptosis through caspase-3/8 activation; the expected results were seen (Fig. 2a). The reason for the apparent MCHB preference for mitochondria remains to be determined.
Figure 1.
Effect of MCHB on cell cycle and AnnexinV. a) Treatment with MCHB (0.1µM) results in increasing accumulation of cells in the S-phase with time. HCT 116 cells were stained with 7-AAD for cell cycle analysis by flow cytometry. b) Induction of AnnexinV as determined by AnnexinV-PE staining of the cells: red graph, untreated; blue graph, MCHB (1.0 µM; 1h)-treated cells.
Figure 2.
Effect of MCHB on caspases, and mitochondria. a) Induction of caspase activities upon treatment of HCT 116 with 1 µM MCHB, relative to untreated cells or doxycycline (known to induce apoptosis through activation of caspase-8 and caspase-3) (*p<0.05); (b) Co-localization of MCHB with mitochondria of HCT 116 (stained with MitoTracker™ green FM) as visualized by confocal microscopy (60× magnification). c) MCHB effect on mitochondrial membrane potential: JC-1 staining of MCHB treated HCT 116 cells Increase in green staining (X-axis) and decrease in red staining (Y-axis) indicate mitochondrial depolarization.
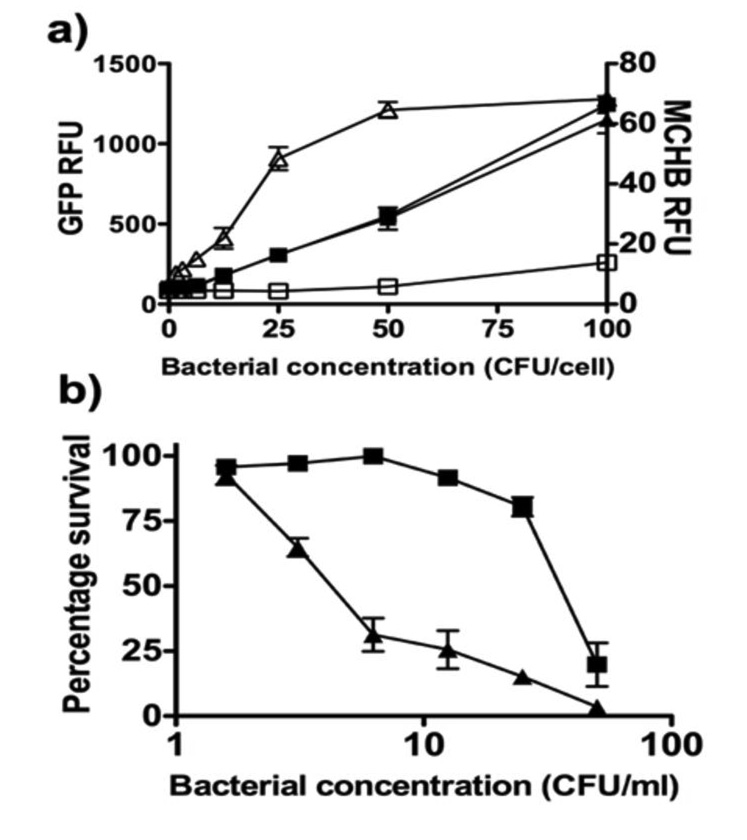
Figure 3.
Correlation between MCHB production from CNOB and JC cell killing. The in vitro GDEPT system involving different doses of GFP-expressing SL7838-chrR6, or SL7838 (control) bacteria was used to infect the JC cells exposed to CNOB. Relative fluorescence units (RFU) of GFP (indicative of the extent of cell infection by the bacteria) and MCHB (indicative of the extent of CNOB reduction) (panel a) and cell killing (panel b) were measured as described in Materials and Methods. Symbols: Panel a): ▲ and ■, JC cells infection indicated by GFP levels by SL7838-chrR6 and SL7838, respectively – note that cancer cells were infected by both strains to the same extent; △ and □ MCHB generation by cells infected with SL7838-chrR6 and SL7838, respectively. Panel b) ▲ and ■, killing of SL7838-chrR6- and SL7838-infected cells, respectively. Note that despite equivalent degree of infection by the two bacterial strains, SL7838-chrR6 infected cells exhibit markedly more MCHB generation and cell killing.
Relationship between reduced CNOB fluorescence and cell killing in vitro
We first determined correlation between MCHB fluorescence and its killing capacity. In the range of 100 to 1000 nM (which is within linear relationship between MCHB concentration and its fluorescence), MCHB fluorescence intensity was directly proportional to its cell killing activity. Thus, 100, 700 and 1000 nM MCHB caused 3, 38, and 65% cell killing, respectively, of JC, MCF-7 and 4T1 cell after 24 hour incubation. Correlation between fluorescence and cell killing was seen also when the SL7838-chrR6 GDEPT system was used to generate MCHB from CNOB in attached JC cells. Increasing doses of GFP-producing SL7838-chrR6 bacteria resulted in a progressive rise in MCHB generation, as detected by GFP and MCHB fluorescence intensities, respectively (Fig. 3a). The increase in MCHB fluorescence generated increasing cell killing (Fig. 3a&b). In contrast, equivalent levels of infection with SL7838 (not encoding ChrR6) used as control, produced little MCHB and were much less lethal: over 10 fold fewer SL7838-chrR6 bacteria were required to kill 50% of the cell layer compared to SL7838 not expressing this enzyme (Fig. 3a&b). SL7838 by itself also generated cancer cell lethality, especially at high doses (Fig. 3b), which is consistent with previous findings [12, 20].
Visualization of CNOB reduction kinetics and bystander effect in vitro
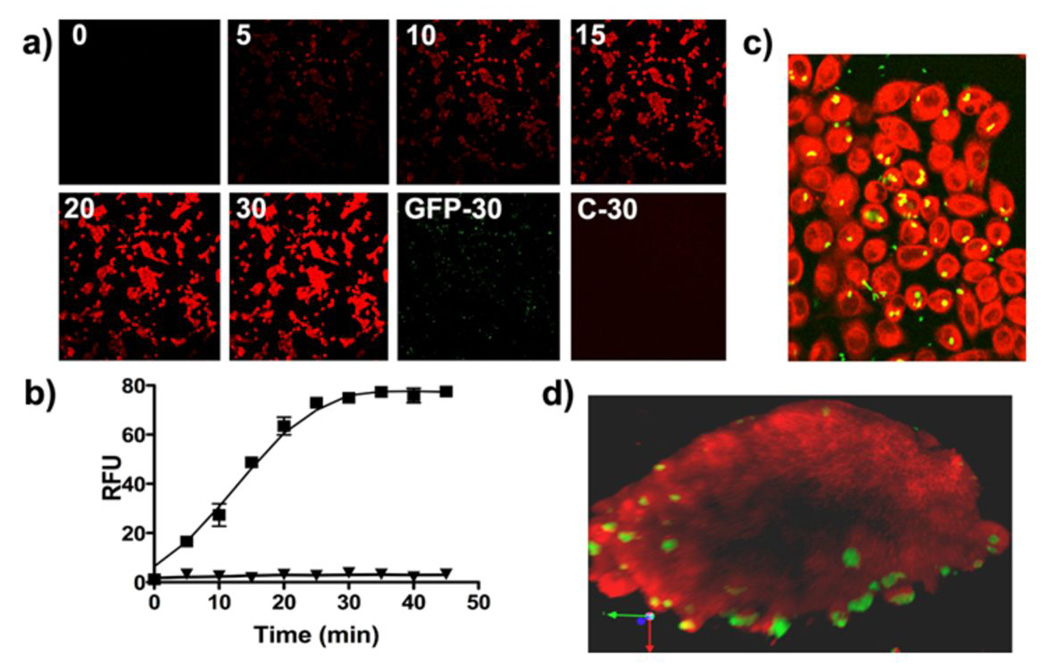
The fluorescence of MCHB afforded a facile means to examine in vitro kinetics of CNOB reduction and the bystander effect of the CNOB/ChrR6 regime. Bystander effect refers to the spread of the activated drug from cells capable of producing it to the neighboring cells lacking this capacity; it is critical to the efficacy of any DEPT therapy because no DEPT approach can transform all the cancer cells in a tumor. The MCHB fluorescent signal in attached JC cells infected with GFP-expressing SL7838-chrR6 bacteria was detectable as early as 10 minutes after CNOB (15 µM) addition (Fig. 4a), and increased in intensity with time, reaching a steady state in some 20 – 30 minutes. Quantification of CNOB conversion to MCHB in a fluorescence plate reader in an equivalent experiment gave similar results (Fig. 4b). Note that when uninfected cancer cells were treated with CNOB (Fig. 4a, Panel ‘C-30’), or when infected cells were not exposed to CNOB (Fig. 4b), no fluorescence was generated. Panel ‘GFP-30’ shows GFP fluorescence of bacteria and their location in a parallel monolayer that did not receive CNOB. Thus, the indigenous nitroreductases of these cells did not activate CNOB, and the red fluorescence seen in Fig. 4 was due to MCHB.
Figure 4.
In vitro imaging of MCHB within JC cancer cells. (a) Kinetics of CNOB reduction as visualized by the generation of MCHB fluorescence in JC cells infected with GFP-expressing SL7838-chrR6 at MOI of 10/cell. Cells were imaged by confocal microscopy at the indicated times (minutes) after CNOB addition (4×magnification). See Materials and Methods for details. The ‘GFP-30’ panel shows GFP fluorescence from bacterial cells at 30 minutes from monolayers to which no CNOB was added. Panel ‘C-30’ represents a control plate of cells treated with CNOB, but not SL7838-chrR6, imaged 30 minutes after the addition of CNOB. (b) An equivalent experiment was run and CNOB conversion to MCHB was quantified at the indicated times following CNOB addition in a fluorescence plate reader for cells with or without CNOB addition (■ and ▲, respectively). (c) The experiment of Fig. 3a was repeated using an MOI of 1.0 CFU/cell, resulting in approximately 20% of cells becoming infected. The representative image shown was taken 30 minutes after CNOB addition (20×magnification). (d) JC cells were used to form multicellular tumor spheroids. SL7838-chrR6 expressing GFP was added at an MOI of approximately 1000 CFU/spheroid and incubated for 30 minutes to permit infection. Images were taken by 2-photon microscope 1 hour after CNOB addition. Z-stack images were reconstructed into a three-dimensional model using Velocity software and snapshots were taken from different angles. Note that the image represents a cross section through the center of the spheroid and 95 µm depth.
To examine the bystander effect, we repeated the above experiment at an MOI of 1 CFU/cell, resulting in approximately 20% of cancer cells becoming infected. It is evident (Fig. 4c) that by 30 minutes after CNOB addition, the red signal of MCHB is present not only in cells infected with the bacteria (green and yellow spots; Fig. 4c), but also in the uninfected ones. We also examined this effect in three dimensional tissue culture cell spheroids, since the latter are more representative of conditions within solid tumors than cell monolayers [21]. There was extensive MCHB presence in the spheroids including in areas away from the bacteria (green/yellow), as visualized by 2-photon microscopy (Fig 4d). However, zones are seen within them where the red fluorescence is absent (Fig 4d and S4). This may be because of penetration barriers to CNOB and/or MCHB within the spheroids.
Visualization of CNOB reduction in vivo
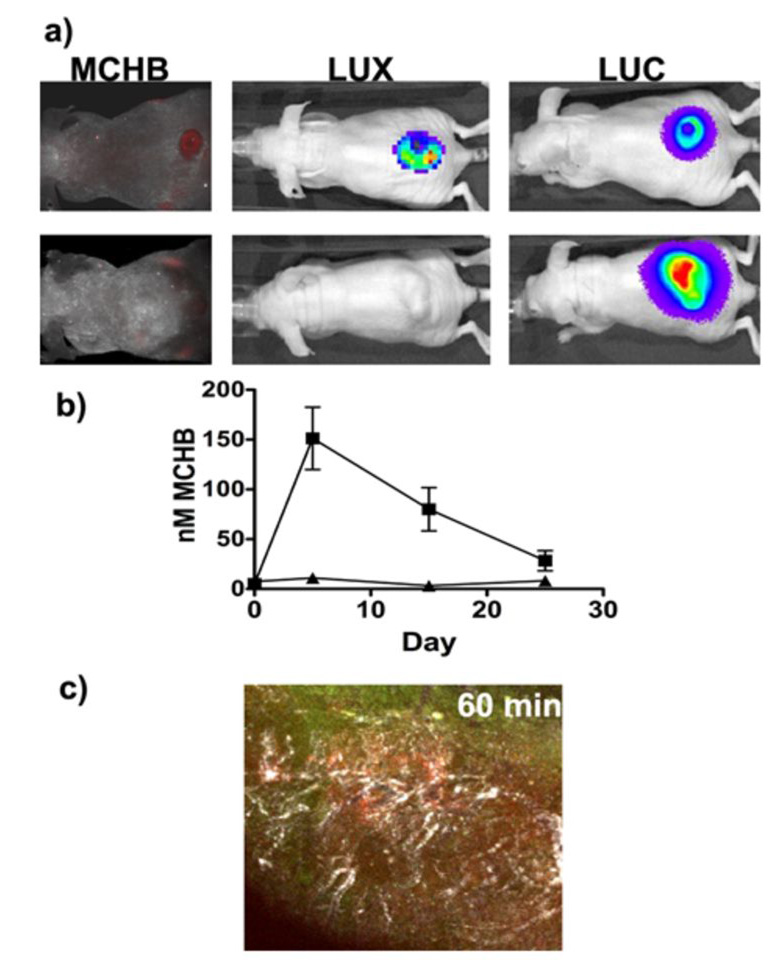
The emission wavelength of MCHB (625 nm) is such that it raises the possibility of its visualization by noninvasive methods in live animals. In order to qualitatively explore this possibility, we established tumors (expressing Luc) in mice followed by intratumoral administration of Lux-expressing SL7838-chrR6, and intravenous injection of CNOB (3.3 mg/kg) 24 hours later. Noninvasive imaging (see Materials and Methods) of living animals revealed the presence of the bacteria (blue, Lux luminescence) and MCHB fluorescence (red), in the infected tumors only (blue; Lux and Luc luminescence) (Fig. 5). Control tumors not infected with the bacteria (Luc luminescence only) do not show the MCHB fluorescence; some fluorescence in one of the kidneys of these mice can however be detected.
Figure 5.
a) Visualization of MCHB generation (red fluorescence) in JC implanted tumors in living mice by noninvasive imaging at 6 hours after CNOB administration. Lux-expressing SL7838-chrR6 bacteria were injected intratumorally, and CNOB (upper panel mouse only) intravenously 24 hours later. Luc and Lux luminescence represent the location of tumors and the bacteria, respectively. (The Luc signal includes Lux, but since the former was more than 50 fold greater, the latter is negligible; see Materials and Methods.) Note the absence of red fluorescence in tumors of mice not injected with bacteria but administered CNOB (lower panels). b) MCHB levels (quantified from fluorescence measurements) in blood samples collected by bleeding mice at indicated time points: ■) CNOB/SL7838-chrR6 treated mice; △) CNOB only treated mice. c) Qualitative visualization of MCHB generation in tumor in a living mouse (intravital microscopy) in relation to vasculature (white) and the ChrR6-generating bacteria (green). CNOB and SL7838-chrR6 were administered as above.
We found that CNOB/SL7838-chrR6 treated mice displayed easily detectable levels of MCHB (as measured by its fluorescence) in their blood in contrast to the control animals injected with CNOB alone (Fig. 5b). To determine if the MCHB in the blood caused hematopoietic toxicity, whole blood counts were performed. Apart from the expected increase in white blood cells due to bacterial infection, no adverse effects were seen (not shown). This raises the possibility that MCHB secreted into the blood might prove useful in assessing the efficacy of CNOB delivery and activation in vivo.
Solid tumors constitute a complex microenvironment [21], and we wondered if MCHB fluorescence would permit the visualization of its distribution within this environment. This was attempted, again only qualitatively, in SL7838-chrR6 and CNOB-inoculated tumors (intratumorally and intravenously, respectively, as above) in living mice using intravital microscopy. Angiosense 680 was also injected intravenously to visualize the vasculature. The CNOB signal was first seen at 10 minutes post-inoculation (not shown). At 60 minutes (Fig. 5c), the red fluorescence was quite marked and was seen primarily in zones of maximal vascularization (white) rather than in association with the ChrR6 producing bacteria (green).
Effect of CNOB/ChrR6 treatment on mice tumors
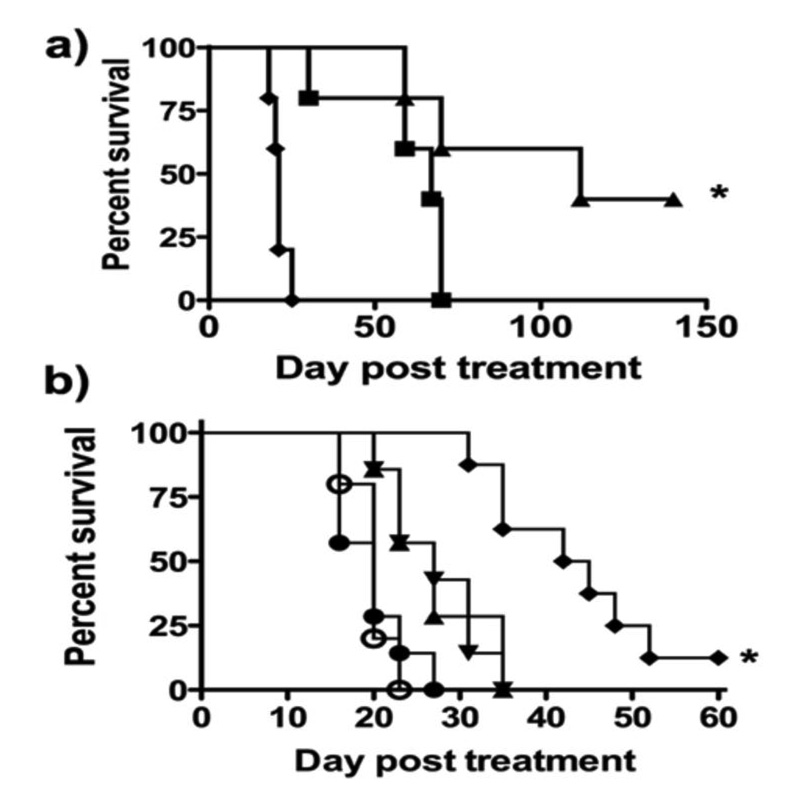
For in vivo studies of CNOB/ChrR6 therapy, 4T1 cells were used to initiate the tumors. These form especially aggressive tumors, so the efficacy of the treatment could be tested in an extreme situation. (Note that, as stated in Materials and Methods, in vitro 4T1 cells respond to CNOB/ChrR6 treatment in the same way as the JC cells; Fig. 4.) To ensure uniform ChrR6 generation within the tumors in these exploratory in vivo studies of the efficacy of the therapy, we made use of 4T1 cells transfected to constitutively express humanized ChrR6 (HChrR6) to initiate subcutaneous tumors in immunocompetent BALB/c mice. Two CNOB concentrations, 3.3 (single dose) or 10 mg/kg (spaced in three doses – see Materials and Methods), were used and administered intravenously. While the control mice injected with PBS were all dead within 25 days, at 3.3 mg/kg, 60% mice were still alive by day 60 post-CNOB injection (Fig. 6a); and at 10 mg/kg, there was 40% complete remission up to 150 days (p=0.0012).
Figure 6.
In vivo Efficacy of CNOB/ChrR6 treatment. Survival (Kaplan-Meier) curves of: a) Mice implanted with 4T1 tumors (50–100 mm3) endogenously expressing humanized ChrR6 (HChrR6) and treated with PBS (◆); 3.3 mg/kg CNOB (■); or 10 mg/kg CNOB (▲) (n=5 animals/group) (*p= 0.0018). b) Mice implanted with non-transfected 4T1 tumors (50–100 mm3) not expressing ChrR6 and treated with PBS (○); CNOB alone (●); SL7838-ChrR6 alone (▲); SL7838 and CNOB (▼); or SL7838-ChrR6 and CNOB 10 mg/kg (♦) (n=8 animals/group). (SL7838 and CNOB compared to SL7838-ChrR6 and CNOB, p=0.0012.) Bacteria (1×105 CFU) were administered via intratumoral injection on day 0; CNOB (3.3mg/kg each time) was given on days 1, 3 and 5 via intravenous injection. (* p<0.05).
In order to test the efficacy of the treatment in a clinically more relevant setting, we iduced tumors in mice using untransfected 4T1 cells and delivered the enzyme using SL7838-chrR6 intratumoral GDEPT system; CNOB (10 mg/kg) was injected intravenously. As expected from previous findings [7, 12, 20] and the data of Fig. 3b, the bacteria alone increased mice survival somewhat, but this was enhanced in mice administered CNOB/SL7838-chrR6 (Fig. 6b; p=0.0012): animals receiving the bacteria alone were all dead by day 35, but in the regime involving CNOB administration, 10% were still alive on day 60. It is clear, however, that the treatment in this setting is less effective than that of Fig. 6a suggesting, in agreement with the data of Fig. 5c, that in this DEPT system, even at the higher CNOB concentration, enzyme and/or the drug/MCHB fail to effectively disseminate in the tumor.
Blood chemistry panel values showed no major toxicity up to a CNOB concentration of 20 mg/kg (Table S3). Some slight muscular damage may be indicated by marginal elevation of markers such as creatine phosphokinase; and that of kidneys by those of blood urea nitrogen, calcium and phosphate. The latter would seem to be consistent with the slight activation of the drug in kidneys of mice not administered ChrR6-encoding bacteria (Fig. 5a; lower panel). However, since the effects are minor, CNOB would appear to be largely non-toxic.
MCHB fluorescence and determination of its biodistribution
Can MCHB biodistribution be determined by simple fluorescence measurements? To address this question we repeated the experiment of Fig 5, except that SL7838-chrR6 bacteria were administered intravenously. Imaging showed that, as expected (12), this resulted in bacterial colonization of organs besides the tumor. The red fluorescence was seen not only in the tumor but also in other organs (not shown). Post mortem determination of MCHB fluorescence intensity in different organs and use of a standard curve made it possible to measure the quantity of MCHB (Fig. S5); most was still generated within the tumor (for which SL7838 has a predeliction), but it was also seen in other organs.
Discussion
We show that the new CNOB/ChrR6 therapy is highly effective in killing different cancer cells lines in vitro. To determine if the therapy is useful in treating cancer in a mouse model, we first implanted mice with subcutaneous tumors that endogenously (and constitutively) generated a humanized form of ChrR6 (HChrR6) in order to ensure uniform generation of the enzyme within the tumor. Intravenously administered CNOB in this situation generated beneficial results, especially at 10 mg/kg dose, which resulted in some 40% mice showing complete remission. (At this concentration the expected peak tissue drug concentration is approximating 14 µM.) We then used a clinically more relevant setting in which mice were implanted with non-transfected 4T1 tumors not generating their own ChrR6, and delivered the enzyme by an intratumorally administered SL7838-chrR6 GDEPT system. Again beneficial results were seen, although they were less impressive. Thus, while on day 60 at 10 mg/kg CNOB, only 10% mice survived in the GPEPT regime, 60% survived in that involving HChrR6, indicating impediments to the reach of one or more components of the therapy within the tumors in the former regime. That there may be barriers to the penetration of MCHB is suggested also cell spheroid (Fig. 4d) and tumor microenvironment imaging (Fig. 5c) results.
This new therapy has certain attractive features. Like CB1954, CNOB most likely kills by intercalating DNA. Thus, like the former, it will target both growing and non-growing cells, which is advantageous as most cells in solid tumors remain quiescent. Also, like CB1954, it was not activated by indigenous nitroreductases of several cancer cell. This appears to be the case also in live mice, as indicated by the absence, by and large, of MCHB generation, and significant toxicity in mice administered CNOB without ChrR6 (Fig. 5a and 5b, Table S3). CNOB like CB 1954 thus holds the promise of affording a generally side effect-free cancer therapy upon successful specific GDEPT targeting of ChrR6 to tumors. However, the most attractive feature of CNOB/ChrR6 therapy may turn out to be the fact that its killing agent, MCHB unlike that of CB1954 [m5-(aziridin-1-yl)-4 hydroxylamino-2-nitrobenzamide], is fluorescent, and its noninvasive visualization in live animals can simplify studies aimed at improving the efficacy of this therapy.
MCHB fluorescence was helpful in in vitro as well as in vivo studies. The kinetics of CNOB activation within the cells, the MCHB killing mechanism, and its bystander effect including the possibility of barriers to its penetration within the cell spheroids, and its biodistribution were determined by simple fluorescence measurements in vitro. Similarly, even though only qualitative, MCHB visualization in living mice by non-invasive imaging afforded a facile means of determining the locus of CNOB reduction, including within the tumor microenvironment. This feature is likely to prove valuable in more detailed studies on MCHB generation within solid tumors since, unlike most other drugs, this might be determined in real time without the need of sacrificing the experimental animals.
Real time imaging of MCHB within the implanted tumors, along with secondary labels such as those against necrotic and hypoxic regions, the vasculature and the tumor matrix material, can reveal the nature of the impediments to activated drug penetration of tumors. Countermeasures might then be investigated, and since their effects can be visualized noninvasively, several treatments may be tested in the same animal model. We are currently studying these aspects as well as methods for quantitative imaging of MCHB in vivo, detailed pharmacokinetics of the CNOB/ChrR6 therapy, further improvement in CNOB activating enzyme [17, 22], and appropriate GDEPT methods [11, 23– 24] to improve this treatment regime and determine its suitability of for translation to the clinic.
Supplementary Material
Acknowledgments
We thank Dr. B. Efron for help with statistical analysis, Dr. David Chu for help with blood chemistry panel analysis, and Dr. Michael Benoit for help with the preparation of this manuscript.
Financial support: This work was supported by grants DE-FG03-97ER-62490, DE-FG03-96ER20228 from the US Department of Energy, 1 RO1 CA125074-01A1 from National Cancer Institute and 1105626 from the Stanford Office of Technology Licensing (to AM); the National Cancer Institute In vivo Molecular Imaging Center at Stanford grant (P50 CA114747) (to SHT, CHC); and the John A and Cynthia Fry Gunn Research Fund (to WL). YB was supported in part by a Lady Davis postdoctoral fellowship.
Abbreviations (arranged alphabetically)
- 7-AAD
7-amino-actinomycin D
- A/G ratio
albumin/globulin ratio
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BUN
blood urea nitrogen
- CB 1954
5-aziridinyl-2,4-dinitrobenzamide
- CNOB
6-chloro-9-nitro-5-oxo-5H-benzo[a]phenoxazine
- CPK
creatine phosphokinase
- GDEPT
gene-delivered enzyme prodrug therapy
- GGT
γ-glutaminyl aminotransferase
- HChrR6
humanized ChrR6
- HPLC
high performance liquid chromatography
- Luc
firefly luciferase
- MCHB
9-amino-6-chloro-5Hbenzo[a]phenoxazine-5-one
- MMC
mitomycin C
- MOI
multiplicity of infection
References
- 1.Patterson AV, Saunders MP, Greco O. Prodrugs in genetic chemoradiotherapy. Curr Pharm Des. 2003;9:2131–2154. doi: 10.2174/1381612033454117. [DOI] [PubMed] [Google Scholar]
- 2.Dachs GU, Tupper J, Tozer GM. From bench to bedside for gene-directed enzyme prodrug therapy of cancer. Anti-cancer drugs. 2005;16:349–359. doi: 10.1097/00001813-200504000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Denny WA. Nitroreductase-based GDEPT. Curr Pharm Des. 2005:1349–1361. doi: 10.2174/1381612023394584. [DOI] [PubMed] [Google Scholar]
- 4.Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 5.Knox RJ, Chen S. Quinone reductase-mediated nitro-reduction: clinical applications. In: Sies H, Packer L, editors. Methods in Enzymology. Vol. 382. Amsterdam: Elsevier; 2004. pp. 194–221. [DOI] [PubMed] [Google Scholar]
- 6.Rooseboom M, Commandeur JN, Vermeulen NP. Enzyme-catalyzed activation of anticancer prodrugs. Pharmacol Rev. 2004;56:53–102. doi: 10.1124/pr.56.1.3. [DOI] [PubMed] [Google Scholar]
- 7.Barak Y, Thorne SH, Ackerley DF, Lynch SV, Contag CH, Matin A. New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Mol Cancer Ther. 2006;5:97–103. doi: 10.1158/1535-7163.MCT-05-0365. [DOI] [PubMed] [Google Scholar]
- 8.Seow HA, Penketh PG, Bauman RP, Sartorelli AC. Bioactivation and resistance to mitomycin C. In: Sies H, Packer L, editors. Methods in Enzymology. Vol. 382. Amsterdam: Elsevier; 2004. pp. 221–233. [DOI] [PubMed] [Google Scholar]
- 9.Chung-Faye GD, Palmer D, Anderson J, Clark MD, Baddeley S, Hussain PI, Murray P, Searle L, Seymour PA, Harris D, Ferry D, Kerr J. Virus-directed, enzyme prodrug therapy with nitroimidazole reductase: a phase I and pharmacokinetic study of its prodrug, CB1954. Clin Cancer Res. 2001;7:2662–2668. [PubMed] [Google Scholar]
- 10.Bagshawe KD. Antibody-directed enzyme prodrug therapy (ADEPT) for cancer. Exp Rev Anticancer Ther. 2006;6:1421–1431. doi: 10.1586/14737140.6.10.1421. [DOI] [PubMed] [Google Scholar]
- 11.Hood JD, Bednarski M, Frausto R, Guccione S, Reisfeld RA, Xiang R, Cheresh DA. Nanoparticle-mediated gene delivery to tumour neovasculature. Science. 2002;296:2404–2407. doi: 10.1126/science.1070200. [DOI] [PubMed] [Google Scholar]
- 12.Thorne SH. Strategies to achieve systemic delivery of therapeutic cells and microbes to tumors. Exp Opinion on Biol Ther. 2007;7:41–51. doi: 10.1517/14712598.7.1.41. [DOI] [PubMed] [Google Scholar]
- 13.O'Callaghan D, Charbit A. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol Gen Genet. 1990;223:156–158. doi: 10.1007/BF00315809. [DOI] [PubMed] [Google Scholar]
- 14.Ackerley DF, Gonzalez CF, Park CH, Blake R, II, Keyhan M, Matin A. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl and Environ Microbiol. 2004;70:873–882. doi: 10.1128/AEM.70.2.873-882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez CF, Ackerley DF, Lynch SV, Matin A. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem. 2005;280:22590–22595. doi: 10.1074/jbc.M501654200. [DOI] [PubMed] [Google Scholar]
- 16.Wilber A, Frandsen JL, Geurts JL, Largaespada DA, Hackett PB, McIvor RS. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Molec Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Barak Y, Nov Y, Ackerley DF, Matin A. Enzyme improvement in the absence of structural knowledge – a novel statistical approach. ISME J. 2008;2:171–179. doi: 10.1038/ismej.2007.100. [DOI] [PubMed] [Google Scholar]
- 18.Cadenas E. Antioxidant and prooxidant functions of DT-diaphorase in quinone metabolism. Biochem Pharmacol. 1995;49:127–140. doi: 10.1016/s0006-2952(94)00333-5. [DOI] [PubMed] [Google Scholar]
- 19.Knox RJ, Friedlos F, Jarman M, Roberts JJ. A new cytotoxic, DNA interstrand crosslinking agent, 5-(aziridin-1-yl)-4-hydroxylamino-2-nitrobenzamide, is formed from 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by a nitroreductase enzyme in Walker carcinoma cells. doi: 10.1016/0006-2952(88)90335-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhao M, Geller J, Ma H, Yang M, Penman S, Hoffman RM. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc Natl Acad Sci U S A. 2007;104(24):10170–10174. doi: 10.1073/pnas.0703867104. Biochem Pharmacol 1988; 37: 4661 – 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 22.Barak Y, Ackerley DF, Dodge CJ, Lal B, Cheng A, Francis AJ, Matin A. Analysis of novel soluble chromate and uranyl reductases and generation of an improved enzyme by directed evolution. Appl and Environ Microbiol. 2006;72:7074–7082. doi: 10.1128/AEM.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schepelmann S, Springer CJ. Viral vectors for gene-directed enzyme prodrug therapy. Curr Gene Ther. 2006;6:647–670. doi: 10.2174/156652306779010679. [DOI] [PubMed] [Google Scholar]
- 24.Thorne SH, Negrin RS, Contag CH. Synergistic antitumor effects of immune cell-viral biotherapy. Science. 2006;311:1780–1784. doi: 10.1126/science.1121411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.