Abstract
OBJECTIVE
We investigated the regulation and involvement of microRNAs (miRNAs) in fat cell development and obesity.
RESEARCH DESIGN AND METHODS
Using miRNA microarrays, we profiled the expression of >370 miRNAs during adipogenesis of preadipocyte 3T3-L1 cells and adipocytes from leptin deficient ob/ob and diet-induced obese mice. Changes in key miRNAs were validated by RT-PCR. We further assessed the contribution of the chronic inflammatory environment in obese adipose tissue to the dysregulated miRNA expression by tumor necrosis factor (TNF)-α treatment of adipocytes. We functionally characterized two adipocyte-enriched miRNAs, miR-103 and miR-143, by a gain-of-function approach.
RESULTS
Similar miRNAs were differentially regulated during in vitro and in vivo adipogenesis. Importantly, miRNAs that were induced during adipogenesis were downregulated in adipocytes from both types of obese mice and vice versa. These changes are likely associated with the chronic inflammatory environment, since they were mimicked by TNF-α treatment of differentiated adipocytes. Ectopic expression of miR-103 or miR-143 in preadipocytes accelerated adipogenesis, as measured both by the upregulation of many adipogenesis markers and by an increase in triglyceride accumulation at an early stage of adipogenesis.
CONCLUSIONS
Our results provide the first experimental evidence for miR-103 function in adipose biology. The remarkable inverse regulatory pattern for many miRNAs during adipogenesis and obesity has important implications for understanding adipose tissue dysfunction in obese mice and humans and the link between chronic inflammation and obesity with insulin resistance.
Adipose tissue is not only a storage depot of triglycerides, but it is also an endocrine organ and an important regulator of whole-body energy homeostasis (1–3). Abnormal fat accumulation in obesity increases risk of life-threatening diseases such as type 2 diabetes, atherosclerosis, and certain types of cancer (4,5). Fundamental for the development of novel therapeutics for obesity and its associated metabolic syndromes is an understanding of the regulation of adipogenesis, which is tightly controlled by a combination of multiple transcription factors and extracellular hormones such as insulin (6–8).
Potential regulators of adipogenesis include microRNAs (miRNAs), which encode an abundant class of ∼22 nucleotide evolutionarily conserved RNAs that control gene expression at the posttranscriptional level by targeting mRNAs for degradation or translational repression or both (9–11). Computational and experimental analyses suggest that miRNAs may regulate expression of ∼30% of human and mouse genes (12). Furthermore, miRNAs are attractive candidates for regulating cell fate decisions and complex diseases such as obesity because the simultaneous coordination of a large number of target genes, potentially accomplished by a single miRNA, may be key to defining specific differentiated or pathogenic cell states.
Although miRNA expression profiles and functions have been extensively investigated in the hematopoietic system and neuronal and muscle tissues (13–15), little is known about the role of miRNAs in metabolic tissues, particularly adipose tissue (16). Of particular relevance, miR-14 and miR-278 in the fat body of flies regulate lipid metabolism (17,18), miR-122 in mouse liver controls triglyceride metabolism and cholesterol biosynthesis (19,20), and experiments using antisense oligonucleotides transfected into cultured human preadipocytes suggested that miR-143 is involved in adipocyte differentiation (21). Using Northern blot analyses, Kajimoto et al. (22) profiled ∼100 miRNAs including three novel miRNAs in 3T3-L1 cells before and after differentiation, and Gu et al. (23) cloned 45 known and 2 novel miRNAs from bovine adipose tissue. Moreover, as based on computational analysis of miRNA target sites in their 3′ untranslated region (UTR) sequences, 71% (282 out of 395) of expressed sequence tags with unique 3′UTRs differentially expressed during 3T3-L1 differentiation are potentially regulated by miRNAs (24).
However, few adipocyte miRNAs have been analyzed, in part because of the low sensitivity and coverage of cloning and Northern blot analyses. Except for miR-143, none of the candidate adipocyte–important miRNAs overlapped in the studies of Esau et al. (21) and Kajimoto et al. (22). Previous studies also used unfractionated primary adipose tissue, which consists of a heterogeneous mixture of cell types (23,25–28). And except for knocking down expression of miR-143, there have been no functional characterizations of adipocyte miRNAs. Additionally, there has been no systematic comparison of miRNA expression levels in normal and obese states, despite the fact that many adipose-important mRNAs are dysregulated in adipocytes from obese animals and humans (25–28).
RESEARCH DESIGN AND METHODS
Cell culture, differentiation, and chemical treatments.
3T3-L1 cells (American Type Culture Collection) were cultured and differentiated as described earlier (29). For tumor necrosis factor (TNF)-α treatment, 1 nmol/l human TNF-α (PeproTech) was added to the growth medium on day 8 after induction of differentiation and incubated for 24 h as described previously (30).
Mouse studies.
Male C57BL/6J and B6.V-Lepob/J (ob/ob) mice (Jackson Laboratory) were maintained at the animal facility of the Whitehead Institute for Biomedical Research. All animal experiments were performed with the approval of the Massachusetts Institute of Technology Committee on Animal Care. For the ob/ob model, all mice were provided with a normal diet. Epididymal fat pads were harvested from mice at 16–17 weeks of age. For the diet-induced obesity (DIO) model, we randomized 10 6-week-old male C57BL/6J littermates to either normal diet or high-fat diet (55% fat, Teklad). After 3 months, epididymal fat pads were harvested. Fat tissues from two to four mice of the same group were pooled together as one biological sample for microarray or RT-PCR experiments. Primary adipocytes were obtained after collagenase/dispase digestion, and enriched preadipocytes (CD11b−CD90−CD31−) were isolated by a negative selection on an autoMACS Separator according to the manufacturer's instruction (Miltenyi Biotec) (Supplementary Fig. S1, available in an online appendix at http://diabetes.diabetesjournals/cgi/content/full/db08-1299/DC1).
miRNA microarray and data analysis.
Total RNA including miRNA was extracted using an miRNeasy Mini kit according to the manufacturer's protocol (Qiagen). Total RNA (6 μg) was size-fractionated, and the small RNAs (<300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase and labeled with Cy3 or Cy5. Hybridizations were performed on a microfluidic biochip platform (LC sciences) as described previously (31). Microarrays with miRNA content corresponding to miRbase v9.0 or v9.1 (nine probes for each miRNA on one chip) were used for profiling during 3T3-L1 adipogenesis and for comparison between wild-type and ob/ob mice, whereas microarrays with miRNA content corresponding to miRbase v10.1 (six probes for each miRNAs on one chip) were used for comparison between control and DIO mice. Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter. The ratio of the two sets of detected signals (log2 transformed, balanced) and P values of the t test were calculated for each probe; differentially regulated miRNAs were those with >50% repeating probes on one chip with P < 0.01 (Student's t test) between the Cy3 and Cy5 signals. Overlapping differentially regulated miRNAs between two biological replicates were considered as regulated miRNAs. Complete miRNA microarray data and detailed protocols are deposited into Array Express (accession nos. E-MEXP-1789, E-MEXP-1930, and E-MEXP-1932). miRNAs described in this article are referred to by their names in miRbase v9.0.
Quantitative real-time RT-PCR assay.
For miRNA RT-PCR, 10 ng RNA was reverse-transcribed, and individual Taqman primers were used for PCR according to the manufacturer's instruction (Applied Biosystems). snoRNA202 was used as the internal control. For mRNA RT-PCR, 1 μg RNA was reverse-transcribed with random primers by SuperScript II Reverse Transcriptase (Invitrogen) and PCR amplified with gene-specific primers and SYBR Green PCR master mix using ABI 7900HT according to the manufacturer's protocol (Applied Biosystems). 18S was used as the internal control. Data were analyzed by the relative quantification (ΔΔCt) method. Primer sequences are available upon request.
miRNA gene cloning and ectopic expression.
Authentic miRNA stem-loop and ∼220 nucleotides flanking sequences on the 5′ and 3′ side of the mature miR-103 or miR-143 was amplified from normal mouse genomic DNA (Clontech) and cloned into the retroviral vector MDH.xdna as described previously (32). Empty vector or expression plasmid with miRNA (8 μg) was co-transfected into 293T cells with retrovirus packaging vector pCL-Eco (4 μg) using FuGene 6 according to the manufacturer's protocol (Roche). Virus supernatant was collected and used to infect 3T3-L1 cells with the addition of polybrene (Sigma) to a final concentration of 4 μg/ml. Infected cells were sorted by fluorescence-activated cell sorting to collect the top 30–40% highly expressing green fluorescent protein (GFP) cells.
Proliferation assay.
3T3-L1 cells expressing miRNA or empty vector alone were seeded in 96-well plates (Corning) at a density of 10,000 cells per well. After 3 days, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed with an MTT Cell Proliferation Assay kit according to the manufacturer's protocol (American Type Culture Collection).
Triglyceride assay.
On days 4 or 7 after differentiation, 3T3-L1 cells were harvested in 300 μl lysis buffer (50 mmol/l Tris-HCl, pH 7.4, 150 mmol/l NaCl, 1% Triton X-100) and sonicated to homogenize the cell suspension. Triglyceride level was measured using a serum triglyceride determination kit according to the manufacturer's instructions (Sigma). Protein was measured by a DC protein assay kit according to the manufacturer's protocol (Bio-Rad).
Statistical analysis.
Data are expressed as means ± SE unless otherwise indicated. Student's t test (unpaired, two-tailed) was used to compare the two groups, and the P value was calculated in Excel (Microsoft) or GraphPad Prism 5 (GraphPad Software). Pearson's correlation analysis and Fisher's exact test were performed in GraphPad Prism 5 (GraphPad Software). For Pearson's correlation analysis, miRNAs from the same genomic cluster (<50 kb) were treated as one entity and their expression levels were averaged. P < 0.05 was considered as statistically significant.
RESULTS
miRNA expression during in vitro adipogenesis of 3T3-L1 preadipocytes.
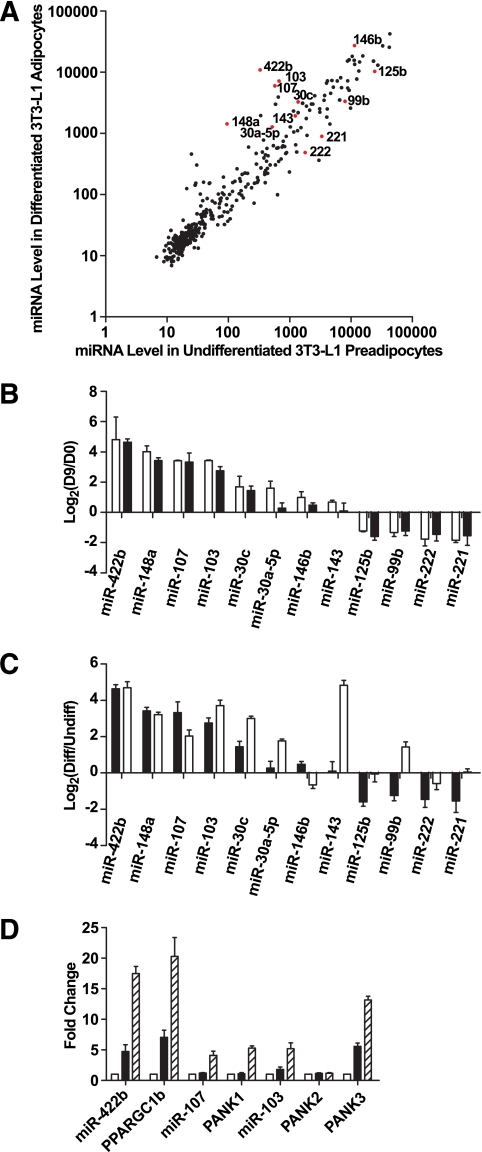
We profiled the expression of miRNAs during 3T3-L1 cell adipogenesis using miRNA microarrays detecting 373 mouse mature miRNAs (miRbase v9.0). Data from two biological replicates were consistent (data not shown). Most miRNAs fell along the diagonal line in the intensity scatter plot, indicating that they were not regulated during the 9 days of adipogenesis (Fig. 1A). Based on criteria described in research design and methods, 28 miRNAs were expressed at significantly different levels between undifferentiated 3T3-L1 preadipocytes (day 0) and differentiated 3T3-L1 adipocytes (day 9) (Fig. 1A, P < 0.01, Student's t test). Expression profiles of miRNA isolated during 3T3-L1 differentiation indicated that major changes occurred as early as days 2–4 (data not shown). Of the miRNAs differentially expressed, the eight miRNAs significantly upregulated during differentiation, namely miR-422b, 148a, 107, 103, 30c, 30a-5p, 146b, and 143, and the four downregulated miRNAs, namely miR-125b, 99b, 222, and 221, were selected for validation by quantitative RT-PCR assays using samples derived from an independent set of experiments. The results confirmed our findings from the microarrays (Fig. 1B).
FIG. 1.
miRNA expression profiling during 3T3-L1 adipogenesis. A: Intensity scatter plot showing comparison of miRNA profiles between undifferentiated 3T3-L1 preadipocytes (day 0) and differentiated 3T3-L1 adipocytes (day 9). The 12 miRNAs that are the focus of our detailed analyses are highlighted in red and indicated by a label. B: Validation of miRNA array results for 12 regulated miRNAs by RT-PCR assays. Expressions for all miRNAs are plotted as fold-change in log2 scale; positive indicates enriched in day 9 vs. day 0. □, Array result (n = 2); ■, RT-PCR result (n = 4). Data are expressed as means ± SE. C: Comparison by RT-PCR of miRNA regulation during 3T3-L1 differentiation and primary fat cell development. Expressions of all miRNAs are normalized to internal control and plotted as fold-change in log2 scale. ■, day 9 vs. day 0, same data as Fig. 1B (n = 4); □, mature adipocytes vs. enriched preadipocytes (n = 4). Data are expressed as mean ± SE. D: Intronic miRNAs and their host genes are co-regulated during adipogenesis. miR-422b is in the intron of PPARGC1b; miR-107 is in the intron of PANK1; the two copies of miR-103 are in the introns of PANK2 and PANK3. miRNA expression levels are measured by RT-PCR and normalized to internal control. Expression levels of mRNAs are also determined by RT-PCR and normalized to internal control. Expressions are shown as fold-change relative to the level at day 0. □, day 0; ■, day 2; ▨, day 4. Data are expressed as mean ± SE (n = 3).
miRNA expression during primary fat cell development.
To further examine the expression profiles during in vivo fat cell development, enriched epididymal adipocytes were purified by floatation after collagenase/dispase digestion and dissociation; enriched preadipocytes (CD11b−CD90−CD31−) in the stromal vascular fraction were depleted of fibroblasts, endothelial cells, macrophages, and erythrocytes (supplementary Fig. S1). Differentiation of these enriched preadipocytes showed enhanced formation and accumulation of lipid droplets compared with the total population of cells in the stromal vascular fraction (data not shown). Expression levels of the same 12 selected miRNAs shown in Fig. 1B were measured by RT-PCR in RNAs from enriched preadipocytes and mature primary adipocytes. As shown in Fig. 1C, 9 of 12 miRNAs exhibited a similar regulation pattern compared with in vitro differentiation. Noteworthy is the fact that miR-143 expression increases to a larger extent in vivo compared with adipogenesis of 3T3-L1 cells; this is one of the miRNAs we selected for overexpression in 3T3-L1 preadipocytes (Fig. 6).
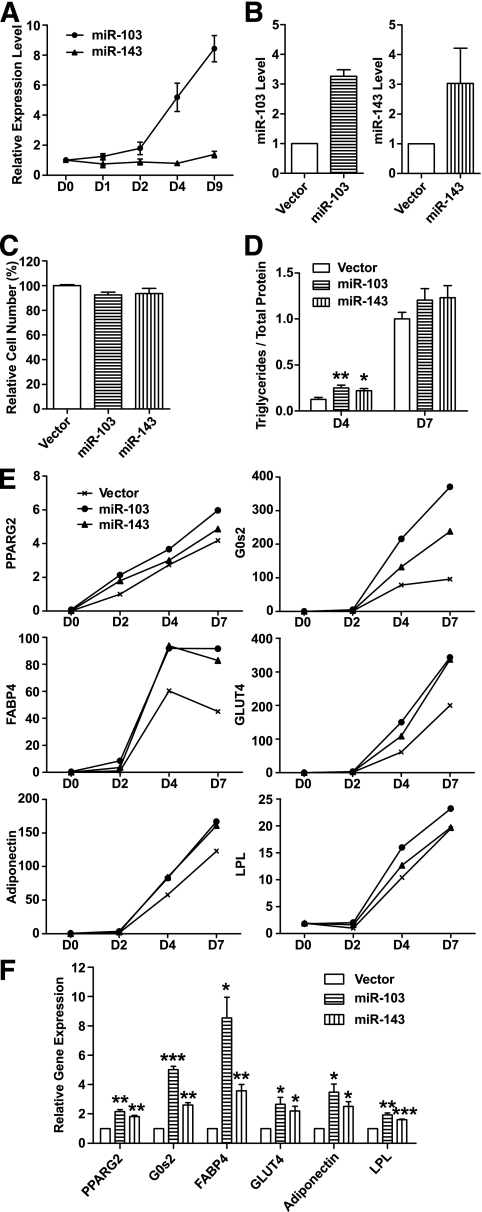
FIG. 6.
Ectopic expression of two adipocyte-induced miRNAs, miR-103 or miR-143, accelerates adipogenesis. A: miR-103 and miR-143 are induced during 3T3-L1 adipogenesis. miRNA expression levels are measured by RT-PCR, normalized to internal control and plotted relative to their respective levels in preadipocyte (day 0). Data are expressed as mean ± SE (n = 3). B: Expression level of miR-103 or miR-143 in infected and sorted cells compared with vector control. Data are expressed as mean ± SE (n = 3). C: Ectopic expression of miR-103 or miR-143 has little effect on 3T3-L1 cell growth by MTT assay. Data are expressed as mean ± SE (n = 6). D: Ectopic expression of miR-103 or miR-143 increases triglyceride accumulation at day 4 (D4) but not day 7 (D7). Triglycerides are normalized to total protein and plotted relative to the level of the control cells expressing vector alone at D7. Data are expressed as mean ± SE (n = 6). **P < 0.01, *P < 0.05. E: Ectopic expression of miR-103 or miR-143 hastens expression of adipogenic markers at an early stage (days 2 and 4) of adipogenesis. mRNA levels are measured by RT-PCR, normalized to internal control and plotted relative to their respective levels at day 2 (D2). Data are expressed as mean only (n = 3). PPARG2, peroxisome proliferator-activated receptor γ 2; G0s2, G0/G1 switch 2; FABP4, fatty acid binding protein 4; GLUT4, glucose transporter 4; LPL, lipoprotein lipase. F: Ectopic expression of miR-103 or miR-143 hastens expression of adipogenic markers at day 2 (D2). Data are expressed as mean ± SE (n = 4). ***P < 0.001, **P < 0.01, *P < 0.05 by one-sample Student's t test.
Coexpression of intronic miRNAs with “host” genes.
Some of the differentially regulated miRNAs originated from introns of known genes. For example, miR-422b is located in the sense orientation in the first intron of peroxisome proliferator–activated receptor (PPAR)-γ coactivator 1β (PPARGC1b). miR-107, miR-103-1, and miR-103-2 reside in the sense orientation in intron 5 of three Pantothenate kinase (PANK) gene family members: PANK1, PANK3, and PANK2, respectively. All of these intronic miRNAs were co-regulated with their respective host genes during 3T3-L1 adipogenesis (Fig. 1D).
miRNA expression in adipocytes from obese mice.
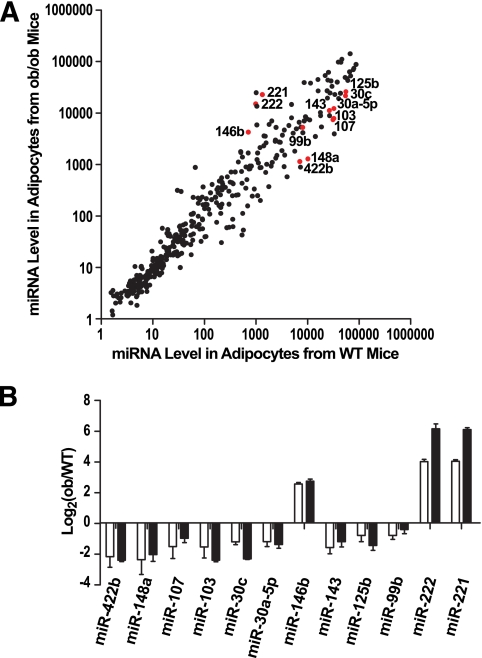
To identify candidate miRNAs associated with abnormal fat accumulation, the same miRNA microarrays were used to compare miRNA levels in normal adipocytes with those of obese mice. We first used leptin-deficient ob/ob mice; at 4 weeks of age, ob/ob mice are obese and exhibit hyperglycemia, glucose intolerance, and insulin resistance, phenotypes that resemble the pathophysiology of human type 2 diabetes (33). A total of 71 miRNAs were expressed at significantly different levels (Fig. 2A, P < 0.01, Student's t test). Twelve regulated miRNAs, including 9 downregulated and 3 upregulated in ob/ob mice, were validated by RT-PCR assays (Fig. 2B).
FIG. 2.
miRNA expression profiling in adipocytes from wild-type (WT) and leptin-deficient ob/ob mice. A: Intensity scatter plot showing comparison of miRNA profiles in adipocytes from WT and ob/ob mice. The 12 miRNAs that are the focus of our detailed analyses are highlighted in red and indicated by a label. B: Validation of miRNA array results for 12 regulated miRNAs by RT-PCR assays. Expression of all miRNAs are normalized to internal control and plotted as fold-change in a log2 scale. □, Array result (n = 2); ■, RT-PCR result (n = 2). Data are expressed as mean ± SE.
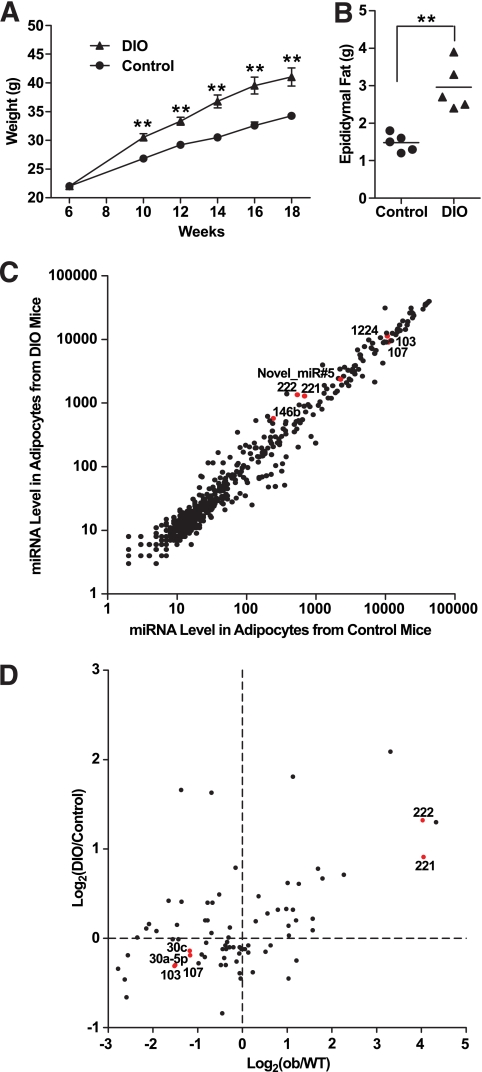
We also used DIO mice as a model of obesity. After being fed with a 55% high-fat diet for 3 months (from weeks 6 to 18), DIO mice became significantly heavier than normal mice and had almost twice the mass of epididymal fat compared with littermate controls (Fig. 3A and B, P < 0.01, Student's t test). Using an updated miRNA microarray capable of detecting 569 mouse miRNAs (miRbase v10.1) and 5 novel miRNAs cloned from adipose tissue by Kajimoto et al. (22) and Gu et al. (23) for this comparison, 35 miRNAs were expressed at significantly different levels (Fig. 3C, P < 0.01, Student's t test). Notably, the recently discovered mirtron miR-1224 and one novel miRNA cloned from bovine adipose tissue (Novel_miR#5: 5′-GGGCCGUGCGCGGGGUCUGC-3′) were expressed above the mean of expression of all miRNAs. Six miRNAs including four upregulated and two downregulated in DIO mice were selected for validation with RT-PCR assays; the results again agreed well with the microarray data (data not shown).
FIG. 3.
miRNA expression profiling in adipocytes from control and DIO mice. A: Weight of control and DIO mice (n = 5). DIO mice were fed with a 55% high-fat diet starting from week 6 to week 18. Controls were fed with a normal diet. Data are expressed as means ± SE. **P < 0.01. B: Weight of epididymal fat pads from control and DIO mice (n = 5) at week 18. **P < 0.01. C: Intensity scatter plot showing comparison of miRNA profiles in adipocytes from control and DIO mice. Mirtron miR-1224, Novel_miR#5 (novel miRNA cloned from bovine adipose tissue described in the text), and some miRNAs used in our detailed analyses are highlighted in red and indicated by a label. D: Scatter plot showing the positive correlation of miRNA regulation in two disease models for obesity: ob/ob and DIO mice. A total of 78 miRNAs, whose levels are above the mean expression of all miRNAs in at least one of the four samples (enriched adipocytes from wild-type [WT], ob/ob, control, and DIO mice), are shown. Fold-changes are based on array results and plotted in a log2 scale. Some miRNAs used in our detailed analyses are highlighted in red and indicated by a label. Pearson's correlation coefficient: r = 0.51, P < 0.0001.
To compare changes in miRNA expression between these two models of obesity, we focused on a set of 78 miRNAs, whose expression levels were above the mean expression of all miRNAs in at least one of the four samples analyzed: enriched adipocytes from wild-type and ob/ob mice (Fig. 2) and adipocytes from control and DIO mice (Fig. 3C). As shown in Fig. 3D, there was a strong positive correlation between changes in miRNA expression levels in the two obesity models (r = 0.51, P < 0.0001, Pearson's correlation). This suggested that similar sets of miRNAs are up- or downregulated in these two models of obesity, although the extent of regulation was usually more striking in adipocytes from ob/ob mice than DIO mice.
Inverse correlation of miRNA expression during adipogenesis and obesity.
Importantly, we noticed that miRNAs upregulated during adipogenesis tended to be downregulated in the obese state, and vice versa. For example, miR-422b, 148a, 107, 103, 30c, 30a-5p, and 143 were induced during adipogenesis but were downregulated in obese adipocytes. Conversely, miR-222 and -221 were decreased during adipogenesis but were upregulated in obese adipocytes (Figs. 1B and 2B).
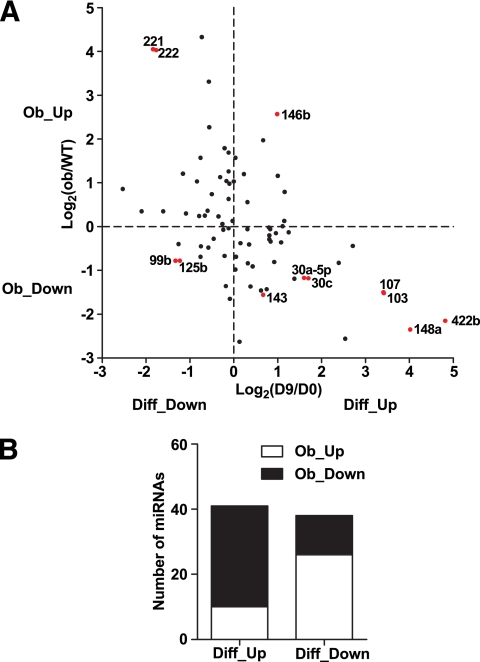
To test our observation more rigorously and on a larger scale, we focused on a set of 79 miRNAs, whose expression levels were above the mean expression of all miRNAs in at least one of the four samples analyzed: undifferentiated 3T3-L1 preadipocytes (day 0), differentiated 3T3-L1 adipocytes (day 9), and adipocytes from wild-type and ob/ob mice. The regulation patterns of these 79 miRNAs are shown as a scatter plot in Fig. 4A (r = −0.51, P < 0.0001, Pearson's correlation) and categorized in Fig. 4B. Among the 41 miRNAs upregulated during differentiation, 31 miRNAs decreased during obesity. In contrast, among the 38 miRNAs downregulated during differentiation, 26 miRNAs were elevated during obesity. The inverse correlation between miRNAs implicated in adipogenesis and obesity was statistically significant (Fig. 4B, P = 0.0001, Fisher's exact test).
FIG. 4.
Inverse correlation of miRNA expression during adipogenesis and obesity. A: Scatter plot showing the inverse correlation of miRNA regulation during adipogenesis and obesity. A total of 79 miRNAs, whose levels are above the mean expression of all miRNAs in at least one of the four samples (undifferentiated 3T3-L1 preadipocytes [day 0], differentiated 3T3-L1 adipocytes [day 9], and adipocytes from wild-type [WT] and ob/ob mice) are shown. Fold-change is based on array result and plotted in a log2 scale. The 12 miRNAs that are the focus of our detailed analyses are highlighted in red and indicated by a label. Pearson's correlation coefficient: r = −0.51, P < 0.0001. B: Fisher's exact test for 79 miRNAs is shown in Fig. 3A. A total of 41 miRNAs were upregulated during differentiation (Diff_Up), among which 31 miRNAs were downregulated during obesity (Ob_Down), whereas 10 miRNAs were upregulated (Ob_Up). A total of 38 miRNAs were downregulated during differentiation (Diff_Down), among which 26 miRNAs were upregulated during obesity (Ob_Up), whereas 12 miRNAs were downregulated (Ob_Down). P = 0.0001 by Fisher's exact test.
miRNA regulation by TNF-α in adipocytes from obese mice.
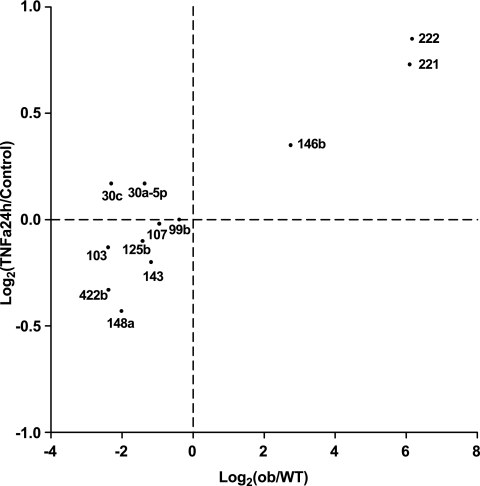
Chronic inflammation by macrophages is a principal feature of obese adipose tissue (3,34–36); TNF-α is a major macrophage-produced cytokine involved in chronic inflammation and is largely responsible for inducing insulin resistance in obese adipose tissue (37). Previously, we reported that in 3T3-L1 adipocytes, TNF-α treatment suppressed expression of many adipocyte-specific genes and reactivated expression of preadipocyte genes, inducing insulin resistance (30). Figure 5 shows, using quantitative RT-PCR assays, that treatment of differentiated 3T3-L1 adipocytes for 24 h with TNF-α reduced the expression of the same miRNAs, including miR-103 and miR-143, which were downregulated in adipose tissue from ob/ob mice. Conversely, TNF-α induced the expression of three miRNAs, including miR-221 and miR-222, which were normally downregulated during adipogenesis and upregulated during obesity. Thus, the changes in miRNA expression we observed in adipocytes from both ob/ob and DIO mice likely were caused by the enhanced expression of TNF-α seen in obese adipose tissue.
FIG. 5.
miRNAs differentially expressed in adipocytes from ob/ob mice compared with wild-type (WT) mice are regulated similarly by TNF-α treatment of 3T3-L1 differentiated adipocytes for 24 h. miRNA levels are measured by RT-PCR, normalized to internal control and plotted as fold-change in log2 scale. x-axis, ob/ob vs. WT, same data as Fig. 2B; y-axis, differentiated adipocyte after TNF-α treatment for 24 h versus untreated control (n = 4). Pearson's correlation coefficient: r = 0.90, P < 0.0001.
miR-103 and miR-143 accelerate adipogenesis when expressed ectopically.
To determine whether the changes in miRNA expression we observed might affect adipogenesis, we focused on two miRNAs, miR-103 and miR-143, which were upregulated during adipogenesis and downregulated during obesity. Both miRNAs are highly conserved and are abundant in adipocytes. The level of mature miR-143 increased slightly during 3T3-L1 adipogenesis, but the rise is much more dramatic in vivo (Figs. 6A and 1C). Expression of miR-103 was induced approximately ninefold during adipogenesis, with the increase occurring mainly after day 2 (Fig. 6A).
To evaluate the effects of these miRNAs on preadipocyte growth and differentiation, we used a retroviral vector to stably express them in 3T3-L1 preadipocytes. GFP was used as a reporter so that infected 3T3-L1 cells could be easily selected by fluorescence-activated cell sorting. We obtained four independent batches of miRNA overexpressing cells. GFP+ cells expressed three- to fourfold higher levels of mature miRNA than control preadipocytes, which expressed the empty vector alone (Fig. 6B). Thus, miR-103 and miR-143 were ectopically expressed at physiological levels.
Figure 6C shows that ectopic expression of miR-103 or miR-143 has little effect on 3T3-L1 cell growth. Without adipogenic stimuli, there were no evident morphological changes in 3T3-L1 cells ectopically expressing these miRNAs (data not shown). However, ectopic expression of miR-103 or miR-143 did accelerate the rate of 3T3-L1 differentiation, as measured by triglyceride accumulation (Fig. 6D) and by the upregulation of many adipocyte-important genes at early stages of adipogenesis (Fig. 6E). Figure 6D shows that cells ectopically expressing either miR-103 or miR-143 accumulated twice the normal level of triglyceride at day 4 of differentiation but a normal level of triglyceride at day 7.
The accelerated differentiation was accompanied by increased expression of key transcription factors such as PPARγ2, key cell cycle regulators such as G0/G1 switch 2 (G0s2), and molecules associated with lipid metabolism (FABP4), glucose homeostasis (GLUT4), and endocrine functions (adiponectin) of adipocytes. Quantitative RT-PCR showed that ectopic expression of miR-103 or miR-143 caused little change in the low level of expression of these genes before the onset of differentiation (day 0). Importantly, ectopic expression of miR-103 or miR-143 did hasten the expression of these pro-adipogenic factors at early stages of adipogenesis (Fig. 6E and F). At day 2, for example, expression of PPARγ2 was doubled by ectopic expression of either miR-103 or miR-143. miR-103 increased the expression level of FABP4 and adiponectin approximately ninefold and fourfold, respectively (Fig. 6F). Gene expression profiling by microarrays (data not shown) also suggested a significant enrichment of other proteins known to be important during adipogenesis or in lipid metabolism pathways in day 2 cells, in which miR-103 or miR-143 was ectopically expressed compared with control cells at day 2. Notably, neither miR-103 nor miR-143 modulated early adipogenic markers such as LPL at day 0 (Fig. 6E and data not shown). Levels of miR-103 and miR-143 are normally induced early in differentiation, and they likely downregulate expression of unknown mRNAs whose encoded protein(s) normally slow the process of adipogenesis.
DISCUSSION
In this study, we have generated a comprehensive database of the expression patterns of over 370 known mouse mature miRNAs during normal fat cell development and, for the first time, in adipocytes from normal and obese mice. Similar changes in miRNA expression occur during in vitro and in vivo adipogenesis. Many of these differentially regulated miRNAs are likely to be important in adipose biology, as we showed that ectopic expression of two normally upregulated miRNAs, miR-103 or miR-143, in preadipocytes accelerates the rate of fat cell formation. Importantly, miRNAs that were induced during adipogenesis were decreased in adipocytes from both types of obese mice and vice versa. We suggest that these changes are linked to the chronic local inflammation environment and enhanced TNF-α levels in obese adipose tissue, since similar changes in the pattern of miRNA expression occurred after TNF-α treatment of differentiated adipocytes. Our work provides an important first step toward construction of the entire RNA regulatory network underlying fat cell development and adipose dysfunction in obesity. An understanding of the role of miRNAs in adipose biology may lead to novel RNA-based therapies that complement current anti-obesity treatments.
Regulated expression of functionally important miRNAs.
Our genome-wide miRNA profiling study identified many differentially regulated miRNAs, including miR-103 and miR-143. miR-143 is likely to be important in adipocyte biology, since transfection of antisense miR-143 oligonucleotides in cultured human preadipocytes inhibits all five adipocyte differentiation markers by at least 40% (21). Our experiments ectopically expressing miR-143 resulted in the opposite effect, upregulation of adipogenesis markers, thus confirming its important role in modulating adipogenesis.
Wang et al. (38) reported that the miR-17-92 cluster is upregulated twofold only during the early clonal expansion stage of adipogenesis and that these miRNAs accelerate adipocyte differentiation by negatively regulating p130. Because we focused only on miRNAs upregulated during adipose differentiation per se, our microarray analyses did not identify the miR-17-92 cluster as significantly upregulated. We did not focus on miRNA changes during the clonal expansion phase in part because there are no primary cell samples corresponding to this stage. In addition, according to our microarray data, the miR-17-92 cluster is not abundantly expressed in differentiated adipocytes.
Importantly, we also studied purified primary adipocytes, which are more homogeneous than the unfractionated primary adipose tissue used in an earlier study (23); this enabled us to identify adipocyte-enriched miRNAs. Comparing the 44 miRNAs most highly expressed in epididymal adipocytes from normal mice with the 29 miRNAs most highly expressed in chondrocytes and osteoblasts (39), expression of 18 miRNAs, including let-7, miR-23, miR-26, and miR-30, are common to all three lineages. This suggests that they are functionally important for differentiation of all three cell types (adipocytes, chondrocytes, and osteoblasts) that are formed from mesenchymal stem cells (40). In contrast, miRNAs preferentially enriched in one particular mesenchymal lineage may have roles in cell fate determination and lineage differentiation of mesenchymal stem cells or be required for the function of these differentiated cells. As an example, miR-140 is enriched in osteoblasts and chondrocytes, and miR-140 was reported previously to be important for bone development by downregulating histone deacetylase 4 (41).
Downstream effectors and targets for miR-103 and miR-143.
A computational study predicted that the miRNA paralogs miR-103 and miR-107 affect multiple mRNA targets in pathways that involve cellular acetyl-CoA and lipid metabolism (42). Supplementary Tables S1 and S2 suggest many potentially important mRNA targets for miR-103 and miR-143, including several anti-adipogenic factors such as ARNT, FZD1, and RUNX1T1/ETO/MTG8, whose levels are normally downregulated during adipogenesis. These genes were selected based on meeting all of the following criteria: 1) expressed in 3T3-L1 cells at day 0 or day 2, 2) a lower mRNA expression level in day 2 cells in which miRNA is ectopically expressed compared with control cells at day 2, and 3) predicted targets based on TargetScan v4.2 (12,43).
Some targets not listed here may have decreased protein levels but unchanged mRNA levels; some may be predicted by computational softwares other than Targetscan. Importantly, miRNAs likely target many mRNAs (10,11), and defining one major target of either miRNA will be difficult and likely impossible. For example, many miR-143 targets shown in supplementary Table S2 are equally if not more interesting than MAPK7/ERK5, which was previously suggested as an miR-143 target (21). Validation of potential targets by luciferase reporter assays and Western blots is ongoing but beyond the scope of this article.
Functionally important adipocyte miRNAs are downregulated in obesity, likely because of macrophage infiltration and TNF-α.
Most strikingly, several miRNAs, including miR-103 and miR-143, exhibit inverse patterns of regulation during adipogenesis compared with those during obesity (Fig. 4A), indicating that obesity leads to a loss of miRNAs that characterize fully differentiated and metabolically active adipocytes. Earlier studies reported that many adipogenic genes whose expression increases during adipogenesis, displayed markedly decreased expression in adipocytes from epididymal fat pads of ob/ob and DIO mice (25–27).
In general, we observed a greater extent of miRNA dysregulation in adipocytes from ob/ob compared with DIO mice. In parallel, several groups reported a greater change of altered mRNA expression in adipocytes from ob/ob mice than DIO mice (25–27). This is likely because ob/ob mice have increased adiposity compared with DIO mice of the same age (data not shown). Alternatively, 3 months of a high-fat diet might not be long enough to induce the same changes as seen in adipocytes from ob/ob mice. Nevertheless, the positive correlation of miRNA regulation we observed in these two different models of obesity (Fig. 3D) suggests that the changes in the adipocyte miRNA expression profile in ob/ob mice cannot result from leptin deficiency alone.
More likely these changes are associated with the chronic inflammatory environment in obese adipose tissue. Increased expression of inflammation-related genes was found in isolated abdominal subcutaneous adipocytes and cultured stromal vascular cells from obese Pima Indians compared with nonobese control subjects (44,45). We hypothesize that one or more miRNAs upregulated upon TNF-α treatment of adipocytes are involved in destabilizing PPARγ or CCAAT/enhancer-binding protein α or another mRNA encoding an adipocyte-important transcription factor.
Ruan et al. (30) reported that TNF-α suppressed adipocyte-specific genes and activated expression of preadipocyte genes in 3T3-L1 cells. In parallel, we showed here that, in 3T3-L1 adipocytes, TNF-α repressed miRNAs that are normally upregulated during adipogenesis. Thus, elevated levels of TNF-α in obese adipose tissue are likely the cause of the changes in miRNA levels we observed in obesity. Our results (Fig. 5) also suggested that obesity had a much stronger impact on the expression of these miRNAs than did exposure of 3T3-L1 cells to TNF-α. These differences are probably due to the relatively long half-life of miRNAs and some delay in transcription in response to TNF-α treatment. Additionally, the chronic inflammatory state of obese adipose tissue likely depends on other inflammatory cytokines such as IL-6, leading to impaired adipose tissue function.
miRNAs are highly connected nodes in regulatory networks underlying adipogenesis and adipose dysfunction in obesity.
Only a small fraction of known miRNAs is significantly regulated during adipogenesis (Fig. 1A). However, each miRNA is thought to regulate, on average, about 200 target genes and has widespread impact on protein output (10,11). Multiple miRNAs can act additively or synergistically at multiple target sites on a single mRNA (46). The potential interaction networks connecting miRNAs and mRNAs are enormous and can be further expanded by feedback or feed-forward loops (47). Whereas miR-103 and miR-143 have been suggested to be important for adipogenesis, little is known about the specific mRNA targets of these miRNAs. Their effects on adipogenesis could be achieved by significant downregulation of one or two “primary” target mRNAs or by more modest downregulation of perhaps hundreds of preadipocyte-important mRNAs. Clearly, we need to identify biologically relevant targets for these key miRNAs. In the reconstructed regulatory network, each miRNA will likely link to multiple target genes and serve as a controlling point and potential target for therapeutic intervention.
Supplementary Material
ACKNOWLEDGMENTS
H.X. was supported by a graduate fellowship from the Singapore-MIT Alliance (SMA). This work was supported by grant C-382-641-001-091 (to B.L. and H.F.L.) from SMA. B.L. was also supported by National Institutes of Health (NIH) grants DK047636 and AI054973, and H.F.L. was also supported by NIH grants DK047618 and DK068348 and grant 5P01 HL066105.
No potential conflicts of interest relevant to this article were reported.
We thank D. Bartel, B. Zhou, I.-H. Shih, M. Fang, and W. Han for their intellectual support; G.W. Wong, J.S. Bogan, S. Wang, H. Ruan, G. Paradis, and M. Le for assistance with techniques and reagents; F. Reinhardt for assistance with animal experiments; G. Bell, B. Yuan, and K. Gurdziel for assistance with microarray data analysis; M. Fang, B. Zhou, A. Kotani, and M. Lam for critical reading of the manuscript; and members of the Lodish and Lim Laboratories for useful discussions.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Rosen ED, Spiegelman BM: Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006; 444: 847– 853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajala MW, Scherer PE: Minireview: the adipocyte: at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology 2003; 144: 3765– 3773 [DOI] [PubMed] [Google Scholar]
- 3.Schaffler A, Muller-Ladner U, Scholmerich J, Buchler C: Role of adipose tissue as an inflammatory organ in human diseases. Endocr Rev 2006; 27: 449– 467 [DOI] [PubMed] [Google Scholar]
- 4.Calle EE, Kaaks R: Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer 2004; 4: 579– 591 [DOI] [PubMed] [Google Scholar]
- 5.Kopelman PG: Obesity as a medical problem. Nature 2000; 404: 635– 643 [DOI] [PubMed] [Google Scholar]
- 6.Feve B: Adipogenesis: cellular and molecular aspects. Best Pract Res Clin Endocrinol Metab 2005; 19: 483– 499 [DOI] [PubMed] [Google Scholar]
- 7.Rosen ED, Spiegelman BM: Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol 2000; 16: 145– 171 [DOI] [PubMed] [Google Scholar]
- 8.Rosen ED, MacDougald OA: Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7: 885– 896 [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP: MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281– 297 [DOI] [PubMed] [Google Scholar]
- 10.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N: Widespread changes in protein synthesis induced by microRNAs. Nature 2008; 455: 58– 63 [DOI] [PubMed] [Google Scholar]
- 11.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP: The impact of microRNAs on protein output. Nature 2008; 455: 64– 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis BP, Burge CB, Bartel DP: Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15– 20 [DOI] [PubMed] [Google Scholar]
- 13.Kluiver J, Kroesen BJ, Poppema S, van den Berg A: The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia 2006; 20: 1931– 1936 [DOI] [PubMed] [Google Scholar]
- 14.Gao FB: Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci 2008; 31: 20– 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callis TE, Deng Z, Chen JF, Wang DZ: Muscling through the microRNA world. Exp Biol Med (Maywood) 2008; 233: 131– 138 [DOI] [PubMed] [Google Scholar]
- 16.Krutzfeldt J, Stoffel M: MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab 2006; 4: 9– 12 [DOI] [PubMed] [Google Scholar]
- 17.Teleman AA, Maitra S, Cohen SM: Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev 2006; 20: 417– 422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Vernooy SY, Guo M, Hay BA: The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 2003; 13: 790– 795 [DOI] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP: miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006; 3: 87– 98 [DOI] [PubMed] [Google Scholar]
- 20.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M: Silencing of microRNAs in vivo with ‘antagomirs.’ Nature 2005; 438: 685– 689 [DOI] [PubMed] [Google Scholar]
- 21.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R: MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 2004; 279: 52361– 52365 [DOI] [PubMed] [Google Scholar]
- 22.Kajimoto K, Naraba H, Iwai N: MicroRNA and 3T3–L1 pre-adipocyte differentiation. RNA 2006; 12: 1626– 1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, Eleswarapu S, Jiang H: Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett 2007; 581: 981– 988 [DOI] [PubMed] [Google Scholar]
- 24.Hackl H, Burkard TR, Sturn A, Rubio R, Schleiffer A, Tian S, Quackenbush J, Eisenhaber F, Trajanoski Z: Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol 2005; 6: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD: The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci U S A 2000; 97: 11371– 11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soukas A, Cohen P, Socci ND, Friedman JM: Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev 2000; 14: 963– 980 [PMC free article] [PubMed] [Google Scholar]
- 27.Moraes RC, Blondet A, Birkenkamp-Demtroeder K, Tirard J, Orntoft TF, Gertler A, Durand P, Naville D, Begeot M: Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology 2003; 144: 4773– 4782 [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Ambrosi J, Catalan V, Diez-Caballero A, Martinez-Cruz LA, Gil MJ, Garcia-Foncillas J, Cienfuegos JA, Salvador J, Mato JM, Fruhbeck G: Gene expression profile of omental adipose tissue in human obesity. FASEB J 2004; 18: 215– 217 [DOI] [PubMed] [Google Scholar]
- 29.Bogan JS, McKee AE, Lodish HF: Insulin-responsive compartments containing GLUT4 in 3T3–L1 and CHO cells: regulation by amino acid concentrations. Mol Cell Biol 2001; 21: 4785– 4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF: Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3–L1 adipocytes: nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes 2002; 51: 1319– 1336 [DOI] [PubMed] [Google Scholar]
- 31.Zhu Q, Hong A, Sheng N, Zhang X, Matejko A, Jun KY, Srivannavit O, Gulari E, Gao X, Zhou X: microParaflo biochip for nucleic acid and protein analysis. Methods Mol Biol 2007; 382: 287– 312 [DOI] [PubMed] [Google Scholar]
- 32.Chen CZ, Li L, Lodish HF, Bartel DP: MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303: 83– 86 [DOI] [PubMed] [Google Scholar]
- 33.Tschop M, Heiman ML: Rodent obesity models: an overview. Exp Clin Endocrinol Diabetes 2001; 109: 307– 319 [DOI] [PubMed] [Google Scholar]
- 34.Wellen KE, Hotamisligil GS: Obesity-induced inflammatory changes in adipose tissue. J Clin Invest 2003; 112: 1785– 1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H: Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003; 112: 1821– 1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW, Jr: Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003; 112: 1796– 1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cawthorn WP, Sethi JK: TNF-alpha and adipocyte biology. FEBS Lett 2008; 582: 117– 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X: miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A 2008; 105: 2889– 2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM: Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A 2008; 105: 1949– 1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR: Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284: 143– 147 [DOI] [PubMed] [Google Scholar]
- 41.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T: The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 2006; 580: 4214– 4217 [DOI] [PubMed] [Google Scholar]
- 42.Wilfred BR, Wang WX, Nelson PT: Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 2007; 91: 209– 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP: MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007; 27: 91– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YH, Nair S, Rousseau E, Allison DB, Page GP, Tataranni PA, Bogardus C, Permana PA: Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia 2005; 48: 1776– 1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair S, Lee YH, Rousseau E, Cam M, Tataranni PA, Baier LJ, Bogardus C, Permana PA: Increased expression of inflammation-related genes in cultured preadipocytes/stromal vascular cells from obese compared with non-obese Pima Indians. Diabetologia 2005; 48: 1784– 1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N: Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495– 500 [DOI] [PubMed] [Google Scholar]
- 47.Shalgi R, Lieber D, Oren M, Pilpel Y: Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 2007; 3: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.