Abstract
Chemokine receptors play fundamental roles in human physiology from embryogenesis to inflammatory response. The receptors belong to the G-protein coupled receptor class, and are activated by chemokine ligands with a range of specificities and affinities that result in a complicated network of interactions. The molecular basis for function is largely a black box, and can be directly attributed to the lack of structural information on the receptors. Studies to date indicate that function can be best described by a two-site model, that involves interactions between the receptor N-domain and ligand N-terminal loop residues (site-I), and between receptor extracellular loop and the ligand N-terminal residues (site-II). In this review, we describe how the two-site model could modulate binding affinity and ligand selectivity, and also highlight some of the unique chemokine receptor features, and their role in function.
Keywords: Chemokine, Chemokine receptor, GPCR, Binding affinity, Selectivity, Receptor activation structure-function, N-terminal domain
The Chemokine System
Chemokine receptors are members of G-protein-coupled receptor (GPCR) superfamily, and are activated on binding their cognate ligands of the chemokine family. Chemokines, or chemotactic cytokines, are a large family of small soluble proteins, and are distinguished and classified, based on the presence of conserved cysteine residues, as ‘CC’, ‘CXC’, ‘C’ or ‘CX3C’ [64]. The receptors, in turn, are classified based on the ligands they bind. These receptors and ligands first shot to prominence for their critical role in the orchestration of inflammation and the immune response, and are now known to play important regulatory roles in diverse biological processes such as organ development, angiogenesis and leukocyte trafficking [56, 62, 63, 86]. Based on their functionality, chemokines are broadly classified as inflammatory or homeostatic, with a few exhibiting dual functions.
The current high level of interest in chemokine biology stems not only from their beneficiary role of immunomodulation and cell trafficking, but also from their harmful role as pathologically dysregulated molecules. The chemokine-receptor system is finely regulated, so any imbalance in the chemokine network has a potentially detrimental effect. Indeed, chemokines and their receptors have been implicated in the pathophysiology of numerous autoimmune and inflammatory diseases, and in cancer progression and metastasis [33, 55, 76]. Chemokine receptors also hold their own unique fascination to researchers of viral diseases, as several viruses have developed ways to manipulate these receptors for their own survival [3, 46]. The best-known examples are those of chemokine receptors CCR5 and CXCR4, which are used by the human immunodeficiency virus (HIV-1) as co-receptors allowing viral entry into the cell [7]. Moreover, chemokine receptors are excellent drug targets as over 40% of the top 200 synthetic drugs in the market target GPCRs. Therefore, the knowledge of how chemokines and chemokine receptors exert their function is essential for rational drug design [42, 76].
A start has been made toward such an understanding, by identifying key steps involved in the process of chemokine-mediated leukocyte recruitment (Fig. 1). Leukocyte migration during conditions of tissue injury or infection is a multi-step process involving the following steps: (i) local upregulation of pro-inflammatory chemokine secretion in response to signaling molecules such as TNF-α and IFN-γ, (ii) presentation of these chemokines on endothelial cell-surface glycosaminoglycans (GAGs), (iii) binding of chemokines to their cognate receptors on the leukocyte cell surface, (iv) activation of the leukocyte receptor resulting, among other effects, in increased adhesiveness, shape change and extravasation of the leukocyte, and (v) migration of the leukocytes along the chemokine gradient within the extracellular matrix toward the site of infection or injury [62, 87]. Each of these processes is a potential checkpoint for regulation of chemokine function, and can be exploited as potential drug targets [42, 76].
Fig. 1.
A schematic showing how chemokines recruit leukocytes. Each step affords an opportunity for regulation and modulation of chemokine function. Chemokine ligands are shown as green circles, and the receptors are shown in brown as a series of stacked lines (corresponding to the 7 transmembrane helices). Chemokine ligand dimerization is shown but receptor dimerization is not shown for simplicity
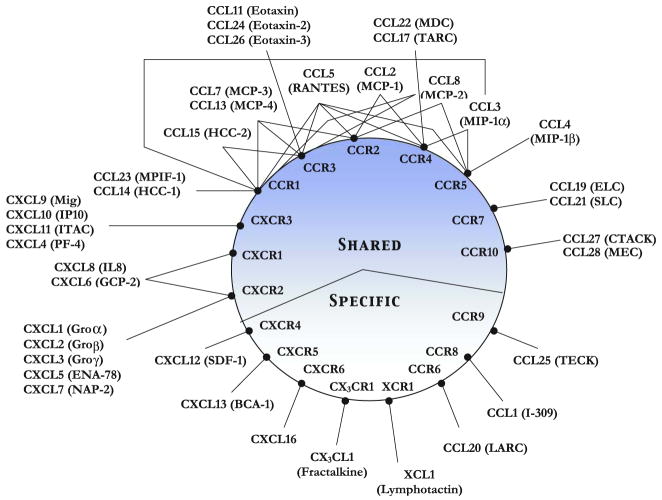
While several chemokine receptors are promiscuous, binding multiple ligands with similar high affinity, some are strictly specific binding only a single ligand. The ligands, in turn, also exhibit a range of diverse specificities, and unraveling the resulting network of interactions appears daunting (Fig. 2). With multiple ligands binding a single receptor and a single ligand binding multiple receptors, and with similar binding affinities and functional activities, there seems to be a high degree of apparent redundancy. Viewed differently, this apparent redundancy could in fact be indicative of complex and fine-tuned regulation, as different ligands binding a single receptor or a single ligand binding multiple receptors could elicit both shared and unique signaling events that orchestrate spatial/temporal regulation of the recruitment process. The chemokine family of ligands and receptors is one of the most intricate biological systems being studied today, so an understanding of the mechanism of action and its relevance to in vivo physiology requires a multi-prong approach and techniques from cell biology, immunology and structural biology to in vivo animal models.
Fig. 2.
Specificity and Promiscuity of Chemokine-receptor interactions. The receptors are shown inside the circle and the ligands outside the circle. The systematic nomenclature for both chemokine ligand and receptors are used, and the commonly used ligand names are shown in brackets. Some chemokines have multiple common names, and we have opted to use just one for simplicity
Structure-function—Complexity and Elegance
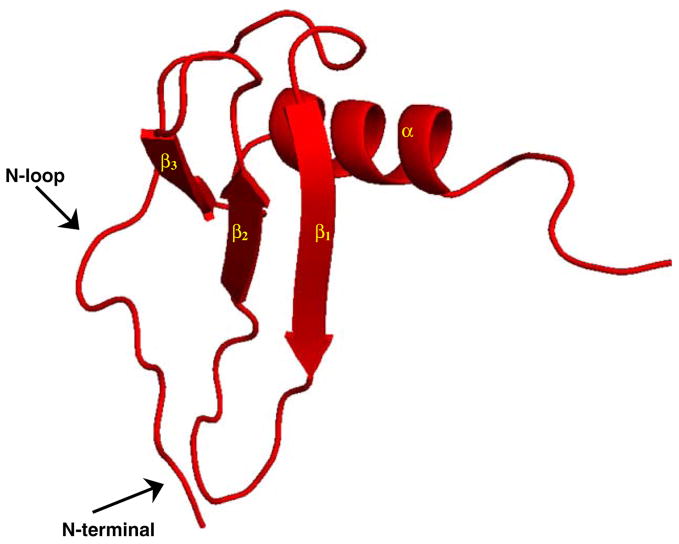
Receptor binding affinity is the first and most basic level of regulation of chemokine function, but affinity is also intimately coupled to both ligand and receptor specificity (Fig. 2). We will initially use both the systematic and the commonly used names of a given chemokine (IL-8/CXCL8), and subsequently confine to the commonly used name (IL-8). An understanding of the structural basis of affinity and selectivity is essential, and with over 40 ligands and 18 receptors identified to date, this is easier said than done. However at the structural level, within this confounding complexity is also an elegant simplicity. Structures of a number of chemokines have been determined, and despite large differences in sequence homology, all exhibit the same basic structural fold known as the chemokine fold [12, 20, 22, 23, 26, 29, 44, 45, 52, 57, 59, 73, 78, 83, 91]. The structure consists of a short N-terminal region, an extended N-loop region, followed by three β-strands and an α-helix (Fig. 3). Structure-function studies indicate all ligands interact with their receptor using the same two regions, N-terminal and N-loop residues (discussed later).
Fig. 3.
A schematic of chemokine structure. The functionally important N-terminal and N-loop residues are highlighted by arrows, and the strands and the helix are labeled
A characteristic property of chemokines is also that all form dimers, and some very high-order oligomers [24]. However, dimerization potency varies by several orders of magnitude, with some chemokines forming very strong dimers (Kd ~ nM), and some forming dimers only under crystallization conditions (Kd ~ mM), and further dimer formation is also sensitive to solution conditions. These observations together suggest that chemokine dimerization should play some fundamental role in its in vivo function. Design of trapped monomers and monomer mutants for several chemokines have shown that the monomer is the active form for in vitro receptor function [49, 67, 82, 84]. Indeed, we have recently observed that the chemokine interleukin (IL-8)/CXCL8 dimer binds its receptor with lower affinity, and have proposed that dimerization negatively regulates receptor function [30, 75, 85]. Interestingly, platelet factor 4 (PF-4)/CXCL4 has been shown to form heterodimers with IL-8 and RANTES/CCL5, and such heterodimers show distinctly different in vitro activities compared to monomers indicating that heterodimerization could also play a role in vivo [65, 95]. In the case of RANTES, it has been shown that higher order oligomers is critical for CCR1-mediated arrest but not for CCR5-mediated spreading/transmigration in flow or transendothelial chemotaxis of leukocytes [5]. These processes involve binding to both GAGs and GPCRs, and in recent years, various studies have shown that dimerization and GAG-binding are coupled, and that such a process plays an essential role in leukocyte recruitment [40, 50]. Several recent excellent reviews have addressed the role of GAG binding, dimerization, and function [37, 48].
While the structural and functional characteristics of chemokine ligands have been intensively investigated, studies on receptors have been few and far between, due to the intrinsic difficulties in studying membrane proteins. Structures of chemokine receptors are not known, and the only GPCR structure known to date is that of bovine rhodopsin [69]. Sequence analysis indicates that all chemokine receptors share a similar fold, and have an extracellular N-terminal domain (N-domain), seven TM segments, three extracellular loops (exoloops), three cytoloops, and a C-terminal segment. By analogy to the structure of rhodopsin, the membrane-spanning segments of all chemokine receptors can be envisaged as being arranged in a circle, with the extracellular domains proximal and held in space by the disulfide bonds. Nearly all members of the GPCR superfamily have a pair of conserved cysteines in exoloops 1 and 2, which are thought to form a disulfide bridge linking these two loops [4]. Most chemokine receptors contain two additional cysteines, one in the N-domain and another on exoloop 3; these two cysteines could form a disulfide bringing the N-domain and this loop into close spatial proximity [10, 89].
Computational methods for structure prediction of membrane proteins have been successful in identifying transmembrane domains as helices or strands, and indeed such methods are critical for classifying membrane proteins as belonging to the GPCR class. However such methods fail to provide an accurate description of the structures of the N-terminal or C-terminal residues and those of the loops linking the transmembrane helices. Currently available computational tools are best suited only for predicting protein structures in a homogeneous environment, such as soluble proteins in an aqueous milieu and membrane spanning proteins in a hydrophobic milieu. N- and C-domains and the loop residues are proximal to the membrane environment, and so the structural properties of these residues are influenced both by the membrane and the aqueous solvent milieu; solution properties such as the dielectric constant in the ‘twilight’ zone will not reflect that of the bulk solvent [99]. Further, these chemokine receptor domains are small in length (< 50 residues), and computational methods in general do not work well for short sequences.
Chemokine-receptor Interactions—Two-site Model
Structure-function studies have consistently shown that the N-terminal and N-loop residues on ligands and the N-terminal domain and one or more exoloops of receptors are involved in the binding interaction [1, 6, 8–19, 22, 32, 34–36, 38, 43, 47, 49, 53, 60, 61, 67, 68, 74, 88, 90, 92, 97, 98, 100–103]. On the basis of structure-function data, a general two-site mechanism of ligand-receptor interaction has been proposed for all chemokines (Fig. 4) Binding involves interactions between the ligand N-loop and receptor N-domain (site-I), and ligand N-terminal and receptor exoloop residues (site-II). However, a comprehensive and quantitative description is currently lacking as to how the two-site interaction mechanism mediates affinity, selectivity, and activation. In this review, we discuss the current knowledge, and also how the two-site model can be used for describing binding affinity and ligand specificity.
Fig. 4.
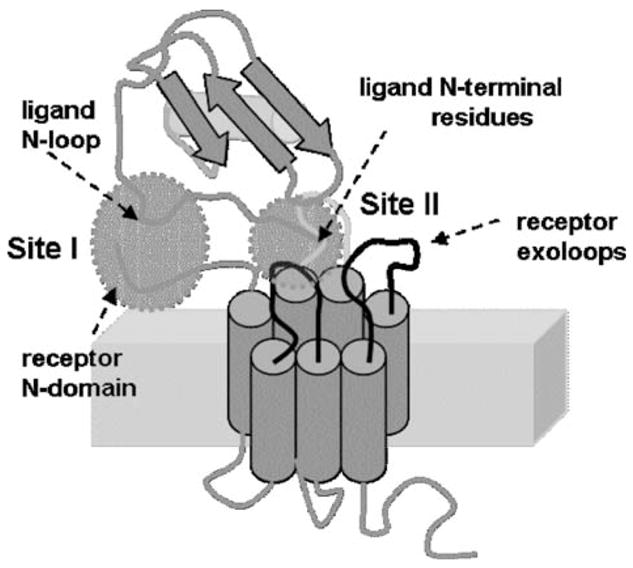
A model of chemokine ligand-receptor interaction (reproduced from ref. 74). The ligand N-loop residues interact with the receptor N-terminal domain residues (site I), and the ligand N-terminal residues interact with the receptor exoloops and trans-membrane residues (site II)
Chemokines are mainly classified into CXC and CC families on the basis of conserved cysteines near the N-terminus. The CXC chemokines can be further divided into two subgroups, ‘ELR’ and ‘non-ELR,’ based on the presence or absence of this motif before the first cysteine. Except for the ELRCXC class, sequence analysis has provided no insights into ligand affinity and receptor selectivity. ELRCXC chemokines bind both CXCR1 and CXCR2 receptors. Among ELRCXC chemokines, IL-8 alone binds both receptors with high affinity. All others including MGSA bind CXCR2 with high affinity, and CXCR1 with low affinity [2, 18]. The structural basis of IL-8 and related chemokine MGSA/CXCL1 binding to CXCR1 and CXCR2 has been well studied [6, 16–19, 32, 35, 38, 39, 43, 54, 74–82, 84, 88, 98, 100]. We will describe the current knowledge on IL-8/MGSA/CXCR1/CXCR2 system to illustrate the structural basis of chemokine receptor function, as the properties of this subset of chemokines and their receptors reflect the complexity seen on a broader level in the entire chemokine-receptor system.
Sequence analysis of the ELRCXC chemokines shows that the N-terminal residues are conserved whereas the N-loop residues are not, suggesting that the differences in binding should be due to site-I interaction (Fig. 4). Structure-function studies have essentially used two approaches, site-specific mutagenesis and generation of chimeric chemokines by swapping identical domains. These studies consistently show that the core residues such as the β-strands and the α-helix function as a scaffold and could be swapped between chemokines without loss of function, and that N-loop (site-I) and N-terminal (site-II) residues mediate receptor binding affinity, selectivity, and activation. The N-terminal residues are shown to be essential for both binding affinity and receptor activation, and that the N-loop residues are essential for binding affinity and receptor selectivity. For instance, IP-10/CXCL10, a chemokine that selectively binds CXCR3 receptor, on grafting the IL-8 N-terminal, N-loop and 30 s turn residues that are adjacent to the N-terminal residues, gained CXCR1 and CXCR2 function [17]. Comparison of IL-8 and MGSA structures also showed the largest structural difference for the N-loop residues [20, 26, 78]. Indeed, MGSA gained IL-8 like function and vice versa by swapping the N-loop residues suggesting that the site-I interaction plays an important role in receptor selectivity [54]. Chemokines bind their receptors with nM affinity, and binding of both N-loop (site-I) and N-terminal residues (site-II) should contribute to the affinity.
If we assume the free energy of binding (ΔG) is the sum of binding energies at site-I (ΔGI) and site-II (ΔGII),
| (1) |
where ΔGtotal is the free energy change from overall ligand binding, ΔGsiteI is from N-loop residues, and ΔGsiteII is from N-terminal residues. IL-8 N-terminal ELR mutants bind with μM affinity and are inactive suggesting that N-loop residues bind their receptors with μM affinity, and that the binding of N-terminal residues is a low-affinity, high specificity interaction and are involved in receptor activation. Mutagenesis and the chimera data for other chemokines also indicate that the site-I and site-II interactions differentially influence binding affinity, selectivity, and activation [22, 34, 53, 68]. For instance, deleting N-terminal residues of RANTES, MCP-1/CCL2 and MCP-3/CCL7 resulted only in marginal loss of binding affinity, but were inactive indicating that N-terminal residues play a major role in activity and not affinity. RANTES N-terminal deletion mutant was no more specific and could bind to additional receptors suggesting that the N-terminal residues also play a direct role in receptor selectivity by negatively regulating binding to other receptors [34]. Further, addition of a single methionine to the N-terminus of RANTES also abolishes its receptor activities but not binding affinities indicating further a fundamental role for the N-terminal residues in determining activity [72]. Deletion/mutation of the first two N-terminal residues in SDF-1α/CXCL12 shows loss of function but native binding affinity indicating that these residues are important for receptor activation but do not contribute to the binding affinity [22]. Interestingly, an SDF-1α -terminal peptide comprising of first nine residues was only 100 fold less potent both in binding affinity and receptor activity indicating that the rest of the N-terminal residues 3–9 contribute to binding affinity, and most importantly that binding to the N-loop is not absolutely essential for receptor activation [53]. These observations at that time were simply described or discussed as change in function either due to binding one or the other site, and in fact are best described by a two-site model where the binding events are coupled.
Structural basis of chemokine receptor function has also been investigated using the approaches of site-specific mutagenesis and generation of chimeric proteins. These studies, though not as extensive as for the chemokine ligands, showed that the N-domain and extracellular loop residues mediate binding affinity, ligand selectivity and activation. Design of CXCR1 and CXCR2 chimeras created to switch ligand specificities show that both the N-domain and the extracellular loop residues play a role in this process, and that binding to site-I and site-II cannot be described as simply additive [1]. In other words, switching the N-domain alone did not switch specificities [32, 47, 94]. It appears from these studies that the site-I interaction is the major contributor to affinity but plays only a partial role for specificity. Similar to the ligand studies, these studies in fact afforded tantalizing glimpses into coupling between the two sites, but were not appreciated at that time.
The most direct evidence for the two-site model comes from the observation that IL-8 binds the isolated CXCR1 N-domain with an affinity similar to that for the N-domain in the intact receptor. A structure of the IL-8-CXCR1 N-domain peptide also shows that only the IL-8 N-loop residues and not the N-terminal residues bind to the receptor N-domain [92]. We also recently made an interesting and unexpected observation that MGSA, like IL-8, also binds the CXCR1 N-domain in micelles with μM affinity under conditions that mimic the native environment [74]. The observation that the binding affinities of IL-8 and MGSA for site-I interaction are similar seems to be inconsistent with the observation that MGSA compared to IL-8 binds CXCR1 with 100-fold lower affinity. To account for these observations, we have proposed a model in which binding to the two sites are not independent (Eq. 1) but coupled (Eq. 2). According to this model, the binding affinity can be described as,
| (2) |
where ΔGcoupling is the coupling energy between the sites. This coupling energy could be positive or negative; in the former case, binding at site-I/II increases the binding affinity at site-II/I, and in the latter, binding at one-site decreases the binding affinity at the other site. In the case of IL-8 and MGSA, we propose that differences between their binding affinities are not due to differences in binding energies at the individual sites but due to differences in coupling energy between the sites. We further propose that the binding is sequential, and that initial binding to site-I results in conformational changes both in the ligand and/or receptor which either facilitates or negates binding at site-II.
As can be seen from Fig. 2, chemokine receptors exhibit a range of ligand binding specificities, and may bind several or all members of a subfamily, in which case a single receptor N-domain sequence binds different ligand N-loop sequences. On the other hand, a single ligand is also known to bind multiple receptors, in which case a single ligand N-loop sequence binds multiple divergent receptor N-domain sequences. The low sequence homology among N-domains of chemokine receptors therefore could explain specificity but not promiscuity. This inconsistency, however, is only inexplicable if we assume that the two sites of interaction are independent and are not coupled. The concept of coupling between the two chemokine-receptor binding sites, is a powerful way to explain the apparent paradox, and reconciles the wide range of specificities seen among the chemokine ligands and receptors. If we assume that binding interactions are not simply additive but coupled, we can rationalize how a single receptor N-domain can bind to different ligand N-loop sequences, or multiple N-domain sequences can bind to a single ligand N-loop with similar apparent affinity. The Site-I interaction may or may not differ in affinity, depending on the ligand-receptor pair, but affinity differences could arise from differential coupling in each case to the corresponding Site-II interactions.
What could be the molecular basis of coupling between Site-I and Site-II? In both the chemokine ligands and receptors, disulfide bonds connect the regions of the protein involved in the two-site interactions. In chemokine ligands, two disulfide bonds ‘tether’ the N-loop and N-terminal residues to the core of the protein. A characteristic feature of chemokines is the conserved disulfides, and deleting these disulfides results in loss of function. In the case of IL-8, it has been shown that modifying but without deleting these disulfides results in lowered receptor-binding affinities indicating that disulfides mediate function in the folded protein [80]. Disulfides could mediate the binding of the individual domains or also could also play a role in communication between the two sites and the protein core. Deleting the disulfides in the receptors also results in loss of function [10, 51], but the mechanisms by which disulfides mediate binding is not known. We propose that the disulfide bonds linking the N-domain and exoloops are involved in similar communication between the two binding sites on receptors. Thus, in addition to the compelling evidence that can be put forth based on functional evidence for two-site coupling, structural features of both ligands and receptors also support evidence for two-site coupling.
A related question is also what is the role of the scaffold (β-strands and the α-helix; Fig. 3). For instance, switching IL-8/MGSA specificities not only required swapping N-loop residues, but also swapping the third β-strand residue Leu49/Ala49. This mutation is not part of the binding site but is adjacent to the 9–50 disulfide bond [54]. Design of chimeras also suggests a role for the scaffold in mediating function. It has been shown that IP-10, MGSA, and IL-8 chimeras containing SDF-1α N-terminal and N-loop residues have highly differential binding affinities and activities, with MGSA chimera showing native SDF-1α function and IL-8 chimera showing no binding or function [22]. These observations are remarkable suggesting that all scaffolds are not equivalent, and that in addition to tethering the functional residues, they could also mediate coupling interactions between the domains and so play a role in function.
There is evidence that structural features such as turns and loops also influence binding. For instance, mutating GAG-binding 40 s loop residues in RANTES also results in reduced CCR1 but not CCR5 binding [58, 71]. These residues are located distal to the N-loop residues, and so most likely do not directly bind to the receptors, but mediate binding by modulating the properties of the N-loop residues [71]. In a similar fashion, mutating IL-8 30 s loop residues (Gly-Pro) results in significant loss of binding and activity [17]. These residues are linked to the N-terminal ELR residues via the 7–34 disulfide bond, and so very likely are not involved in direct binding but mediate binding of the ELR residues. Interestingly, it has been shown recently that a tripeptide, Pro-Gly-Pro, activates CXCR1 and CXCR2 receptors [96]. This tripeptide, derived from degradation of the extracellular matrix, has been shown to play an important role in chronic obstructive pulmonary disease (COPD). It has been proposed that the tripeptide activates the receptors by mimicking the 30 s turn ‘GP’ motif observed in IL-8 structure. However, we feel that the tripeptide and the chemokines activate receptors using completely different mechanisms, and the observation of the GP motif in IL-8 is unrelated the chemotactic activity of the tripeptide. We propose that this tripeptide, like most small molecule agonists for GPCR receptors, binds to a site in the transmembrane region, which is distinctly different from chemokine binding to the N-domain and extracellular loop region.
Sequence analysis of the chemokine receptors show overall high homology, except for the N-terminal and C-terminal residues. Low sequence homology among the receptor N-terminal residues suggests that the N-domain is involved in binding, and play a direct role in imparting ligand specificity. Sequence analysis of the ligands also shows that the N-loop and N-terminal residues are least conserved. Several studies have indicated a major determinant role for the N-domain of chemokine receptors in ligand affinity and selectivity [13, 25, 32, 47, 74, 92, 94]. The N-domain of GPCR receptors can vary from a few to as many as 600 amino acids, and there seems to be a weak correlation between the length of the N-domain and ligand size [41]. Chemokine receptors are a notable exception to this rule, possessing in general very short N-domains (~50 residues), which do not correlate to the size (~8–10 kDa) of their ligands. This anomaly can be explained by the two-site interaction; N-domain and the extracellular loop together constitute a binding site that is larger and comparable in size to that of the ligands they bind.
We have confined our discussion so far to the structural characteristics of the chemokine ligand and receptors, and have not considered whether the immediate milieu such as the membrane environment plays a role in the coupling interactions. We observe that the CXCR1 and CXCR2 N-domains are unstructured in buffer but adopt specific structure in detergent micelles, that mimic the native membrane environment (74; unpublished observations), highlighting the significant contribution of the membrane environment to structure-function of these domains. NMR studies in micelles indicate that the CXCR1 N-domain adopts a defined secondary structure, but is also conformationally flexible. This structured yet flexible conformation could support interactions with multiple ligands, enhance coupling between the two interaction sites, and thus play a major role in selectivity. It has been shown that the chemokine receptor CCR5 is preferentially associated with lipid rafts and that depleting cholesterol reduces MIP-1α/CCL3 binding [66]. These observations suggest that association with lipid rafts and cholesterol is important for receptor folding and activity, and so could play a role in mediating the coupling interactions between the ligand the receptor.
An interesting feature of chemokine receptor N-domains is the predominance of charged and hydrophobic residues. The exact role of the amino acid composition for N-domain binding is not clear. Most chemokines carry a net positive charge (pI > 9), so it was initially thought electrostatic interactions mediate binding. However, mutational studies show that none of the charged residues in CXCR1 are critical for binding. Some of the chemokine receptor (CCR5, CCR2, CXCR4, CX3CR1) N-domain tyrosines are also post-translationally modified by sulfation, and it has been shown that sulfation is essential in these receptors for high-affinity ligand binding [21, 27, 28, 31, 70]. Sequence analysis does suggest that most chemokine receptors carry signature sequences for tyrosine sulfation. There is also evidence from in vitro studies that some of the chemokine receptors form homodimers and heterodimers, and such dimerization modulates function [93]. These observations add additional dimensions to the complexity of receptor structure-function, and future studies should reveal how these different but interconnected properties of receptor structure mediate affinity, selectivity, and activation.
Summary
In retrospect, a family as large and complex as the chemokine/chemokine receptor system would have to possess mechanism(s) to allow maximum flexibility in interactions between different ligand-receptor pairs. Binding involves interactions between the receptor N-domain and ligand N-terminal loop residues (site-I), and between receptor extracellular loop and the ligand N-terminal residues (site-II). Structure-function data indicate that binding affinity and specificity cannot be unequivocally assigned to interactions at one or other site, so we propose that the binding to the individual sites is not independent but coupled. Further, the observation that the sequence diversity or similarity of individual interacting sites do not correlate with the ligand specificity is also consistent with a coupled two-site model. Several unique features of chemokine receptor extracellular domains combine to provide a perfect template for subtle and fine regulation of affinity, selectivity, and activity. We propose that the defined and unique conformation of the binding domains, combined with their sequence and structural flexibility, their communication through intramolecular contacts and/or disulfide bonds, endows chemokine receptors with their characteristic ability to bind multiple ligands with a range of affinities, creating the network of interactions that gives the chemokine family its versatility.
Acknowledgments
This work was supported by the National Institutes of Health, American Heart Association, and the John Sealy Endowment Grant (to K. R.), and by a McLaughlin Predoctoral Fellowship (to L. R.).
References
- 1.Ahuja SK, Lee JC, Murphy PM. CXC chemokines bind to unique sets of selectivity determinants that can function independently and are broadly distributed on multiple domains of human interleukin-8 receptor B. Determinants of high affinity binding and receptor activation are distinct. J Biol Chem. 1996;271:225–232. doi: 10.1074/jbc.271.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Ahuja SK, Murphy PM. The CXC chemokines growth-regulated oncogene (GRO)α, GROβ, GROγ, Neutrophil-activating peptide-2, and epithelial cell-dervived neutrophil-activating peptide-78 are potent agonists for the type B, but not the type A, Human interleukin-8 receptor. J Biol Chem. 1996;271:20545–20550. doi: 10.1074/jbc.271.34.20545. [DOI] [PubMed] [Google Scholar]
- 3.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin JM. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 5.Baltus T, Weber KS, Johnson Z, Proudfoot AE, Weber C. Oligomerization of RANTES is required for CCR1-mediated arrest but not CCR5-mediated transmigration of leukocytes on inflamed endothelium. Blood. 2003;102:1985–1988. doi: 10.1182/blood-2003-04-1175. [DOI] [PubMed] [Google Scholar]
- 6.Baly DL, Horuk R, Yansura DG, Simmons LC, Fairbrother WJ, Kotts C, Wirth CM, Gillece-Castro BL, Toy K, Hesselgesser J, Allison DE. A His19 to Ala mutant of melanoma growth-stimulating activity is a partial antagonist of the CXCR2 receptor. J Immunol. 1998;161:4944–4949. [PubMed] [Google Scholar]
- 7.Berger EA, Murphy PM, Farber JM. Chemokine Receptors as HIV-1 Coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 8.Blanpain C, Doranz BJ, Bondue A, Govaerts C, De Leener A, Vassart G, Doms RW, Proudfoot A, Parmentier M. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem. 2003;278:5179–5187. doi: 10.1074/jbc.M205684200. [DOI] [PubMed] [Google Scholar]
- 9.Blanpain C, Doranz BJ, Vakili J, Rucker J, Govaerts C, Baik SS, Lorthioir O, Migeotte I, Libert F, Baleux F, Vassart G, Doms RW, Parmentier M. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 Env protein. J Biol Chem. 1999;274:34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- 10.Blanpain C, Lee B, Vakili J, Doranz BJ, Govaerts C, Migeotte I, Sharron M, Dupriez V, Vassart G, Doms RW, Parmentier M. Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J Biol Chem. 1999;274:18902–18908. doi: 10.1074/jbc.274.27.18902. [DOI] [PubMed] [Google Scholar]
- 11.Bondue A, Jao SC, Blanpain C, Parmentier M, LiWang PJ. Characterization of the role of the N-loop of MIP-1 beta in CCR5 binding. Biochemistry. 2002;41:13548–13555. doi: 10.1021/bi026087d. [DOI] [PubMed] [Google Scholar]
- 12.Booth V, Keizer DW, Kamphuis MB, Clark-Lewis I, Sykes BD. The CXCR3 binding chemokine IP-10/CXCL10: structure and receptor interactions. Biochemistry. 2002;41:10418–10425. doi: 10.1021/bi026020q. [DOI] [PubMed] [Google Scholar]
- 13.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736–23744. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 14.Campanella GS, Lee EM, Sun J, Luster AD. CXCR3 and heparin binding sites of the chemokine IP-10 (CXCL10) J Biol Chem. 2003;278:17066–17074. doi: 10.1074/jbc.M212077200. [DOI] [PubMed] [Google Scholar]
- 15.Chung IY, Kim YH, Choi MK, Noh YJ, Park CS, Kwon do Y, Lee DY, Lee YS, Chang HS, Kim KS. Eotaxin and monocyte chemotactic protein-3 use different modes of action. Biochem Biophys Res Commun. 2004;314:646–653. doi: 10.1016/j.bbrc.2003.12.134. [DOI] [PubMed] [Google Scholar]
- 16.Clark-Lewis I, Dewald B, Geiser T, Moser B, Baggiolini M. Platelet factor 4 binds to interleukin 8 receptors and activates neutrophils when its N terminus is modified with Glu-Leu-Arg. Proc Natl Acad Sci USA. 1993;90:3574–3577. doi: 10.1073/pnas.90.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark-Lewis I, Dewald B, Loetscher M, Moser B, Baggiolini M. Structural requirements for IL8 function identified by design of analogs and CXC chemokine hybrids. J Biol Chem. 1994;269:16075–16081. [PubMed] [Google Scholar]
- 18.Clark-Lewis I, Kim K-S, Rajarathnam K, Gong J-H, Dewald B, Moser B, Baggiolini M, Sykes BD. Structure-activity relationships of chemokines. J Leukoc Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 19.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-activity relationship of interleukin-8 determined using chemically synthesized analogs. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 20.Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 1990;29:1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 21.Cormier EG, Persuh M, Thompson DA, Lin SW, Sakmar TP, Olson WC, Dragic T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc Natl Acad Sci USA. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crump MP, Gong J-H, Loetscher P, Rajarathnam K, Amara A, Aranzana-Seisdedos F, Virelizier J-L, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crump M, Rajarathnam K, Kim K-S, Clark-Lewis I, Sykes BD. Solution structure of eotaxin: a chemokine that selectively recruits eosinophils in allergic inflammation. J Biol Chem. 1998;273:22471–22479. doi: 10.1074/jbc.273.35.22471. [DOI] [PubMed] [Google Scholar]
- 24.Czaplewski LG, McKeating J, Craven CJ, Higgins LD, Appay V, Brown A, Dudgeon T, Howard LA, Meyers T, Owen J, Palan SR, Tan P, Wilson G, Woods NR, Heyworth CM, Lord BI, Brotherton D, Christison R, Craig S, Cribbes S, Edwards RM, Evans SJ, Gilbert R, Morgan P, Hunter MG, et al. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants. J Biol Chem. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 25.Datta-Mannan A, Stone MJ. Chemokine-binding specificity of soluble chemokine-receptor analogues: identification of interacting elements by chimera complementation. Biochemistry. 2004;43:14602–14611. doi: 10.1021/bi048990e. [DOI] [PubMed] [Google Scholar]
- 26.Fairbrother WJ, Reilly D, Colby TJ, Hesselgesser J, Horuk R. The solution structure of melanoma growth stimulatory activity. J Mol Biol. 1994;242:252–270. doi: 10.1006/jmbi.1994.1577. [DOI] [PubMed] [Google Scholar]
- 27.Farzan M, Babcock GJ, Vasilieva N, Wright PL, Kiprilov E, Mirzabekov T, Choe H. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1 alpha association and HIV-1 entry. J Biol Chem. 2002;277:29484–29489. doi: 10.1074/jbc.M203361200. [DOI] [PubMed] [Google Scholar]
- 28.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 30.Fernando H, Chin C, Rösgen J, Rajarathnam K. Dimer dissociation is essential for interleukin-8 (IL8) binding to CXCR1 receptor. J Biol Chem. 2004;279:36175–36178. doi: 10.1074/jbc.C400283200. [DOI] [PubMed] [Google Scholar]
- 31.Fong AM, Alam SM, Imai T, Haribabu B, Patel DD. CX3CR1 tyrosine sulfation enhances fractalkine-induced cell adhesion. J Biol Chem. 2002;277:19418–19489. doi: 10.1074/jbc.M201396200. [DOI] [PubMed] [Google Scholar]
- 32.Gayle RB, Sleath PR, Srinivason S, Birks CW, Weerawarna KS, Cerretti DP, Kozlosky CJ, Nelson N, Vanden Bos T, Beckmann MP. Importance of the amino terminus of the interleukin-8 receptor in ligand interactions. J Biol Chem. 1993;268:7283–7289. [PubMed] [Google Scholar]
- 33.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:123–128. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 34.Gong JH, Uguccioni M, Dewald B, Baggiolini M, Clark-Lewis I. RANTES and MCP-3 antagonists bind multiple chemokine receptors. J Biol Chem. 1996;271:10521–10527. doi: 10.1074/jbc.271.18.10521. [DOI] [PubMed] [Google Scholar]
- 35.Hammond ME, Shyamala V, Siani MA, Gallegos CA, Feucht PH, Abbott J, Lapointe GR, Moghadam M, Khoja H, Zakel J, Tekamp-Olsen P. Receptor recognition and specificity of interleukin-8 is determined by residues that cluster near a surface-accessible hydrophobic pocket. J Biol Chem. 1996;271:8228–8235. doi: 10.1074/jbc.271.14.8228. [DOI] [PubMed] [Google Scholar]
- 36.Han KH, Green SR, Tangirala RK, Tanaka S, Quehenberger O. Role of the first extracellular loop in the functional activation of CCR2. The first extracellular loop contains distinct domains necessary for both agonist binding and transmembrane signaling. J Biol Chem. 1999;274:32055–32062. doi: 10.1074/jbc.274.45.32055. [DOI] [PubMed] [Google Scholar]
- 37.Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE. Regulation of protein function by glycosaminoglycans—as exemplified by chemokines. Annu Rev Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 38.Hebert CA, Chuntharapai A, Smith M, Colby T, Kim J, Horuk R. Partial functional mapping of the human interleukin-8 type A receptor. Identification of a major ligand binding domain. J Biol Chem. 1993;268:18549–18553. [PubMed] [Google Scholar]
- 39.Hesselgesser J, Chitnis CE, Miller LH, Yansura DG, Simmons LC, Fairbrother WJ, Kotts C, Wirth C, Gillece-Castro BL, Horuk R. A mutant of melanoma growth stimulating activity does not activate neutrophils but blocks erythrocyte invasion by malaria. J Biol Chem. 1995;270:11472–11476. doi: 10.1074/jbc.270.19.11472. [DOI] [PubMed] [Google Scholar]
- 40.Hoogewerf AJ, Kuschert GSV, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, Wells TN. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 41.Ji TH, Grossmann M, Ji I. G protein-coupled receptors. I. Diversity of receptor-ligand interactions. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 42.Johnson Z, Power CA, Weiss C, Rintelen F, Ji H, Ruckle T, Camps M, Wells TNC, Schwarz MK, Proudfoot AEI, Rommel C. Chemokine inhibition—why, when, where, which and how? Bioch Soc Trans. 2004;37:366–377. doi: 10.1042/bst0320366. [DOI] [PubMed] [Google Scholar]
- 43.Katancik JA, Sharma A, de Nardin E. Interleukin 8:neutrophil-activating peptide-2 and GRO-alpha bind to and elicit cell activation via specific and different amino acid residues of CXCR2. Cytokine. 2000;12:1480–1488. doi: 10.1006/cyto.2000.0742. [DOI] [PubMed] [Google Scholar]
- 44.Kim KS, Clark-Lewis I, Sykes BD. Solution structure of GRO/melanoma growth stimulatory activity determined by 1H NMR spectroscopy. J Biol Chem. 1994;269:32909–32915. [PubMed] [Google Scholar]
- 45.Kim KS, Rajarathnam K, Clark-Lewis I, Sykes BD. Structural characterization of a monomeric chemokine: monocyte chemoattractant protein-3. FEBS Lett. 1996;395:277–282. doi: 10.1016/0014-5793(96)01024-1. [DOI] [PubMed] [Google Scholar]
- 46.Lalani AS, McFadden G. Evasion and exploitation of chemokines by viruses. Cytokine Growth Factor Rev. 1999;10:219–233. doi: 10.1016/s1359-6101(99)00018-0. [DOI] [PubMed] [Google Scholar]
- 47.LaRosa GJ, Thomas KM, Kaufmann ME, Mark R, White M, Taylor L, Gray G, Witt D, Navarro J. Amino terminus of the interleukin-8 receptor is a major determinant of receptor subtype specificity. J Biol Chem. 1992;267:25402–25406. [PubMed] [Google Scholar]
- 48.Lau EK, Allen S, Hsu AR, Handel TM. Chemokine-receptor interactions: GPCRs, glycosaminoglycans and viral chemokine binding proteins. Adv Protein Chem. 2004;68:351–391. doi: 10.1016/S0065-3233(04)68010-7. [DOI] [PubMed] [Google Scholar]
- 49.Laurence JS, Blanpain C, Burgner JW, Parmentier M, LiWang PJ. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry. 2000;39:3401–3409. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- 50.Laurence JS, Blanpain C, De Leener A, Parmentier M, LiWang PJ. Importance of basic residues and quaternary structure in the function of MIP-1 beta: CCR5 binding and cell surface sugar interactions. Biochemistry. 2001;40:4990–4999. doi: 10.1021/bi002593w. [DOI] [PubMed] [Google Scholar]
- 51.Limatola C, Di Bartolomeo S, Catalana M, Trettel F, Fucile S, Castellani L, Eusebi F. Cysteine residues are critical for chemokine receptor CXCR2 functional properties. Exp Cell Res. 2005;307:65–75. doi: 10.1016/j.yexcr.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 52.Lodi PJ, Garrett DS, Kuszewski J, Tsang ML-S, Weatherbee JA, Leonard WJ, Gronenborn AM, Clore GM. High-resolution solution structure of the β chemokine hMIP-1β by multidimensional NMR. Science. 1994;263:1762–1767. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
- 53.Loetscher P, Gong JH, Dewald B, Baggiolini M, Clark-Lewis I. N-terminal peptides of stromal cell-derived factor-1 with CXC chemokine receptor 4 agonist and antagonist activities. J Biol Chem. 1998;273(35):22279–22283. doi: 10.1074/jbc.273.35.22279. [DOI] [PubMed] [Google Scholar]
- 54.Lowman HB, Slagle PH, DeForge LE, Wirth CM, Gillece-Castro BL, Bourell JH, Fairbrother WJ. Exchanging interleukin-8 and melanoma growth-stimulating activity receptor binding specificities. J Biol Chem. 1996;271:14344–14352. doi: 10.1074/jbc.271.24.14344. [DOI] [PubMed] [Google Scholar]
- 55.Luster AD. Chemokines—chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 56.Luster AD. The role of chemokines in linking innate and adaptive immunity. Curr Opin Immunology. 2002;14:129–135. doi: 10.1016/s0952-7915(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 57.Mayer KL, Stone MJ. NMR solution structure and receptor peptide binding of the CC chemokine eotaxin-2. Biochemistry. 2000;39:8382–8395. doi: 10.1021/bi000523j. [DOI] [PubMed] [Google Scholar]
- 58.Martin L, Blanpain C, Garnier P, Wittamer V, Parmentier M, Vita C. Structural and functional analysis of the RANTES-glycosaminoglycans interactions. Biochemistry. 2001;40:6303–6318. doi: 10.1021/bi002670n. [DOI] [PubMed] [Google Scholar]
- 59.Mizoue LS, Bazan JF, Johnson EC, Handel TM. Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry. 1999;38:1402–1414. doi: 10.1021/bi9820614. [DOI] [PubMed] [Google Scholar]
- 60.Monteclaro FS, Charo IF. The amino-terminal extracellular domain of the MCP-1 receptor, but not the RANTES/MIP-1alpha receptor, confers chemokine selectivity. Evidence for a two-step mechanism for MCP-1 receptor activation. J Biol Chem. 1996;271:19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 61.Moser B, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. Interleukin-8 antagonists generated by N-terminal modification. J Biol Chem. 1993;268:7125–7128. [PubMed] [Google Scholar]
- 62.Moser B, Wolf M, Walz A, Loetscher P. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 2004;25:75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Murphy PM. Chemokines and the molecular basis of cancer metastasis. N Engl J Med. 2001;345:833–835. doi: 10.1056/NEJM200109133451113. [DOI] [PubMed] [Google Scholar]
- 64.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology. nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 65.Nesmelova IV, Sham Y, Dudek AZ, van Eijk LI, Wu G, Slungaard A, Mortari F, Griffioen AW, Mayo KH. Platelet factor 4 and interleukin-8 CXC chemokine heterodimer formation modulates function at the quaternary structural level. J Biol Chem. 2005;280:4948–4958. doi: 10.1074/jbc.M405364200. [DOI] [PubMed] [Google Scholar]
- 66.Nguyen DH, Taub D. Cholesterol is essential for macrophage inflammatory protein 1 beta binding and conformational integrity of CC chemokine receptor 5. Blood. 2002;99:4298–4306. doi: 10.1182/blood-2001-11-0087. [DOI] [PubMed] [Google Scholar]
- 67.Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, Mulkins M, Bhakta S, McCarley D, Wiesent L, Wong B, Jarnagin K, Handel TM. Monomeric MCP-1 binds and activates the MCP-1 receptor CCR2b. J Biol Chem. 1998;273:33157–33165. doi: 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- 68.Pakianathan DR, Kuta EG, Artis DR, Skelton NJ, Hebert CA. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry. 1997;36:9642–9648. doi: 10.1021/bi970593z. [DOI] [PubMed] [Google Scholar]
- 69.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 70.Preobrazhensky AA, Dragan S, Kawano T, Gavrilin MA, Gulina IV, Chakravarty L, Kolattukudy PE. Monocyte chemotactic protein-1 receptor CCR2B is a glycoprotein that has tyrosine sulfation in a conserved extracellular N-terminal region. J Immunol. 2000;165:5295–5303. doi: 10.4049/jimmunol.165.9.5295. [DOI] [PubMed] [Google Scholar]
- 71.Proudfoot AE, Fritchley S, Borlat F, Shaw JP, Vilbois F, Zwahlen C, Trkola A, Marchant D, Clapham PR, Wells TN. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J Biol Chem. 2001;276:10620–10626. doi: 10.1074/jbc.M010867200. [DOI] [PubMed] [Google Scholar]
- 72.Proudfoot AE, Power CA, Hoogewerf AJ, Montjovent MO, Borlat F, Offord RE, Wells TN. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 73.Qian YQ, Johanson KO, McDevitt P. Nuclear magnetic resonance solution structure of truncated human GRObeta [5–73] and its structural comparison with CXC chemokine family members GROalpha and IL-8. J Mol Biol. 1999;294:1065–1072. doi: 10.1006/jmbi.1999.3333. [DOI] [PubMed] [Google Scholar]
- 74.Rajagopalan L, Rajarathnam K. Ligand selectivity and affinity of chemokine receptor CXCR1: role of N-terminal domain. J Biol Chem. 2004;279:30000–30008. doi: 10.1074/jbc.M313883200. [DOI] [PubMed] [Google Scholar]
- 75.Rajagopalan L, Rösgen J, Bolen DW, Rajarathnam K. Novel use of an osmolyte to dissect thermodynamic linkages between receptor N-domain folding, ligand binding, and ligand dimerization in a chemokine-receptor system. Biochemistry. 2005;44:12932–12939. doi: 10.1021/bi051219z. [DOI] [PubMed] [Google Scholar]
- 76.Rajarathnam K. Designing decoys for chemokine-chemokine receptor interaction. Curr Pharm, Des. 2002;8:2159–2169. doi: 10.2174/1381612023393233. [DOI] [PubMed] [Google Scholar]
- 77.Rajarathnam K, Clark-Lewis I, Sykes BD. 1H NMR studies of interleukin-8 analogs: characterization of the domains essential for function. Biochemistry. 1994;33:6623–6630. doi: 10.1021/bi00187a032. [DOI] [PubMed] [Google Scholar]
- 78.Rajarathnam K, Clark-Lewis I, Sykes BD. 1H NMR solution structure of an active inter-leukin-8 monomer. Biochemistry. 1995;34:12893–12990. doi: 10.1021/bi00040a008. [DOI] [PubMed] [Google Scholar]
- 79.Rajarathnam K, Clark-Lewis I, Dewald B, Baggiolini M, Sykes BD. 1H NMR evidence that Glu-38 interacts with the N-terminal domain in Interleukin-8. FEBS Lett. 1996;399:43–46. doi: 10.1016/s0014-5793(96)01277-x. [DOI] [PubMed] [Google Scholar]
- 80.Rajarathnam K, Dewald B, Baggiolini M, Sykes BD, Clark-Lewis I. Disulfide bridges in interleukin-8 probed using non-natural disulfide analogs: dissociation of roles in structure and function. Biochemistry. 1999;38:7653–7658. doi: 10.1021/bi990033v. [DOI] [PubMed] [Google Scholar]
- 81.Rajarathnam K, Kay CM, Clark-Lewis I, Sykes BD. Characterization of quaternary structure of Interleukin-8 and functional implications. Methods Enzymol. 1997;287:89–105. doi: 10.1016/s0076-6879(97)87009-7. [DOI] [PubMed] [Google Scholar]
- 82.Rajarathnam K, Kay CM, Dewald B, Wolf M, Baggiolini M, Clark-Lewis I, Sykes BD. Neutrophil activating peptide-2 (NAP-2) and Melanoma growth stimulatory activity (MGSA) are functional as Monomers for Neutrophil activation. J Biol Chem. 1997;272:1725–1729. doi: 10.1074/jbc.272.3.1725. [DOI] [PubMed] [Google Scholar]
- 83.Rajarathnam K, Li Y, Rohrer T, Gentz R. Solution structure and dynamics of Myeloid progenitor inhibitor factor-1 (MPIF-1): a novel monomeric CC chemokine. J BiolChem. 2001;276:4909–4916. doi: 10.1074/jbc.M005085200. [DOI] [PubMed] [Google Scholar]
- 84.Rajarathnam K, Sykes BD, Kay CM, Dewald B, Geiser T, Baggiolini M, Clark-Lewis I. Neutrophil activation by monomeric IL8. Science. 1994;264:90–92. doi: 10.1126/science.8140420. [DOI] [PubMed] [Google Scholar]
- 85.Rajarathnam K, Prado G, Fernando H, Clark-Lewis I, Navarro J. Probing receptor binding activity of CXCL8 Dimer using a disulfide ‘trap’. Biochemistry. 2006;45:7882–7888. doi: 10.1021/bi0605944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richmond A, Fan GH, Dhawan P, Yang J. How do chemokine/chemokine receptor activations affect tumorigenesis? Novartis Found Symp. 2004;256:74–89. [PubMed] [Google Scholar]
- 87.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 88.Schraufstatter IU, Ma M, Oades ZG, Barritt DS, Cochrane CG. The role of Tyr13 and Lys15 of interleukin-8 in the high affinity interaction with the interleukin-8 receptor type A. J Biol Chem. 1995;270:10428–10431. doi: 10.1074/jbc.270.18.10428. [DOI] [PubMed] [Google Scholar]
- 89.Shi XF, Liu S, Xiangyu J, Zhang Y, Huang J, Liu S, Liu CQ. Structural analysis of human CCR2b and primate CCR2b by molecular modeling and molecular dynamics simulation. J Mol Model (Online) 2002;7:217–222. doi: 10.1007/s00894-002-0089-6. [DOI] [PubMed] [Google Scholar]
- 90.Shinkai A, Komuta-Kunitomo M, Sato-Nakamura N, Anazawa H. N-terminal domain of eotaxin-3 is important for activation of CC chemokine receptor 3. Protein Eng. 2002;15:923–929. doi: 10.1093/protein/15.11.923. [DOI] [PubMed] [Google Scholar]
- 91.Skelton NJ, Aspiras F, Ogez J, Schall TJ. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry. 2002;34:5329–5342. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- 92.Skelton NJ, Quan C, Reilly D, Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure Fold Des. 1999;7:157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 93.Springael JY, Urizar E, Parmentier M. Dimerization of chemokine receptors and its functional consequences. Cytokine Growth Factor Rev. 2005;16:611–623. doi: 10.1016/j.cytogfr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki H, Prado GN, Wilkinson N, Navarro J. The N terminus of interleukin-8 (IL8) receptor confers high affinity binding to human IL8. J Biol Chem. 1994;269:18263–18266. [PubMed] [Google Scholar]
- 95.von Hundelshausen P, Koenen RR, Sack M, Mause SF, Adriaens W, Proudfoot AE, Hackeng TM, Weber C. Heterophilic interactions of platelet factor 4 and RANTES promote monocyte arrest on endothelium. Blood. 2005;105:924–930. doi: 10.1182/blood-2004-06-2475. [DOI] [PubMed] [Google Scholar]
- 96.Weathington NM, van Houwelingen AH, Noerager BD, Jackson PL, Kraneveld AD, Galin FS, Folkerts G, Nijkamp FP, Blalock JE. A novel peptide CXCR ligand derived from extra-cellular matrix degradation during airway inflammation. Nat Med. 2006;12:317–323. doi: 10.1038/nm1361. [DOI] [PubMed] [Google Scholar]
- 97.Wells TN, Power CA, Lusti-Narasimhan M, Hoogewerf AJ, Cooke RM, Chung CW, Peitsch MC, Proudfoot AE. Selectivity and antagonism of chemokine receptors. J Leukoc Biol. 1996;59:53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 98.Williams G, Borkakoti N, Bottomley GA, Cowan I, Fallowfield AG, Jones PS, Kirtland SJ, Price GJ, Price L. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J Biol Chem. 1996;271:9579–9586. doi: 10.1074/jbc.271.16.9579. [DOI] [PubMed] [Google Scholar]
- 99.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 100.Wu L, Ruffing N, Shi X, Newman W, Soler D, Mackay CR, Qin S. Discrete steps in binding and signaling of interleukin-8 with its receptor. J Biol Chem. 1996;271:31202–31209. doi: 10.1074/jbc.271.49.31202. [DOI] [PubMed] [Google Scholar]
- 101.Yan Z, Zhang J, Holt JC, Stewart GJ, Niewiarowski S, Poncz M. Structural requirements of platelet chemokines for neutrophil activation. Blood. 1994;84:2329–2339. [PubMed] [Google Scholar]
- 102.Ye J, Kohli LL, Stone MJ. Characterization of binding between the chemokine eotaxin and peptides derived from the chemokine receptor CCR3. J Biol Chem. 2000;275:27250–27257. doi: 10.1074/jbc.M003925200. [DOI] [PubMed] [Google Scholar]
- 103.Zhou N, Luo Z, Luo J, Liu D, Hall JW, Pomerantz RJ, Huang Z. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J Biol Chem. 2001;276:42826–42833. doi: 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]