Abstract
Contemporary models in the field of pharmacokinetic–pharmacodynamic (PK–PD) modeling often incorporate the fundamental principles of capacity limitation and operation of turnover processes to describe the time course of pharmacological effects in mechanistic terms. This permits the identification of drug- and system-specific factors that govern drug responses. There is considerable interest in utilizing mechanism-based PK–PD models in translational pharmacology, whereby in silico, in vitro, and preclinical data may be effectively coupled with relevant models to streamline the discovery and development of new therapeutic agents. These translational PK–PD models form the subject of this review.
BASIC TENETS OF PHARMACODYNAMICS
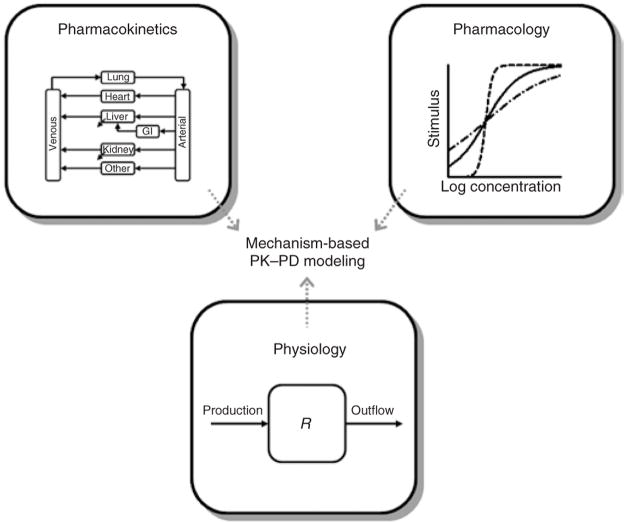
The basic principles of pharmacokinetics, pharmacology, and physiology form the foundation of mechanism-based pharmacokinetic–pharmacodynamic (PK–PD) modeling. A summary of these components is shown as a diagram in Figure 1. Pharmacokinetics encompasses the factors affecting the time course of drug/metabolite concentrations in relevant biological fluids and tissues after various routes of administration and represents the driving force for pharmacological and most toxicological effects. Noncompartmental (i.e., area/moment analysis) and mammillary plasma-clearance models that quantitatively assess pharmacokinetic processes (i.e., absorption, distribution, metabolism, and excretion) are the most common methods used for PK data analysis. At the very minimum, the primary parameters of drug distribution and elimination should be identified (volume of distribution and clearance). Despite the widespread use of these assessment techniques in studies using various animal species, the relatively empirical and hybrid nature of the parameters derived from such techniques do not readily allow for extrapolation of the PK properties across species and compounds, a highly desirable feature of translational models. In contrast, physiology-based PK (PBPK) models seek to emulate physiological pathways and processes that control plasma and tissue drug concentrations, and this approach is regarded as the state-of-the-art technique in advanced PK systems analysis.1,2 As stated by Dedrick, “Physiologic modeling enables us to examine the joint effect of a number of complex interrelated processes and assess the relative significance of each.”3 The compartments in PBPK models represent organs and tissues of interest and are arranged and connected according to anatomical and physiological relationships (Figure 1, top left). A series of mass-balance differential equations that extend from Fick’s law of perfusion/diffusion describe the rate of change of drug concentrations within each tissue. Other major processes may be incorporated, including drug metabolism and/or excretion, partitioning, binding, and transport. Most PK–PD models utilize the values of either free or total drug concentrations in plasma for driving PD, but there are increasing efforts to use techniques applicable across species, such as microdialysis and imaging, to capture the drug at or closer to its sites of action, that is, in the biophase.4
Figure 1.
Major components contributing to assembly of mechanism-based pharmacokinetic–pharmacodynamic (PK–PD) models. GI, gastrointestinal.
The law of mass action and the relatively low concentration of pharmacological receptors or targets impart capacity limitation in most drug responses. This concept is reflected in the traditional Hill function or sigmoidal Emax model of drug effects:5
| (1) |
where capacity or efficacy (Emax) and sensitivity or potency (EC50) parameters define the nonlinear relationship between drug effect (E) and concentration in plasma or at a biophase (C). Several curves defined by Equation 1 are shown in Figure 1 (top right) for three different values of the Hill coefficient (γ). Whereas Equation 1 represents a linear transduction of Clark’s receptor occupancy theory, more complex functions of receptor occupancy, including the operational model of agonism,6 can be used for characterizing many pharmacological effects. Nevertheless, capacity limitation is a hallmark feature of quantitative pharmacology and, as a consequence, a wide range of suitable dose levels is typically required to characterize the drug-specific parameters of the system.
The third major component of pharmacodynamics is physiological turnover and homeostasis. For a simple open system (as shown at the bottom of Figure 1), the turnover of a substance, R, can be described by:
| (2) |
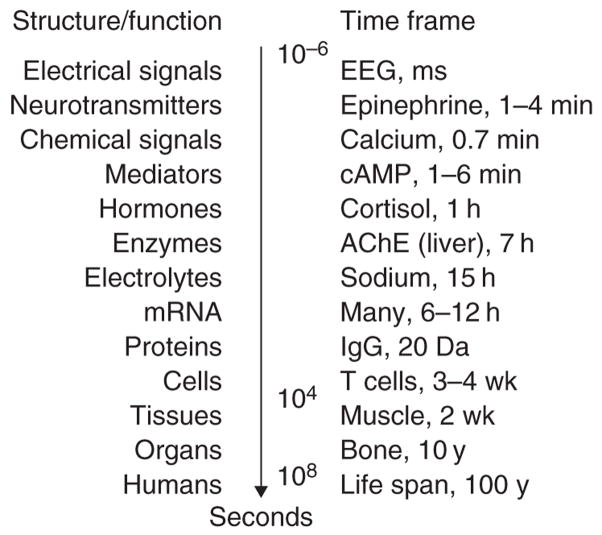
where the rate of change of R is determined by a zero-order production rate (kin) and a first-order removal rate constant (kout), and R0 is the initial value (kin/kout, assuming the steady-state value is time invariant). Indirect response models reflect inhibition or stimulation of either kin or kout.7 Biological materials, structures, or functions, many of which are used as biomarkers of drug effects and disease processes, exhibit turnover rates over a large range of temporal scales (Figure 2). A knowledge of the turnover rates for physiological system components at the desired level of organization is important for identifying the rate-limiting steps for specific pharmacological responses and for assisting in the design of studies. Such information might also impact the characterization of feedback mechanisms which are abundant in physiology, given that both drugs and diseases often interfere with the normal biological cascades that are responsible for regulating the homeostasis of physiological systems. Time-dependent transduction steps can be factored into models; these are often a series of turnover processes that assemble into systems biology models.7
Figure 2.
Time frames of turnover, life span, and half-life of various physiological materials, structures, and functions in humans. AChE, acetylcholinesterase; cAMP, adenosine 3′,5′-monophosphate; EEG, electroencephalogram; IgG, immunoglobulin G.
Mechanism-based PD models, therefore, frequently reflect an integration of the basic components to describe and understand the complex interplay between the pharmacology of drug action and the (patho-)physiological control systems.7,8 One example is the target-mediated PK–PD model developed for interferon-β1a in monkeys.9 This model includes receptor binding as a key factor in both PK and PD processes and utilizes a precursor-dependent indirect response model to capture the induction of neopterin (a classic biomarker of interferon-β receptor agonism) in concordance with known mechanisms. Two feedback signals account for altered drug and neopterin concentrations after multiple dosing, based on adaptation processes for receptor downregulation and reduced neopterin production. Relatively complex models are appearing with increasing frequency, fueled by advanced analytical methods for measuring biomarkers, intermediary biosignals, and system components with high specificity and sensitivity, as well as by increased industrial, regulatory, and academic interest in using these models for drug development and pharmacological studies. The mechanistic assessment of the biological systems (e.g., calcium/bone metabolism) also offers the opportunity to extrapolate knowledge from one drug class to another and more quickly address new therapeutic targets.
TRANSLATIONAL PK/PD MODELING
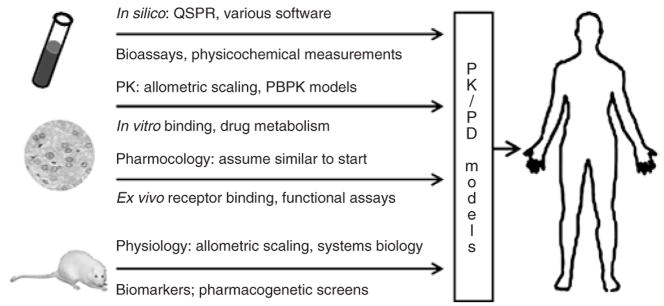
Translational PK–PD modeling, shown in Figure 3, is the integration of in silico, in vitro, and in vivo preclinical data with mechanism-based models to anticipate the effects of new drugs in humans and across levels of biological organization. Translational models hold promise to facilitate design and/or selection of lead compounds, selection of the first-in-human dose, early clinical trial design, and proof-of-concept studies of experimental drugs and drug combinations.10,11 This discussion will be limited to the scaling-up of PK–PD models developed in animals for application in humans. A recent review of quantitative structure–PK/PD relationships (QSPRs) describes approaches to predicting PK/PD profiles from in silico and in vitro experiments.12 Incidentally, considerable progress has been made in the field of toxicology, with QSPR models being combined with PBPK and PBPK–PD models to predict the exposure and dynamics of toxic chemicals in animals and humans.13,14 Implementing translational PK–PD methodology in the discovery and development of biotherapeutics has also been reviewed.15
Figure 3.
Components of mechanism-based pharmacokinetic–pharmacodynamic (PK–PD) models for translation of animal data to human clinical pharmacology. Predictive techniques (top of arrows) can be augmented by selective measurements (bottom of arrows). PBPK, physiology-based PK; QSPR, quantitative structure–PK/PD relationship.
In any modeling endeavor, one begins by defining the goals and objectives of the analysis. These benchmarks will guide model development, determine the appropriate level of precision and model detail (parsimony), and reveal methods to be used to qualify or validate the model. In addition, the successful use of PK–PD modeling and biomarker data is predicated on: (i) selection of mechanism-based biomarkers and their link with clinical end points, (ii) quantification of drug and/or metabolites in biological fluids under Good Laboratory Practices (GLP) conditions, (iii) GLP-like assay methods for biomarkers, and (iv) mechanism-based PK–PD modeling and validation.16 Ideally, measurements of responses to drugs should be sensitive, gradual, reproducible, objective, and meaningful. Measurements in animal models need to reflect relevant processes in humans, thereby facilitating “proof-of-concept” studies.
In order to scale up PK–PD models to anticipate outcomes in humans, structural models developed on the basis of data obtained from lower species should be applicable in humans, and the likelihood of this condition being met may or may not be known a priori. For example, the model developed for interferon-β1a in monkeys was shown to characterize its PK–PD properties well in human male volunteers.9,17 This was not unexpected, given that most of the mechanisms and processes emulated by the model appear to be largely conserved across species, a feature often shared for many macromolecules.15 Although the structural nature of PBPK models makes them uniquely suited for scaling and predicting human drug exposures, the extrapolation of PK–PD models from animals to humans is primarily based on classical allometric relationships. Notwithstanding the controversies surrounding the prospective use of allometry18,19 or the rationale for allometric correction factors20 for predicting PK properties in humans, there are general expectations that many physiological processes and organ sizes (θ) tend to obey a power law:21
| (3) |
where W is body weight and a and b are drug/process coefficients. The allometric exponent, b, tends to be around 0.75 for clearance processes, 1.0 for organ sizes or physiological volumes, and 0.25 for physiological times or the duration of physiological events (e.g., heartbeat and breath duration, cell life span, and turnover times of endogenous substances or processes).22 West and colleagues describe a theoretical basis for allometry founded on the fractal nature of biological systems and energy balance.23 In order to improve the translational potential of empirical PK models, nonlinear mixed-effects modeling has been coupled with allometric relationships24 and in vitro metabolism experiments as well.25
The basic expectation for allometry in pharmacodynamics is that biological turnover rates in mechanistic models for most general structures and functions should be predictable among species on the basis of allometric principles, whereas intrinsic capacity (Emax) and sensitivity (EC50) to drugs tend to be similar across species. However, many genetic differences are also found. Brodie and colleagues were the first to examine some PK–PD properties across species, demonstrating interspecies differences in global terms such as duration of action and half-life, but similarities in plasma concentration on awakening following hexobarbital administration.26 There has long been a case made for the usefulness of studying drug effects in preclinical models, and a general belief that the plasma drug concentration required for eliciting a certain (intensity of) action (e.g., EC50) is often similar in experimental animals and humans.27 While interspecies differences in relative receptor affinity and plasma protein binding often exist,28 several examples show reasonable concordance of such properties between rats and humans for congeneric series of drugs. Ito and colleagues demonstrated a linear correlation between the logarithm of equilibrium dissociation constants of benzodiazepines in the cerebral cortex tissue of rats and humans, of more than four orders of magnitude.29 Cox and co-workers also showed a similar relationship for the EC50 values of four synthetic opioids between these same two species.30 A retrospective analysis of S(+)-ketoprofen PK–PD parameters obtained from mechanistic modeling for two response biomarkers supports these basic expectations. Allometric scaling showed that PK parameters changed proportionally to body weight (albeit with unusual power coefficients) and PD parameters exhibited limited ranges in essentially a weight-independent manner.31
Interspecies scaling has been applied to complex PK–PD models, including the hypothermic and cortisol responses to buspirone and flesinoxan (two 5-HT1A receptor agonists)32 and the effects of erythropoietin on reticulocytes, red blood cells, and hemoglobin levels in humans.33 Despite the relative complexity of the models, their diverse structural components, and the differences in the molecular sizes of the drugs, the prevailing observations were that: (i) PK and physiological turnover parameters obeyed allometric principles, and (ii) pharmacological capacity and sensitivity parameters were essentially species-independent. Clinical trial simulations using the scaled models for buspirone and flesinoxan32 also suggest that such an approach may be useful for predicting responses in humans.
CONCLUSIONS
Major advances have been made in mechanism-based modeling of drug responses in animals and humans based on the integration of fundamental pharmacokinetic, pharmacological, and physiological processes. At present, the most common approach for transforming mechanistic models into translational PK–PD models is to utilize allometric principles for PK and turnover parameters, whereas pharmacological terms are often fixed across species. In addition to the assessment of drug metabolism rates, receptor binding or functional assays are needed in situations where genetic differences are expected. New theoretical and experimental approaches will be needed in order to identify the conditions under which allometry is appropriate, to screen efficiently for key differences, and to provide techniques for scaling-up complex biological and pharmacological systems, analogous to the enabling QSPR–PBPK methodology for intermolecular and interspecies PK predictions. Research is also needed for testing whether inclusion of disease state and progression in preclinical models is able to facilitate the prediction of the disease-modifying properties of drugs in early human testing. In any event, translational PK–PD modeling has the potential to direct and integrate pharmaceutical sciences toward the efficient design and development of novel drugs based on first principles.
Acknowledgments
This work was supported by Grant GM 57980 from the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.Gerlowski LE, Jain RK. Physiologically based pharmacokinetic modeling: principles and applications. J Pharm Sci. 1983;72:1103–1127. doi: 10.1002/jps.2600721003. [DOI] [PubMed] [Google Scholar]
- 2.Nestorov I. Whole body pharmacokinetic models. Clin Pharmacokinet. 2003;42:883–908. doi: 10.2165/00003088-200342100-00002. [DOI] [PubMed] [Google Scholar]
- 3.Dedrick RL. Animal scale-up. J Pharmacokinet Biopharm. 1973;1:435–461. doi: 10.1007/BF01059667. [DOI] [PubMed] [Google Scholar]
- 4.Brunner M, Langer O. Microdialysis versus other techniques for the clinical assessment of in vivo tissue drug distribution. AAPS J. 2006;8:E263–E271. doi: 10.1007/BF02854896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariens EJ. Affinity and intrinsic activity in the theory of competitive inhibition. I Problems and theory. Arch Int Pharmacodyn Ther. 1954;99:32–49. [PubMed] [Google Scholar]
- 6.Black JW, Left P. Operational models of pharmacological agonism. Proc R Soc Lond B Biol Sci. 1983;220:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 7.Mager DE, Wyska E, Jusko WJ. Diversity of mechanism-based pharmacodynamic models. Drug Metab Dispos. 2003;31:510–518. doi: 10.1124/dmd.31.5.510. [DOI] [PubMed] [Google Scholar]
- 8.Danhof M, de Jongh J, De Lange EC, Della Pasqua O, Ploeger BA, Voskuyl RA. Mechanism-based pharmacokinetic-pharmacodynamic modeling: biophase distribution, receptor theory, and dynamical systems analysis. Annu Rev Pharmacol Toxicol. 2007;47:357–400. doi: 10.1146/annurev.pharmtox.47.120505.105154. [DOI] [PubMed] [Google Scholar]
- 9.Mager DE, Neuteboom B, Efthymiopoulos C, Munafo A, Jusko WJ. Receptor-mediated pharmacokinetics and pharmacodynamics of interferon-β1a in monkeys. J Pharmacol Exp Ther. 2003;306:262–270. doi: 10.1124/jpet.103.049502. [DOI] [PubMed] [Google Scholar]
- 10.Aarons L, et al. Role of modelling and simulation in Phase I drug development. Eur J Pharm Sci. 2001;13:115–122. doi: 10.1016/s0928-0987(01)00096-3. [DOI] [PubMed] [Google Scholar]
- 11.Meno-Tetang G, Lowe PJ. On the prediction of the human response: a recycled mechanistic pharmacokinetic/pharmacodynamic approach. Basic Clin Pharmacol Toxicol. 2005;96:182–192. doi: 10.1111/j.1742-7843.2005.pto960307.x. [DOI] [PubMed] [Google Scholar]
- 12.Mager DE. Quantitative structure-pharmacokinetic/pharmacodynamic relationships. Adv Drug Delivery Rev. 2006;58:1326–1356. doi: 10.1016/j.addr.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Béliveau M, Lipscomb J, Tardif R, Krishnan K. Quantitative structure-property relationships for interspecies extrapolation of the inhalation pharmacokinetics of organic chemicals. Chem Res Toxicol. 2005;18:475–485. doi: 10.1021/tx049722k. [DOI] [PubMed] [Google Scholar]
- 14.Timchalk C, Nolan RJ, Mendrala AL, Dittenber DA, Brzak KA, Mattsson JL. A physiologically based pharmacokinetic and pharmacodynamic (PBPK/PD) model for the organophosphate insecticide chlorpyrifos in rats and humans. Toxicol Sci. 2002;66:34–53. doi: 10.1093/toxsci/66.1.34. [DOI] [PubMed] [Google Scholar]
- 15.Agoram BM, Martin SW, van der Graaf PH. The role of mechanism-based pharmacokinetic-pharmacodynamic (PK-PD) modelling in translational research of biologics. Drug Discov Today. 2007;12:1018–1024. doi: 10.1016/j.drudis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Colburn WA, Lee JW. Biomarkers, validation and pharmacokinetic-pharmacodynamic modelling. Clin Pharmacokinet. 2003;42:997–1022. doi: 10.2165/00003088-200342120-00001. [DOI] [PubMed] [Google Scholar]
- 17.Mager DE, Jusko WJ. Receptor-mediated pharmacokinetic/pharmacodynamic model of interferon-β1a in humans. Pharm Res. 2002;19:1537–1543. doi: 10.1023/a:1020468902694. [DOI] [PubMed] [Google Scholar]
- 18.Bonate PL, Howard D. Prospective allometric scaling: does the emperor have clothes? J Clin Pharmacol. 2000;40:335–340. doi: 10.1177/00912700022009017. [DOI] [PubMed] [Google Scholar]
- 19.Mahmood I. Critique of prospective allometric scaling: does the emperor have clothes? J Clin Pharmacol. 2000;40:341–344. doi: 10.1177/00912700022009026. [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Mayersohn M. A mathematical description of the functionality of correction factors used in allometry for predicting human drug clearance. Drug Metab Dispos. 2005;33:1294–1296. doi: 10.1124/dmd.105.004135. [DOI] [PubMed] [Google Scholar]
- 21.Adolph EF. Quantitative relations in the physiological constitutions of mammals. Science. 1949;109:579–585. doi: 10.1126/science.109.2841.579. [DOI] [PubMed] [Google Scholar]
- 22.Boxenbaum H. Interspecies scaling, allometry, physiological time, and the ground plan of pharmacokinetics. J Pharmacokinet Biopharm. 1982;10:201–227. doi: 10.1007/BF01062336. [DOI] [PubMed] [Google Scholar]
- 23.West GB, Brown JH, Enquist BJ. A general model for the origin of allometric scaling laws in biology. Science. 1997;276:122–126. doi: 10.1126/science.276.5309.122. [DOI] [PubMed] [Google Scholar]
- 24.Cosson VF, Fuseau E, Efthymiopoulos C, Bye A. Mixed effect modeling of sumatriptan pharmacokinetics during drug development. I: interspecies allometric scaling. J Pharmacokinet Biopharm. 1997;25:149–167. doi: 10.1023/a:1025728028890. [DOI] [PubMed] [Google Scholar]
- 25.Proost JH, et al. Predictions of the pharmacokinetics of succinylated human serum albumin in man from in vivo disposition data in animals and in vitro liver slice incubations. Eur J Pharm Sci. 2006;27:123–132. doi: 10.1016/j.ejps.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Quinn GP, Axelrod J, Brodie BB. Species, strain and sex difference in metabolism of hexobarbitone, amidopyrine, antipyrine and aniline. Biochem Pharmacol. 1958;1:152–159. [Google Scholar]
- 27.Levy G. The case for preclinical pharmacodynamics. In: Yacobi A, Skelly JP, Shah VP, Benet LZ, editors. Integration of Pharmacokinetics, Pharmacodynamics, and Toxicokinetics in Rational Drug Development. Plenum; New York: 1993. [Google Scholar]
- 28.Chien JY, Friedrich S, Heathman MA, de Alwis DP, Sinha V. Pharmacokinetics/pharmacodynamics and the stages of drug development: role of modeling and simulation. AAPS J. 2005;7:E544–E559. doi: 10.1208/aapsj070355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito K, Asakura A, Yamada Y, Nakamura K, Sawada Y, Iga T. Prediction of the therapeutic dose for benzodiazepine anxiolytics based on receptor occupancy theory. Biopharm Drug Dispos. 1997;18:293–303. doi: 10.1002/(sici)1099-081x(199705)18:4<293::aid-bdd24>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 30.Cox EH, Langemeijer MWE, Gubbens-Stibbe JM, Muir KT, Danhof M. The comparative pharmacodynamics of remifentanil and its metabolite, GR90291, in a rat electroencephalographic model. Anesthesiology. 1999;90:535–544. doi: 10.1097/00000542-199902000-00030. [DOI] [PubMed] [Google Scholar]
- 31.Lepist EI, Jusko WJ. Modeling and allometric scaling of s(+)-ketoprofen pharmacokinetics and pharmacodynamics: a retrospective analysis. J Vet Pharmacol Ther. 2004;27:211–218. doi: 10.1111/j.1365-2885.2004.00579.x. [DOI] [PubMed] [Google Scholar]
- 32.Zuideveld KP, van der Graaf PH, Peletier LA, Danhof M. Allometric scaling of pharmacodynamic responses: application to 5-Ht1A receptor mediated responses from rat to man. Pharm Res. 2007;24:2031–2039. doi: 10.1007/s11095-007-9336-y. [DOI] [PubMed] [Google Scholar]
- 33.Woo S, Jusko WJ. Interspecies comparisons of pharmacokinetics and pharmacodynamics of recombinant human erythropoietin. Drug Metab Dispos. 2007;35:1672–1678. doi: 10.1124/dmd.107.015248. [DOI] [PubMed] [Google Scholar]