Abstract
OBJECTIVE
The evolutionary conservation of transcriptional mechanisms has been widely exploited to understand human biology and disease. Recent findings, however, unexpectedly showed that the transcriptional regulators hepatocyte nuclear factor (HNF)-1α and -4α rarely bind to the same genes in mice and humans, leading to the proposal that tissue-specific transcriptional regulation has undergone extensive divergence in the two species. Such observations have major implications for the use of mouse models to understand HNF-1α– and HNF-4α–deficient diabetes. However, the significance of studies that assess binding without considering regulatory function is poorly understood.
RESEARCH DESIGN AND METHODS
We compared previously reported mouse and human HNF-1α and HNF-4α binding studies with independent binding experiments. We also integrated binding studies with mouse and human loss-of-function gene expression datasets.
RESULTS
First, we confirmed the existence of species-specific HNF-1α and -4α binding, yet observed incomplete detection of binding in the different datasets, causing an underestimation of binding conservation. Second, only a minor fraction of HNF-1α– and HNF-4α–bound genes were downregulated in the absence of these regulators. This subset of functional targets did not show evidence for evolutionary divergence of binding or binding sequence motifs. Finally, we observed differences between conserved and species-specific binding properties. For example, conserved binding was more frequently located near transcriptional start sites and was more likely to involve multiple binding events in the same gene.
CONCLUSIONS
Despite evolutionary changes in binding, essential direct transcriptional functions of HNF-1α and -4α are largely conserved between mice and humans.
Changes in gene transcription are central for evolution (1,2). At the same time, the conservation of a large body of gene regulatory mechanisms has enabled the use of genetic models and comparative genomics to provide a wealth of insights into the role of gene regulation in human biology and disease (3–7).
Recent studies have challenged preconceived ideas concerning the extent of conservation of gene regulation. A systematic comparison of ∼4,000 orthologous genes showed that the transcription factors hepatocyte nuclear factor (HNF)-1α, HNF-4α, FOXA2 (forkhead box A2), and HNF-6 frequently bind to different genes in mice and humans, leading to the conclusion that tissue-specific transcriptional regulation has significantly diverged across these two species (8). An analogous striking divergence of regulator binding sites has been observed across related yeast species (9). Such results have major implications for human disease. For example, of all mouse genes bound by HNF-1α, a regulator encoded by the most frequently mutated gene in human monogenic diabetes (MODY3) (10), only 20% showed binding to human orthologs (8). This finding questions the value of mouse models of human MODY3 (maturity-onset diabetes of the young 3). By extension, this notion affects other diseases caused by defects in genes encoding for transcriptional regulators, including several susceptibility variants recently implicated in type 2 diabetes (11–13).
The significance of such observations, however, is uncertain, because many genomic binding events could be functionally dispensable. Only essential functions of regulators are expected to be under strong evolutionary constraints. Essential regulatory functions are also the most relevant to the phenotypic consequences of human disease. We have now assessed the conservation of HNF-1α and -4α binding in genes where we could document that these regulators are required for transcription. In contrast to the previous global comparative study (8), our results reveal a high conservation of the essential functions of HNF-1α and -4α in mice and humans.
RESEARCH DESIGN AND METHODS
Gene expression analysis.
Mouse gene expression datasets from Hnf1a- and Hnf4α-deficient liver are available in ArrayExpress (accession numbers: E-MEXP-1733 and E-MEXP-1709, respectively). A more comprehensive analysis of the Hnf1a-deficient expression datasets is reported elsewhere (14). Briefly, Affymetrix Mouse Genome 430 2.0 arrays were used for the comparison of RNA from liver from C57BL6/J Hnf1a−/− and wild-type 4-week-old male mice (14), or from liver-specific Hnf4α deletion (albumin Cre+/− / Hnf4 fl/fl) and wild-type controls. Hnf1a−/− and albumin Cre+/− / Hnf4 fl/fl mouse models have been previously described (15,16). Affymetrix expression data were normalized with RMA, and the LIMMA package was used for statistical analysis to identify downregulated genes in triplicate hybridizations using an adjusted P value <0.05. For genes with multiple probes, we selected a single most informative probe showing the lowest P value in mutant/wild-type comparisons. For human expression studies, we used the results of a published microarray analysis of human hepatocellular adenomas with biallelic HNF-1α mutations, and we used the entire set of genes that were downregulated relative to normal tissue as listed in the supplementary data of the report by Rebouissou et al. (17). To relate expression ratios of bound genes versus all genes, we reprocessed the published human hepatocellular adenoma and control tissue HG-U133A Affymetrix chip dataset (GEO GSE7473) with RMA using identical conditions as for the mouse chip datasets.
Genomic binding analysis.
We used the genomic binding datasets in human hepatocytes and mouse liver genes reported by Odom et al. (8). Unless otherwise stated, we used the default (P < 0.01) criteria based on the JBD (joint binding deconvolution) algorithm that was reported in that study to select bound genes (8). Analogous results were obtained with the alternate binding criteria that were presented in the same study (8).
To assess independent binding datasets, we used mouse hepatocyte HNF-1α and HNF-4α ChIP/chip experiments obtained with β-Cell Biology Consortium (BCBC) promoter arrays. A more detailed description of BCBC HNF-1α binding studies is described elsewhere (14). Data for BCBC HNF-1α and -4α binding studies are available in Arrayexpress (accession numbers E-MEXP-1714 and E-MEXP-1730, respectively). Briefly, freshly isolated mouse hepatocytes were used for chromatin immunoprecipitation as described (18,19). After reverse cross-linking, immunoprecipitated DNA was amplified with ligation-mediated PCR and used for hybridization of BCBC promoter microarrays. For HNF-1α, we used version BCBC 5A0, and for HNF-4α we used version BCBC 5A1. Six microarrays were used for each antibody with dye swapping. Normalized data were analyzed with the LIMMA package. Unless otherwise stated we used a stringent threshold to define genes as bound (P < 0.001 and Log2 immunoprecipitate/input binding ratios/M >0.8), although alternate ratios ranging from M >0.3 to 1 did not alter the conclusions. Control experiments with IgG showed negligible binding with these criteria. We used antibodies SC-6556 for HNF-4α and SC-8986 for HNF-1 (Santa Cruz Biotechnology). The HNF-1 antibody cross-reacts with HNF-1β. However, in our experience the low abundance of HNF-1β in wild-type hepatocytes is insufficient to elicit detectable binding when using an HNF-1β–specific antibody that shows robust enrichment in experimental conditions in which HNF-1β is induced (14). Thus, HNF-1β cross-reactivity in our studies was negligible.
Integration of binding and expression datasets.
Of the 4,022 genes reported by Odom et al. (8), we matched 3,665 genes to probes represented in the Affymetrix Mouse Genome 430 2.0 arrays based on either identical Refseq or mouse gene symbols linked to the Refseqs; in the latter instance, we verified genomic positions of Refseq and gene symbols to eliminate errors caused by equivocal nomenclature. An analogous approach was used for matching other gene sets described in this analysis. A compilation of the gene expression and binding findings can be found in an online appendix, available at http://diabetes.diabetesjournals.org/cgi/content/full/db08-0812/DC1.
In silico promoter analysis.
We extracted 5′ flanking sequences (−500 to +1 bp) from mouse (mm8 assembly) and human (hg17 assembly) genomes based on annotations from Ensembl release 49. After the recovery of sequences in one species, we extracted the aligned sequence in the other species based on the multiple genome alignments from the University of California at Santa Cruz using the Galaxy platform (20). We considered the latter sequence as the putative orthologous promoter if at least 50% of the nucleotides aligned. We then scanned sequences with the HNF-1α (M00132) matrices from Transfac Professional using Patser (21). We considered hits above a threshold of 90% of the matrix score range, which corresponds to high-affinity HNF-1α binding sequences (22).
Statistical analysis.
Statistical significance was calculated with two-sided Fisher's exact test, or by testing the hypergeometric distribution as stated. To assess whether HNF-1α binding enrichment among downregulated genes differed in mouse versus human samples, we used binary logistic regression implemented with SPSS 14.0.2.
Microarray data presented in this article have been deposited in ArrayExpress (http://www.ebi.ac.uk) under the accession numbers E-MEXP-1733, E-MEXP-1709, E-MEXP-1714, and E-MEXP-1730.
RESULTS
Conservation of essential functions of HNF-1α.
We first integrated the mouse and human liver HNF-1α binding results reported in a systematic comparison of ∼4,000 orthologous genes (8) with gene expression studies in HNF-1α–deficient mouse and human tissues. We studied expression profiles from Hnf1a−/− versus wild-type mouse liver and from a previously reported study comparing gene expression in human hepatocellular adenomas carrying biallelic mutations of HNF1A versus control tissue (17). The results showed that most genes bound by HNF-1α in mouse or human chromatin did not exhibit changes in gene expression in HNF-1α–deficient mouse and human tissues (Fig. 1).
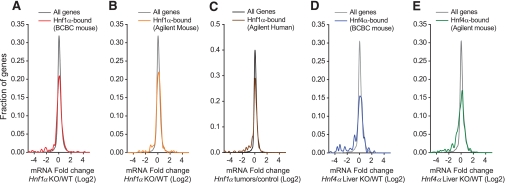
FIG. 1.
HNF-1α and -4α are only essential for transcription in a subset of the genes to which they bind. Dark lines depict the distribution of liver gene expression ratios for all genes in the experimental models described in the title of the horizontal axis. Colored lines depict expression ratios for the subset of genes that are bound in liver by either HNF-1α or -4α using different platforms indicated in the upper legends. KO, knockout; WT, wild type. (A high-quality digital representation of this figure is available in the online issue.)
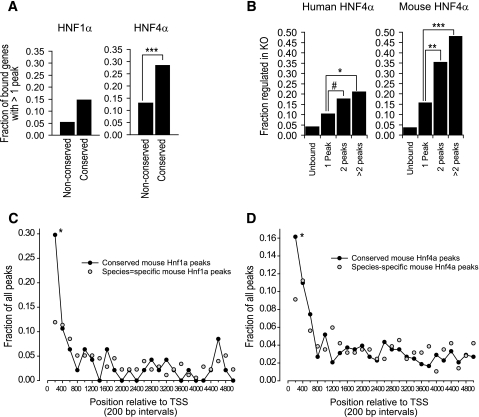
The reasons for the lack of perturbation of many HNF-1α–bound genes in cells lacking HNF-1α are currently unknown (see discussion). However, for a subset of HNF-1α–bound genes, we could clearly ascertain that HNF-1α plays an essential regulatory role in liver because they showed significant downregulation in the loss-of-function models. HNF-1α binding frequency was significantly enriched 2.7-fold in genes that were downregulated in Hnf1a−/− liver (P < 0.0001) and 4.9-fold in human genes downregulated in HNF1A-deficient tumors (P < 0.0001). This enrichment reflects the essential transactivating function of HNF-1α in a subset of its direct targets.
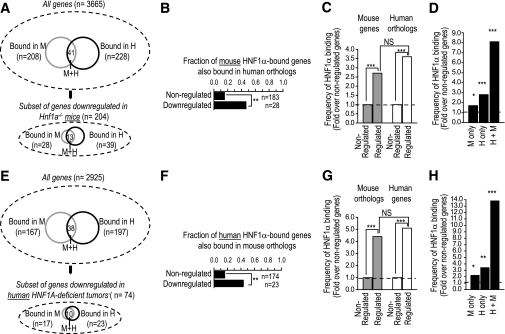
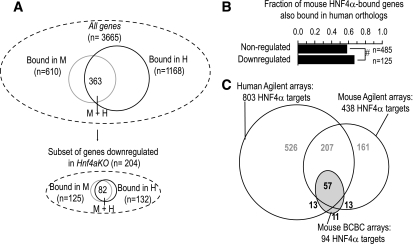
We next assessed HNF-1α binding conservation specifically in the subset of genes where the mouse and human expression studies could document that HNF-1α function is essential (Fig. 2A and E). Of note, throughout this analysis we focused on binding conservation in gene orthologs irrespective of whether this occurred in precisely aligned sequences because it is thought that regulatory functions can be conserved through compensatory sequence changes (8,23,24). Only 17% of HNF-1α–bound mouse genes that were not downregulated in Hnf1a−/− mice showed conserved binding in human orthologs, as opposed to 46% of downregulated targets (P < 0.0001) (Fig. 2B). We estimated that binding was conserved in as many as 65% of the genes that accounted for the increase in binding frequency among HNF-1α–dependent genes. Similarly, HNF-1α binding was conserved in only 15% of cases among genes that were not downregulated in HNF1A-deficient tumors, in contrast to 43% conservation of downregulated targets (P < 0.0001) (Fig. 2F). Thus, HNF-1α binding exhibits much greater human-mouse conservation in genes in which it is essential for transcription.
FIG. 2.
Conservation of HNF-1α function. A and E: HNF-1α binding in mice (M) and humans (H) in the study by Odom et al. (8). The larger Venn diagrams represent binding in all studied genes; smaller diagrams below represent the subset of genes that were downregulated in Hnf1a−/− liver (A) or HNF1A-deficient hepatocellular adenomas (E) (17). Only genes represented in both binding and expression arrays were analyzed. B and F: Binding conservation was 3-fold higher in genes that were significantly downregulated in Hnf1a−/− liver (B) and 2.7-fold higher in genes downregulated in HNF1A-deficient adenomas (F) (17), in comparison with nonregulated genes. C and G: HNF-1α binding was enriched in mouse genes that were downregulated in Hnf1a−/− liver (C) and human genes downregulated in HNF1A-deficient tumors (G). In contrast to the expectation if HNF-1α function is divergent, HNF-1α binding enrichment was comparable in the orthologs of such HNF-1α–dependent genes. D and H: Genes downregulated in Hnf1a−/− mice or HNF1A-deficient adenomas showed a marked enrichment of conserved binding events (mice and humans). Species-specific binding (mice only, human only) was also moderately enriched, but this was not selective for the species where gene regulation is experimentally verified. *P < 0.01, **P < 0.001, ***P < 0.0001, Fisher's exact test. NS, nonsignificant effect of species on binding enrichment in downregulated genes using logistic regression analysis.
Limitations of binding studies to quantify binding conservation.
Even among target genes where HNF-1α was functionally essential, binding was not conserved in all cases (Fig. 2B and F). However, the extent to which this reflects true species-specific regulation or the effect of experimental variables is uncertain. Significant false-negative and false-positive binding results in both species can theoretically lead to a marked overestimation of binding divergence. This notion is important because even in optimized chromatin immunoprecipitation microarray (ChIP-chip) protocols, the reported false-negative rate is >20% (25,26).
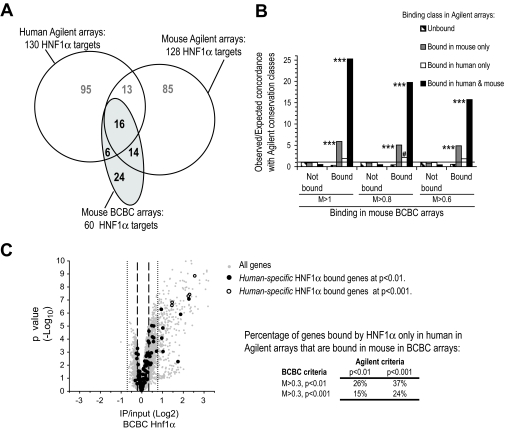
To provide an independent test of HNF-1α binding accuracy, we compared data published by Odom et al. (8), based on Agilent 10-Kb tiles surrounding transcription start sites, with another mouse liver HNF-1α binding experiment based on BCBC promoter arrays containing 1- to 2-Kb PCR product tiles. Despite major platform and analytical differences, there was a considerable overlap of targets (Fig. 3A). This analysis also confirmed species-specific binding because HNF-1α binding in mouse BCBC arrays showed a higher overlap with mouse-specific rather than human-specific binding events (Fig. 3A and B).
FIG. 3.
Comparison of HNF-1α occupancy in different platforms. A: Venn diagrams depicting HNF-1α–bound genes in mouse BCBC arrays versus human and mouse Agilent arrays from Odom et al. (8). We analyzed 2,150 genes with data in both platforms. Note that Agilent arrays cover 10-Kb surrounding transcription start sites, whereas BCBC arrays cover 1- to 2-Kb 5′ flanking regions, and thus complete binding overlap is not expected. B: Concordance of HNF-1α occupancy in mouse BCBC arrays at different Log2 binding ratio thresholds (M >1, >0.8, and >0.6; P < 0.001) with that in Agilent arrays expressed as the fold increase over the random expectation. Statistical significance was calculated with the hypergeometric distribution. ***P < 0.0001, #P < 0.05, only values for overrepresented classes are shown. C: Volcano plot of HNF1 binding ratios for all probes in mouse BCBC arrays ( ) and for genes classified as human-specific binding events using either default or stringent criteria in the report by Odom et al. (8) (● and ○). The results show that 15–37% of genes classified as human-specific HNF1α targets are bound in mouse BCBC arrays using low or moderate stringency criteria. Dashed and dotted lines depict lenient and stringent binding criteria in the BCBC arrays. IP, immunoprecipitate.
) and for genes classified as human-specific binding events using either default or stringent criteria in the report by Odom et al. (8) (● and ○). The results show that 15–37% of genes classified as human-specific HNF1α targets are bound in mouse BCBC arrays using low or moderate stringency criteria. Dashed and dotted lines depict lenient and stringent binding criteria in the BCBC arrays. IP, immunoprecipitate.
We furthermore observed that binding in mouse BCBC arrays overlapped disproportionately with the conserved subset of mouse Agilent targets, in contrast to mouse-specific Agilent targets (Fig. 3B). This could result from false-positive mouse-specific events and/or, as discussed below, if species-specific events have distinct properties that are captured less efficiently by the BCBC platform.
Importantly, several HNF-1α targets classified as human-specific in the report by Odom et al. (8) were strongly bound in mouse BCBC arrays (Fig. 3A), and up to 26–37% were bound in mouse chromatin at less stringent thresholds (Fig. 3C and D). This demonstrates false-negative binding in ChIP-chip studies and indicates that overlaps of lists of bound genes from different species do not provide an unequivocal measure of HNF-1α binding conservation.
Other factors can overestimate binding divergence and were not tested, yet they remain plausible. This includes the extremely different experimental conditions inherent to the mouse-human binding comparison, and the likelihood that in at least some instances, regulator binding selectively relocates in one species to a region that is not interrogated in array platforms. Thus, documented and presumed factors can collectively lead to an overestimation of the interspecific binding divergence.
HNF-1α binding enrichment is conserved in orthologs of HNF-1α–dependent genes.
To overcome the nonexhaustive nature of binding conservation estimates, we undertook an alternate analytical approach that does not make assumptions about the completeness of binding detection. The increased frequency with which HNF-1α binds to genes that are downregulated in HNF-1α deficiency, compared with nonregulated genes, provides a measure of the direct essential function of HNF-1α within those genes. It follows that if the function of HNF-1α is conserved in only ∼20% of its target genes, as implied in the study by Odom et al. (8), then the enrichment of HNF-1α binding events that is observed in the HNF-1α–dependent gene set from one species should be diluted in the gene set that is composed of orthologous genes from the other species. The results failed to show differences in binding enrichment between genes that are shown to be downregulated in HNF-1α deficiency and their orthologs (Fig. 2C and G). Thus, human orthologs of the gene set that was downregulated in Hnf1a−/− mice had a similar increase in HNF-1α binding frequency as the regulated mouse gene set, and the same occurred for mouse orthologs of genes that are HNF-1α–dependent in human tissues (Fig. 2C and G).
Further inspection of regulated genes revealed a remarkable enrichment of conserved binding events (Fig. 2D and H). A more moderate enrichment of species-specific binding was also observed (Fig. 2D and H). However, this was not restricted to the species where regulation was observed (as would be expected if it reflected species-specific regulation). For example, human-specific binding was paradoxically enriched in genes that were HNF-1α–dependent in mouse liver (Fig. 2D). This is consistent with the incomplete detection of binding outlined above (Fig. 2D and H). Taken together, these findings fail to detect evidence for major human-mouse divergence of functionally essential HNF-1α binding events.
HNF1 motif enrichment is conserved in orthologs of HNF-1α–dependent genes.
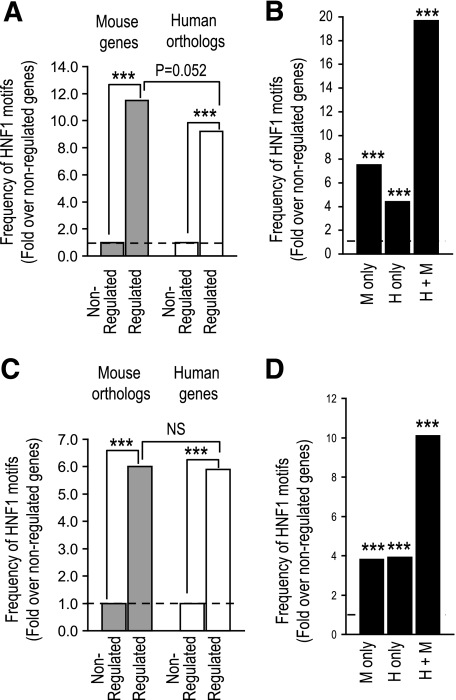
The analysis of HNF-1α binding was focused on a large but incomplete subset of genes. To provide an independent confirmation of the binding studies, we analyzed computational high-affinity HNF-1α binding sequence motifs (22). We did not assess the degree of conservation of precisely aligned motifs because its significance may be obscured by the high degree of interspecies binding site turnover (factor A binds to gene X in both species, but in different regions) (8,23,24). We therefore studied the conservation of HNF1 motif enrichment among HNF-1α–dependent genes. HNF1 motifs were enriched 11.5- and 6-fold in the immediate 5′ flanking regions of experimentally defined mouse and human HNF-1α–dependent genes, respectively (Fig. 4). We thus used the enrichment of HNF1 motifs in regulated genes as a surrogate quantitative measure of direct HNF-1α functional effects within such genes. In analogy to the binding analysis, we asked whether the enrichment of HNF1 motifs was absent or markedly decreased in promoter regions of orthologs of HNF-1α–dependent genes, as predicted from the hypothesis that HNF-1α function has undergone a major evolutionary divergence. The results showed that high-affinity HNF-1α binding motifs were highly enriched in human orthologs of genes that showed HNF-1α dependence in mice (albeit at a marginally lower rate than the mouse orthologs) and in mouse orthologs of genes that showed HNF-1α dependence in human tumors (Fig. 4). This finding further supports that a substantial fraction of functional HNF-1α targets is conserved in mice and humans.
FIG. 4.
Conservation of high-affinity HNF1 motifs in HNF-1α–dependent genes. A and C: We identified HNF1 motifs with scores >0.9 in the immediate (500 bp) 5′ flanking regions of all mouse and human genes. Motifs were strongly enriched in mouse and human genes that are experimentally determined to be HNF-1α dependent in Hnf1a−/− liver (A) and HNF1A-deficient tumors (C) (17). In contrast to the expectation if HNF-1α function is divergent, high-affinity HNF1 motifs were also enriched in the orthologs of such HNF-1α–dependent genes. B and D: Genes downregulated in Hnf1a−/− mice or HNF1A-deficient adenomas showed a marked enrichment of conserved HNF1 motifs (mouse and human). Species-specific binding (mouse only, human only) was also moderately enriched, but this was not selective for the species where gene regulation is experimentally verified. The effect of species on HNF1 motif enrichment in downregulated genes was studied with logistic regression analysis. ***P < 0.0001, Fisher's exact test. H, human; M, mouse.
HNF-4α binding conservation among HNF-4α–dependent genes.
We also studied HNF-4α, another regulator involved in human diabetes (27). In analogy to HNF-1α, most HNF-4α–bound genes were not perturbed in Hnf4a-deficient liver (Fig. 1). Among the subset of genes that did show decreased expression in Hnf4a-deficient liver, a similar number was bound by HNF-4α in mice and humans, in contrast to the expectation if these genes were selectively regulated in mice (Fig. 5A). The overall conservation of mouse HNF-4α binding was in reality quite high: even among nonregulated mouse genes there was 58% conservation, and this increased to 66% in Hnf4a–dependent genes (Fig. 5B). The true extent of conservation is likely to be higher because several genes classified as human-specific targets were also bound in mice in an independent experiment (Fig. 5C). This analysis therefore also failed to support an extensive divergence of HNF-4α function across mice and humans.
FIG. 5.
Conservation of HNF-4α binding among HNF-4α–dependent genes. A: Venn diagrams depict HNF-4α–bound genes in mice (M) and humans (H) from the study by Odom et al. (8) in all genes and in the subset that is downregulated in liver-specific Hnf4a-deficient mice. Only genes represented in both binding and expression arrays were analyzed. Note that the overall binding frequency of HNF-4α is twofold higher in human chromatin, and therefore binding enrichment comparisons in ortholog pairs are uninformative because even if there is 100% conservation, the enrichment will be twofold higher in mouse genes. HNF-4α binding was nevertheless significantly enriched in mouse Hnf4α-dependent genes and their human orthologs (3.7- and 1.8-fold, respectively). B: Fraction of HNF-4α–bound mouse genes that exhibit conserved binding in human orthologs, according to their expression changes in Hnf4a-deficient liver. Statistical significance was calculated with Fisher's exact test. #P < 0.05. C: Venn diagrams depicting HNF-4α–bound genes in mouse BCBC arrays versus human and mouse Agilent arrays from the study by Odom et al. (8). We analyzed 2,495 genes with data in both platforms. Note that Agilent arrays cover 10-Kb surrounding transcription start sites, whereas BCBC arrays cover 1- to 2-Kb 5′ flanking regions, and thus complete binding overlap is not expected.
Distinct properties of conserved and nonconserved binding.
Because conserved and species-specific binding showed different functional properties, we predicted that they should also differ in other properties. We studied binding multiplicity and observed that conserved HNF-1α and -4α targets were more frequently bound at multiple sites on the same gene, as compared with genes that were bound in a species-specific manner (Fig. 6A). Interestingly, HNF-4α dependence strongly correlated with HNF-4α binding multiplicity, suggesting that this may represent a critical attribute of functional HNF-4α binding (Fig. 6B). Conserved binding was also more likely to be located in proximal promoter regions than species-specific binding (Fig. 6C). Because BCBC arrays are built with large proximal PCR fragments rather than oligonucleotide tiles, these two properties could theoretically partly explain the abovementioned differential detection of conserved events by the two platforms. The data presented by Odom et al. (8) also indicate that genes with conserved HNF-1α binding were twice as likely to contain a canonical HNF1 sequence motif. Collectively, these findings showed that conserved and nonconserved binding events may differ not only in functionality, but also in location, multiplicity, and binding site sequence.
FIG. 6.
Distinct binding properties of species-specific versus conserved binding. A: Fraction of genes with two or more binding peaks among mouse-specific versus conserved HNF-1α and -4α targets. B: Fraction of downregulated genes in Hnf4a-deficient mouse liver according to the number of HNF-4α peaks in human or mouse orthologs. A similar analysis is not shown for HNF-1α because the frequency of multiple binding events is low. The results show that HNF-4α peak multiplicity correlates with both binding conservation and regulation in Hnf4a-deficient cells. C and D: Spatial distribution of mouse HNF-1α and -4α binding events that are either species-specific ( ) or conserved (●). Circles represent the fraction of peaks that are located within 200-bp intervals relative to the transcriptional start site (TSS). Results show that proximal binding is more frequently conserved. *P < 0.01; **P < 0.001; ***P < 0.0001; #P < 0.05.
) or conserved (●). Circles represent the fraction of peaks that are located within 200-bp intervals relative to the transcriptional start site (TSS). Results show that proximal binding is more frequently conserved. *P < 0.01; **P < 0.001; ***P < 0.0001; #P < 0.05.
DISCUSSION
The results presented here are consistent with a recent report indicating that HNF-1α and -4α binding has undergone evolutionary divergence across mice and humans (8), yet they qualify this information in two critically important ways. First, the data suggest that current large-scale binding assays overestimate the evolutionary divergence of transcription factor binding. Second, and more importantly, we show that binding to gene targets where HNF-1α and -4α exert essential functions is considerably conserved between mice and humans.
Our analysis rests on the observation that only a small portion of HNF-1α and -4α binding events are affected in loss-of-function studies. This result is striking, but entirely consistent with several recent studies that compared gene expression models with binding patterns for Oct4, Nanog, glucocorticoid receptor, and p63 (28–30). This is central to our analysis because high evolutionary conservation is not expected among binding events that are not functionally essential. Consistent with this prediction, we observed that binding conservation was markedly dependent on the gene expression phenotype in loss-of-function studies.
There are several likely causes for the lack of functional dependence on HNF-1α and -4α for numerous direct targets of these factors. First, HNF-4α or -1α are expected to be dispensable in many bound genes because of redundant regulatory factors. Second, in an undetermined number of genes, binding could simply have limited functional consequences, as recently proposed for many binding sites of several Drosophila regulators (31). On the other hand, some bound genes with unperturbed expression may be dependent on HNF-1α or -4α only in specific physiological or developmental settings. For example, functional dependence of HNF-1α– or HNF-4α–bound genes is highly tissue specific, although most bound genes show no changes in gene expression in either liver or pancreatic islets of mice lacking these factors (J.M.S., S.F.B., J.F., unpublished observations). Even though some unperturbed targets in null mutant cells are likely to be truly functionally dependent on HNF-1α or -4α in other settings, the observed differences in binding conservation between perturbed or unperturbed genes suggests that this classification is largely correct. In fact, we predict that binding conservation differences between gene expression classes would be larger if all functionally significant targets were correctly classified.
Our results highlight that the comparison of two incomplete binding datasets from different species can lead to an overestimation of evolutionary divergence. One expected cause of incomplete detection is the high false-negative rate in ChIP-chip (25,26). In part, this is because it relies on the en masse amplification of thousands of DNA templates and unavoidably results in poor amplification of a subset of sequences in each of the two species. Failure to detect binding conservation can also result from transcription factors binding outside of the interrogated regions in only one species. Furthermore, extreme differences in experimental conditions in the two species can differentially affect the binding measurements. This includes differences in age, leanness, nutritional status, recent exposure to drug therapies, cause of death, and use of cultured cells versus freshly isolated tissue in the mouse models and human organ donors (8).
To circumvent the limitation that current assays do not capture all binding events, we studied the extent to which the increased frequency with which HNF-1α binds to HNF-1α–regulated genes in one species is conserved in orthologous genes. Because HNF-1α has a complex well-characterized DNA binding sequence motif (22), we also studied whether the enrichment of high-affinity HNF1 motifs is conserved among regulated ortholog pairs. Both comparisons independently tested the hypothesis that functional binding is divergent between mice and humans. Neither approach makes assumptions about the fraction of binding events that are detected, or the extent of turnover of evolutionary conserved transcription factor binding sites. For both experimental and computational sites, we observed no evidence to support an evolutionary divergence of functional HNF-1α binding between mice and humans.
Taken together, these results suggest that functionally important binding events exhibit a much stronger evolutionary conservation than anticipated from studies that only measure the conservation of binding. Similar conclusions were drawn in a recent study that related binding of muscle regulators with the conservation of bound sequences in 12 Drosophila genomes (32). That study concluded that binding to conserved sequences was more likely to be biologically significant because it occurred more frequently in the proximity of muscle genes than binding events occurring in nonconserved sequences (32).
We expect that the degree of conservation will vary for different regulators, depending on the nature of the cellular functions they regulate. Comparative studies using accurate genome-wide sequencing approaches are warranted to fully understand the evolutionary conservation of different regulators, but, importantly, such studies should not be restricted to assaying genomic occupancy.
Our findings also showed that compared with conserved binding, species-specific binding events differed not only in function, but also in several binding properties. This suggests that a subset of species-specific binding events could be fundamentally distinct from conserved, functionally relevant binding events. We speculate that such species-specific binding events may be less exposed to evolutionary pressure, but they could be instrumental in the acquisition of new functions.
Recent data proposing that transcriptional regulation has diverged between mice and humans questioned the value of mouse genetic models (8). Our findings therefore have important implications for the use of mouse models of human monogenic diabetes and more generally for the use of animal models and comparative genomics to understand transcriptional regulation and human disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Ministerio de Educación y Ciencia and the E.U. VI Framework program. J.M.S. was supported by the Ramon y Cajal Programme.
No potential conflicts of interest relevant to this article were reported.
We thank the Instituto Nacional de Bioinformatica de Genoma España for support, Frank Gonzalez (National Cancer Institute) for Hnf1a mice, Jose Antonio Rios for statistical advice, Duncan Odom for helpful insights, Natalia del Pozo for animal assistance, Thien Vu Manh for initial database development, and Pedro Jares (Institut d'Investigacions Biomèdiques August Pi i Sunyer) and Lauro Sumoy (Centre de Regulació Genòmica) for microarray hybridizations and processing.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Carroll SB: Endless forms: the evolution of gene regulation and morphological diversity. Cell 101: 577– 580, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, de Carvalho AB, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia ACL, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li RQ, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long MY, Low L, Lozovsky E, Lu J, Luo MH, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MAF, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VLS, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GKS, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang PL, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M: Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203– 218, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE: Human-mouse genome comparisons to locate regulatory sites. Nat Genet 26: 225– 228, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Bedell MA, Largaespada DA, Jenkins NA, Copeland NG: Mouse models of human disease. Part II: recent progress and future directions. Genes Dev 11: 11– 43, 1997 [DOI] [PubMed] [Google Scholar]
- 5.Francis GA, Fayard E, Picard F, Auwerx J: Nuclear receptors and the control of metabolism. Ann Rev Physiol 65: 261– 311, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Pearson ER, Boj SF, Steele AM, Barrett T, Stals K, Shield JP, Ellard S, Ferrer J, Hattersley AT: Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLoS Med 4: e118, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pontoglio M, Sreenan S, Roe M, Pugh W, Ostrega D, Doyen A, Pick AJ, Baldwin A, Velho G, Froguel P, Levisetti M, Bonner-Weir S, Bell GI, Yaniv M, Polonsky KS: Defective insulin secretion in hepatocyte nuclear factor 1 alpha-deficient mice. J Clin Invest 101: 2215– 2222, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E: Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730– 732, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borneman AR, Gianoulis TA, Zhang ZDD, Yu HY, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M: Divergence of transcription factor binding sites across related yeast species. Science 317: 815– 819, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox RD, Lathrop GM, Boriraj VV, Chen XN, Cox NJ, Oda Y, Yano H, Lebeau MM, Yamada S, Nishigori H, Takeda J, Fajans SS, Hattersley AT, Iwasaki N, Hansen T, Pedersen O, Polonsky KS, Turner RC, Velho G, Chevre JC, Froguel P, Bell GI: Mutations in the hepatocyte nuclear factor-1 alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384: 455– 458, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40: 638– 645, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K: Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320– 323, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881– 885, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Servitja JM, Pgnatelli M, Maestro M, Cardalda C, Boj SF, Lozano J, Blanco E, Lafuente A, McCarthy MI, Sumoy L, Guigo R, Ferrer J: Hnf1α (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YH, Sauer B, Gonzalez FJ: Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1alpha knockout mouse. Mol Cell Biol 18: 3059– 3068, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I: Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 20: 2293– 2305, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebouissou S, Imbeaud S, Balabaud C, Boulanger V, Bertrand-Michel J, Terce F, Auffray C, Bioulac-Sage P, Zucman-Rossi J: HNF1 alpha inactivation promotes lipogenesis in human hepatocellular adenoma independently of SREBP-1 and carbohydrate-response element-binding protein (ChREBP) activation. J Biol Chem 282: 14437– 14446, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Boj SF, Parrizas M, Maestro MA, Ferrer J: A transcription factor regulatory circuit in differentiated pancreatic cells. Proc Natl Acad Sci U S A 98: 14481– 14486, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrizas M, Maestro MA, Boj SF, Paniagua A, Casamitjana R, Gomis R, Rivera F, Ferrer J: Hepatic nuclear factor 1-alpha directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol Cell Biol 21: 3234– 3243, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A: Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451– 1455, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertz GZ, Stormo GD: Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15: 563– 577, 1999 [DOI] [PubMed] [Google Scholar]
- 22.Tronche F, Ringeisen F, Blumenfeld M, Yaniv M, Pontoglio M: Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J Mol Biol 266: 231– 245, 1997 [DOI] [PubMed] [Google Scholar]
- 23.Dermitzakis ET, Clark AG: Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114– 1121, 2002 [DOI] [PubMed] [Google Scholar]
- 24.Ludwig MZ, Bergman C, Patel NH, Kreitman M: Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564– 567, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA: Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799– 804, 2002 [DOI] [PubMed] [Google Scholar]
- 26.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA: Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947– 956, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI: Mutations in the hepatocyte nuclear factor-4 alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384: 458– 460, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH: The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431– 440, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K: Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24: 593– 602, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH: Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet 1: e16, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, Chu HC, Ogawa N, Inwood W, Sementchenko V, Beaton A, Weiszmann R, Celniker SE, Knowles DW, Gingeras T, Speed TP, Eisen MB, Biggin MD: Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, Ruby JG, Brennecke J, Hodges E, Hinrichs AS, Caspi A, Park SW, Han MV, Maeder ML, Polansky BJ, Robson BE, Aerts S, van Helden J, Hassan B, Gilbert DG, Eastman DA, Rice M, Weir M, Hahn MW, Park Y, Dewey CN, Pachter L, Kent WJ, Haussler D, Lai EC, Bartel DP, Hannon GJ, Kaufman TC, Eisen MB, Clark AG, Smith D, Celniker SE, Gelbart WM, Kellis M: Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219– 232, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.