SUMMARY
Selenocysteine (Sec) is incorporated into proteins in response to UGA codons. This residue is frequently found at the catalytic sites of oxidoreductases. In the present study, we characterized the selenoproteome of an anaerobic bacterium, Clostridium sp. (also known as Alkaliphilus oremlandii) OhILA, and identified 13 selenoprotein genes, 5 of which have not been previously described. One of the detected selenoproteins was methionine sulfoxide reductase A (MsrA), an antioxidant enzyme that repairs oxidatively damaged methionines in a stereospecific manner. To date, little is known about MsrA from anaerobes. We characterized this selenoprotein MsrA which had a single Sec residue at the catalytic site but no cysteine (Cys) residues in the protein sequence. Its SECIS (Sec insertion sequence) element did not resemble those in Escherichia coli. Although with low translational efficiency, the expression of the Clostridium selenoprotein msrA gene in E. coli could be demonstrated by 75Se metabolic labeling, immunoblot analyses, and enzyme assays, indicating that its SECIS element was recognized by the E. coli Sec insertion machinery. We found that the Sec-containing MsrA exhibited at least a 20-fold higher activity than its Cys mutant form, indicating a critical role of Sec in the catalytic activity of the enzyme. Furthermore, our data revealed that the Clostridium MsrA was inefficiently reducible by thioredoxin, which is a typical reducing agent for MsrA, suggesting the use of alternative electron donors in this anaerobic bacterium that directly act on the selenenic acid intermediate and do not require resolving Cys residues.
Keywords: MsrA, selenoproteins, oxidoreductase, SECIS elements, thioredoxin
Introduction
Selenocysteine (Sec) is inserted into proteins when UGA (normally a stop codon) is recoded to serve as Sec codon 1,2. This amino acid is typically found at the catalytic sites of oxidoreductase selenoproteins and is responsible for high catalytic activity of selenoenzymes as compared with their cysteine (Cys)-containing homologs 3-6. Selenoproteins are found in all three domains of life: bacteria, archaea and eukaryotes. The glycine reductase A from Clostridium sticklandii was discovered as the first selenoprotein 7,8. A complex machinery is required for translational insertion of Sec in response to UGA codon 2,9,10. A stem-loop structure in selenoprotein mRNAs, called SECIS (Sec insertion sequence) element, is essential for Sec insertion in both prokaryotes and eukaryotes. SECIS elements are located immediately downstream of Sec-encoding UGA codons in bacteria, whereas they are present in 3′-untranslational regions in archaea and eukaryotes. In addition to this cis-acting factor, several trans-acting factors, such as Sec-specific elongation factor and tRNASec, are required for Sec incorporation.
There has been significant progress recently in identifying selenoprotein genes in organisms from bacteria to mammals owing to both a dramatic increase in genomic sequence information and development of bioinformatics methods that allow searches for selenoprotein genes11-16. These methods include searches for 1) SECIS elements; and 2) Sec/Cys pairs in predicted homologs.
Methionine residues are easily oxidized by various reactive oxygen species to methionine sulfoxides; however, the oxidized methionines can be reduced back to methionine by repair enzymes, the methionine sulfoxide reductases (Msrs) 17. Two distinct families of these enzymes, MsrA and MsrB, have evolved for stereospecific reduction of methionine sulfoxide residues in proteins. MsrA is specific for the reduction of the S-form of methionine sulfoxide, whereas MsrB is specific for the R-form. Msrs are present in most organisms in all three domains of life (they have been even found in anaerobic bacteria), but are absent in some hyperthermophiles and parasites 18,19. Msrs play a pivotal role in the repair of oxidatively damaged proteins and defend cells from oxidative stress. Therefore, these proteins are viewed as protein repair and antioxidant enzymes 20-22. These enzymes have been implicated in regulation of physiological and pathological processes such as aging and neurodegenerative diseases 23,24.
In this study, we characterized the selenoproteome of an anaerobic gram-positive bacterium, Clostridium sp. (also known as Alkaliphilus oremlandii) OhILAs and found that this bacterium is predicted to be rich in selenoproteins and may contain at least 13 selenoproteins, including MsrA. We further experimentally characterized Clostridium selenoprotein MsrA. We found that its SECIS element on the mRNA is recognized by the Escherichia coli Sec insertion machinery, and that Sec plays a critical role in the catalytic activity of this selenoprotein.
Materials and Methods
Identification of selenoprotein genes in Clostridium sp. OhILAs
The complete genome of Clostridium sp. OhILAs (NC_009922) was downloaded from the NCBI ftp server (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). We used bSECISearch program developed previously 15 to search this genome for all candidate selenoprotein sequences containing in-frame UGA codons and bacterial SECIS (bSECIS) elements. The resulting dataset was then divided into homologs of previously known selenoproteins and selenoprotein candidates. All candidates were manually analyzed for the location of the UGA codon, occurrence of Sec- and Cys-containing homologs in Sec-utilizing or other organisms, and presence of SECIS elements in Sec-containing homologs. In parallel, independent BLAST homology searches for Sec-containing homologs of all known selenoprotein families were performed using default parameters. Finally, both known and candidate selenoprotein sets were generated. Multiple sequence alignments were performed using ClustalW program 25 with default parameters and visualized with BoxShade program v3.21.
Cloning, expression, and purification of wild-type selenoprotein and its Cys mutant of Clostridium MsrA
A coding region of the selenoprotein msrA gene was amplified by PCR using Clostridium sp. OhILAs genomic DNA (kindly provided from Dr. John Stolz at the Duquesne University, Pittsburgh, USA) and cloned into NdeI/XhoI sites of pET21b (Novagen). To amplify the gene, forward (5′-GCGCCATATGGATACCAATCAGAAGTTG-3′) and reverse (5′-CGCGCTCGAGTTTAATTTCTTCGACGATAG-3′) primers were used. The resulting construct, designated pET-CLOS-MsrA, coded for full-length selenoprotein MsrA with a C-terminal His-tag (LEHHHHH). A SECIS element was found to be located immediately downstream of UGA Sec codon (see Figure 4). A Cys mutant form was also generated in which Sec16 was replaced with Cys by site-directed mutagenesis. The resulting construct was named pET-CLOS-MsrA/U16C.
Figure 4.
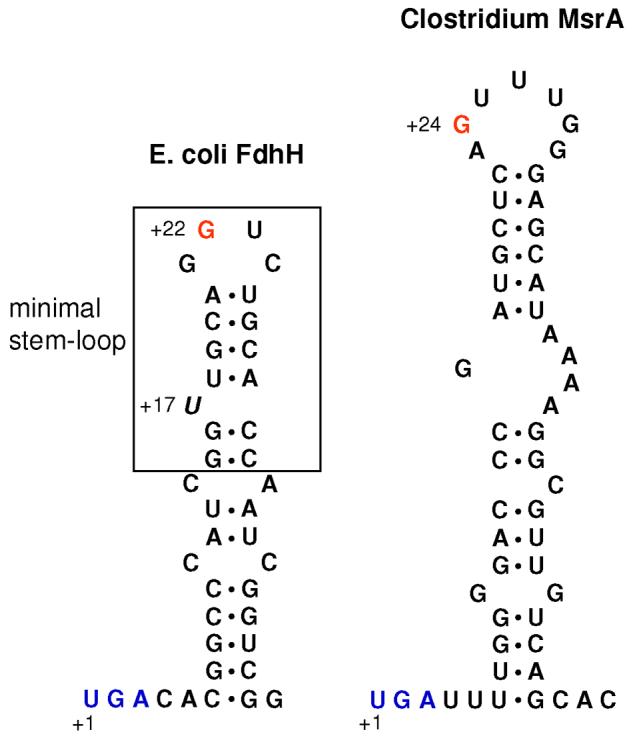
Structures of SECIS elements in E. coli fdhH (left) and Clostridium sp. OhILAs msrA (right). In the E. coli fdhH SECIS element, the minimal step-loop structure is boxed and the bulged U is shown in italics. The Clostridium msrA SECIS element lacks the bulged U. Conserved G in the apical loops is shown in red in both SECIS elements. Sec UGA codons are shown in blue.
To express selenoprotein msrA in E. coli, the plasmid pET-CLOS-MsrA was transformed into BL21(DE3) cells that also harbored a plasmid pSUABC 26, which encodes E. coli selA, selB, and selC. The transformed cells were grown in LB media containing 4 μM sodium selenite, 100 μg/ml ampicillin, and 25 μg/ml chloramphenicol at 37°C with shaking until optical density at 600 nm reached 0.6∼0.8; then 0.2 mM IPTG was added and the cells were further cultured for 4 h at 30°C, harvested, and stored at −20°C until use. A typical expression level of the selenoprotein MsrA was 0.6 μg per mg crude protein.
The cell pellets were resuspended in the extraction buffer (50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 10 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride) and lysed by sonication. The selenoprotein MsrA was further purified using a Talon-metal affinity resin (Clontech) according to the manufacturer's protocol. The eluted proteins were dialyzed against 50 mM sodium phosphate, pH 7.5, and 50 mM NaCl. Typical yield of the purified selenoprotein MsrA was 3-5 μg from 250 ml culture broth, as estimated by Western blot assays. Although the protein was not pure, this preparation was sufficient to determine kinetic parameters of the enzyme.
To express the Cys mutant form (U16C), BL21(DE3) cells transformed with pET-CLOS-MsrA/U16C were cultivated in LB media containing 100 μg/ml ampicillin at 37°C with shaking until optical density at 600 nm reached 0.6∼0.8, 0.1 mM IPTG was added, and the cells were further cultured for 6 h at 30°C. The cells were harvested and stored at −20°C until use. Procedures for purification of this protein were as described above for the selenoprotein form. The purity of the Cys mutant protein was analyzed by SDS-PAGE and found to exceed 95%.
Metabolic labeling with 75Se
To prepare cells expressing Clostridium selenoprotein MsrA with metabolically labeled 75Se, E. coli BL21(DE3) cells were transformed with pET-CLOS-MsrA and grown at 37°C in 5 ml LB media containing ampicillin until optical density at 600 nm reached ∼0.6. Then, 0.05 mCi of freshly neutralized [75Se]selenous acid (specific activity 1,000 Ci/mmol, University of Missouri Research Reactor, Columbia, USA) was added to the cell culture with 1 mM IPTG for induction. The cells were further grown at 37°C for 5 h, harvested, washed with PBS buffer and lysed. Cell extracts (40 μg of total protein) were applied to a 10% Bis-Tris gel (Invitrogen), electrophoresed, and transferred onto a PVDF membrane. The 75Se radioactivity pattern on the membrane was visualized by using a PhosphorImager (GE Health Care).
Determination of protein concentration
Concentration of recombinant selenoprotein MsrA was determined by western blotting using His antibodies and Cys mutant as an internal standard, followed by quantifying the blot signals with a densitometer. Concentration of purified Cys mutant was determined by Bradford method using a Bio- Rad protein assay reagent (Bio-Rad) and bovine serum albumin as a standard.
MsrA enzyme assay and analysis of kinetics
MsrA activity was measured in the presence of dithiothreitol (DTT) or thioredoxin (Trx) as a reducing agent. In the DTT-dependent reaction, reaction mixture (100 μl) contained 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 20 mM DTT, 200 μM DABSyl-methionine-S-sulfoxide, and purified (or crude) proteins. In the Trx-dependent reaction, the reaction mixture (100 μl) contained 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 0.2 mM NADPH, 6.8 μM E. coli Trx (Sigma) or human Trx1 27, 0.8 μM human Trx reductase 1 27, 200 μM DABSyl-methionine-S-sulfoxide, and purified proteins. The reactions were carried out at 37°C for 30 min and analyzed by HPLC as described previously 28. A full-length mouse MsrA was purified from E. coli as described previously 29 and used for comparison.
Km and kcat values were determined from Lineweaver-Burk plots using 50-400 μM substrate in the DTT-dependent reaction.
Results and Discussion
Clostridium sp. OhILAs is a selenoprotein-rich organism
By searching the Clostridium sp. OhILAs genome against a set of all known selenoprotein genes, we identified 8 selenoproteins in Clostridium sp. OhILAs, including formate dehydrogenase alpha subunit (FdhA), which is the most widespread prokaryotic selenoprotein that might be responsible for maintaining the Sec utilization trait in sequenced bacteria 30, and selenophosphate synthetase (SelD), another widespread selenoprotein which is a key component in prokaryotic selenoprotein biosynthesis. Other detected selenoproteins included proline reductase, HesB-like protein, glutaredoxin, glycine reductase selenoproteins A (GrdA) and B (GrdB), and MsrA. Previously, selenoprotein MsrA has only been characterized in a eukaryotic organism, Chlamydomonas reinhardtii 31,32. The selenoproteome of Clostridium sp. OhILAs is shown in Table 1 and all detected selenoprotein sequences are included in the supplementary sequence data.
Table 1.
Known and new selenoproteins detected in Clostridium sp. OhILAs
| Protein family | Accession no. | COG/Pfam | Occurrence | Sec position (protein length) |
|---|---|---|---|---|
| Known selenoproteins | ||||
| FdhA* | YP_001513778 | COG3383 | 1 | 349 (891) |
| SelD* | YP_001513274 | COG0709 | 1 | 16 (346) |
| MsrA* | YP_001513481 | COG0225 | 1 | 16 (209) |
| Proline reductase* | YP_001511596 | pfam07355 (low homology) |
1 | 151 (241) |
| HesB-like |
YP_001511838 (partial sequence) |
COG0316 | 1 | 32 (95) |
| Glutaredoxin* | YP_001513661 | COG0695 | 1 | 13 (76) |
| GrdA* | YP_001512500 | pfam04723 | 1 | 44 (158) |
| GrdB* | YP_001512501 | pfam07355 | 1 | 350 (435) |
| New selenoprotein candidates | ||||
| Putative anaerobic dehydrogenase* | YP_001511765 | COG0243 | 1 | 257 (1021) |
| BFD-like (2Fe-2S)-binding domain protein |
YP_001512630 (partial sequence) |
COG2906 (low homology) |
1 | 11 (162) |
| Split soret cytochrome c precursor* |
YP_001512574 YP_001512575 |
pfam09719 (low homology) |
2 | 83 (223) 81 (221) |
| Predicted NADH:ubiquinone oxidoreductase, subunit RnfC |
YP_001511593 (partial sequence) |
COG4656 | 1 | 389 (424) |
annotated as selenoproteins in the current genome annotation
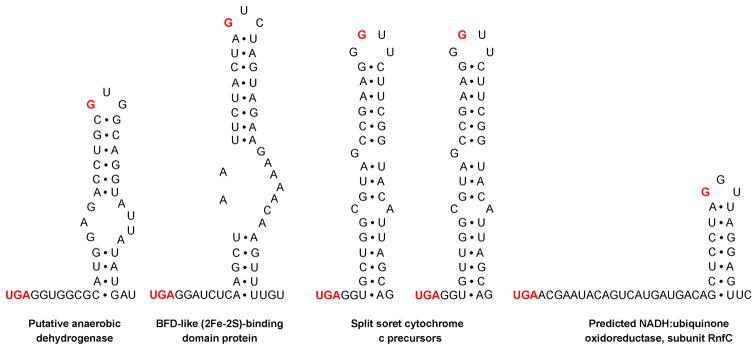
Next, we used bSECISearch to analyze Clostridium sp. OhILAs for full complement of selenoprotein genes. In addition to the 8 homologs of known selenoproteins, we detected 5 new selenoproteins that represented 4 protein families (Table 1). The mRNAs for all new selenoproteins contained predicted stable stem-loop structures downstream of Sec-encoding UGA codons that shared common features of bacterial SECIS elements 15, e.g., a stable stem with a small apical loop containing a G nucleotide located at a certain distance from the Sec UGA codon (Figure 1). No additional common features could be found in those Clostridium SECIS elements and all of them were predicted to lack a bulged U which is conserved in E. coli SECIS elements. Selenoprotein homologs of the new selenoproteins were also found in several other anaerobic bacteria, such as Desulfitobacterium hafniense, Alkaliphilus metalliredigens, and Clostridium botulinum (see Figure 2). In addition, conserved Cys-containing homologs could be detected for all new selenoproteins in other organisms. Multiple alignments of new selenoprotein families and their Cys homologs are shown in Figure 2 and highlight conserved Sec/Cys pairs in homologous sequences that support predicted redox functions of these proteins 33.
Figure 1.
Predicted bacterial SECIS elements in new selenoprotein genes. Only sequences downstream of in-frame UGA codons are shown. In-frame UGA codons and conserved G nucleotides in the apical loop are shown in red.
Figure 2.
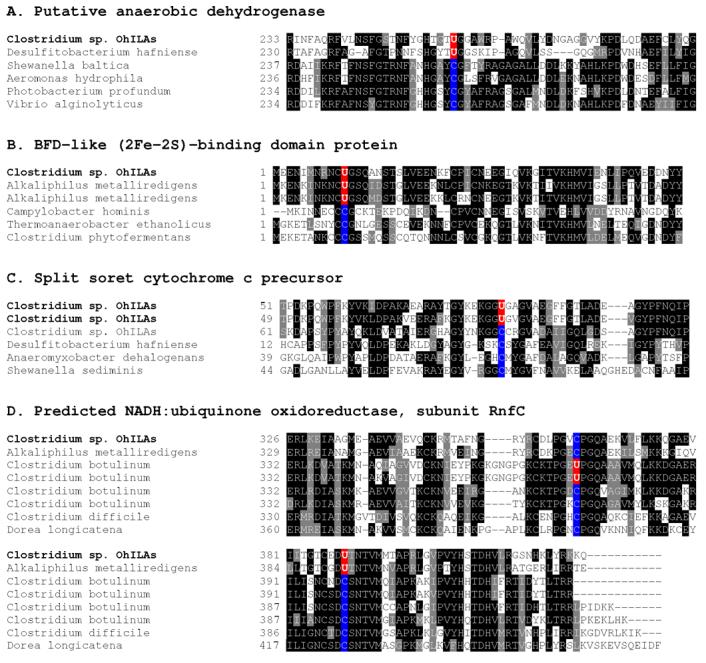
Multiple sequence alignments of new selenoproteins and their Cys homologs. The alignments show Sec-flanking regions in detected selenoproteins. Both selenoprotein sequences detected in Clostridium sp. OhILAs and their Sec/Cys-containing homologs present in indicated organisms are shown. Conserved residues are highlighted. Predicted Sec (U) and the corresponding Cys (C) residues in other homologs are shown in red and blue, respectively.
Although the functions of new selenoproteins are not clear, they either contained a domain of known function or were homologous to protein families with known functions. However, in some of them Sec was not part of the detected domains, suggesting the presence of functional redox-active sites in the locations outside of known domains. For example, Sec was located in the N-terminal region of BFD-like (2Fe-2S)-binding protein in which the BFD-like domain (COG2906) was present in the C-terminal region. It was also found in a CxxU (x, any amino acid residue; U, Sec) redox motif in the C-terminal region of a predicted NADH:ubiquinone oxidoreductase subunit RnfC, whereas the known RnfC domain (COG4656) was located in the N-terminal region. The other three new selenoproteins (putative anaerobic dehydrogenase and two cytochrome c precursor sequences) were annotated as putative selenoproteins in this genome although the criteria used in their detection/annotation are not clear. Since these sequences have passed the stringent criteria employed by bSECISearch in our study, they should be viewed as excellent candidate selenoproteins.
While this study was prepared for publication, the sequences of 3 new selenoproteins were released at GenBank database (putative anaerobic dehydrogenase, 158139457; split soret cytochrome c precursor 1, 158320067; split soret cytochrome c precursor 2, 158320068). It is interesting that the Sec positions differed in the predicted NADH:ubiquinone oxidoreductase subunit RnfC family between Clostridium sp. OhILAs and several strains of Clostridium botulinum (Figure 2). Taken together, our data suggested that novel Se-dependent pathways are present in this selenoprotein-rich organism. Further experimental verification is needed for the newly identified selenoproteins.
Clostridium selenoprotein MsrA sequence
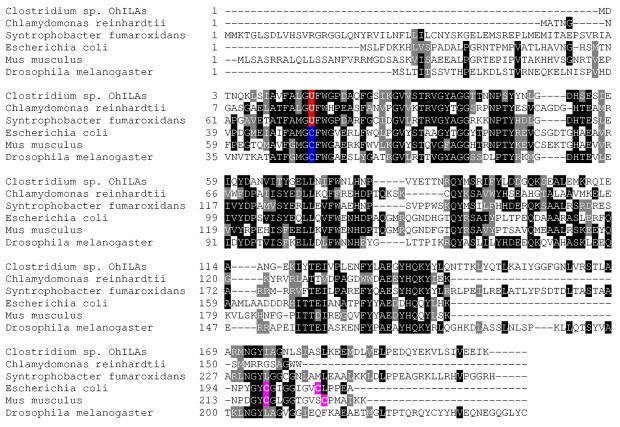
Msr genes have been found in most organisms, even in anaerobes. The presence of Msr genes in anaerobic bacteria may be due to their roles in protein repair and antioxidant defense when transiently exposed to oxygen. To date, little is known about such anaerobe-based Msrs. We selected the anaerobic bacterial selenoprotein MsrA for further experimental examination. Interestingly, sequence context revealed lack of any Cys residues in this protein. Since Sec corresponded to the catalytic residue, this observation suggested the lack of resolving residues during catalysis (Figure 3). The catalytic mechanism of Cys-containing MsrA is well characterized and involves a sulfenic acid intermediate 34-36. During the reaction, the catalytic Cys attacks sulfoxide moiety of methionine sulfoxide resulting in sulfenic acid on this residue with a concomitant release of product, methionine. Then, the sulfenic acid intermediate is rearranged into a disulfide bond by reacting with another Cys, known as resolving (or recycling) Cys, and finally the disulfide bond is reduced by a two-electron reductant. Trx is typically an in vivo reducing agent while DTT can be used in vitro. In the case of Sec-containing forms, the selenenic acid intermediate may form a selenenylsulfide bond with a resolving Cys; however, neither of the previously identified selenoprotein MsrAs possesses conserved Cys that could function as resolving residues 31. Furthermore, we previously reported that Chlamydomonas selenoprotein MsrA does not use a resolving Cys residue 31.
Figure 3.
Multiple sequence alignment of MsrAs. Sec (U) residues are highlighted in red in the catalytic GUFW(G/H) motif and the corresponding Cys residues are highlighted in blue. The known resolving Cys residues in the C-terminal region are highlighted in pink. Genbank accession numbers are as follows: Clostridium sp. OhILAs, 106894182; Chlamydomonas reinhardtii, 23452038; Syntrophobacter fumaroxidans, 71547479; E. coli, 1790665; Mus musculus, 31981013; Drosophila melanogaster, 23338220.
Expression of Clostridium selenoprotein MsrA in E. coli
The mechanism of Sec insertion has been well elucidated in E. coli 3,37-39. The SECIS element in mRNA is essential for Sec insertion and is located immediately downstream of in-frame UGA codons in bacteria. Studies on E. coli SECIS elements revealed that only a minimal stem-loop is required for Sec insertion in which a GU sequence on the tip of apical loop and a bulged U present at the bottom of the stem are essential for Sec insertion (Figure 4, left) 40-42. We recently reported a consensus bacterial SECIS structural model in which a conserved G is present in the apical loop, whereas the bulged U is an E. coli adaptation that is missing in most other bacterial SECIS elements 15. Consistent with this consensus, the SECIS element of Clostridium msrA had a G in the apical loop, but not the bulged U in the stem (Figure 4, right). The differences between E. coli SECIS elements and corresponding structures in other bacteria result in barriers in heterologous selenoprotein expression 43.
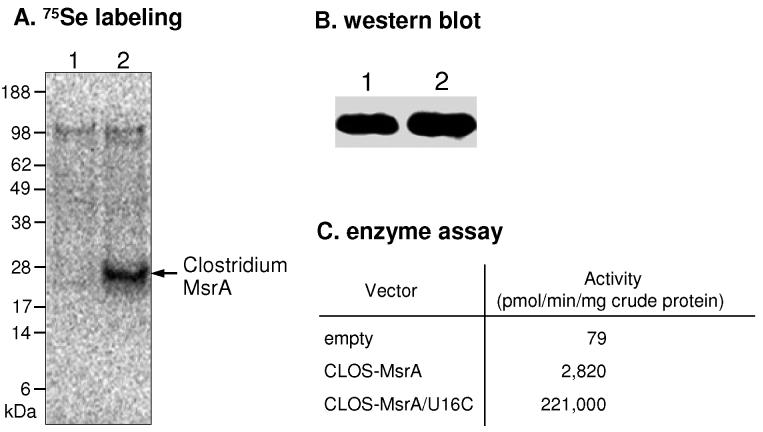
To test whether Clostridium selenoprotein MsrA can be expressed in E. coli as recombinant protein, we metabolically labeled cells transformed with Clostridium msrA gene with 75Se. As shown in Figure 5A, a 25 kDa radioactive band corresponding in size to the calculated molecular weight of recombinant Clostridium selenoprotein MsrA, was detected. We also verified the expression of this protein by western blot analysis (Figure 5B). In addition, enzyme assays revealed that crude extracts from cells expressing the Clostridium msrA gene had a 36-fold increased activity compared with that from cells containing an empty vector (Figure 5C). Thus, these data showed that Clostridium selenoprotein msrA could be expressed in E. coli, indicating that its SECIS element is recognized by the E. coli Sec insertion machinery. To estimate the efficiency in Sec insertion (UGA readthrough), we analyzed the amounts of wild-type and Cys mutant (U16C) forms of the enzyme in crude extracts by Western blotting using the purified U16C as an internal standard. It should be noted that most of U16C protein (>95%) expressed in BL21(DE3) cells was present in the soluble fraction. The yields of the wild-type selenoprotein and the U16C mutant were 0.6 and 380 μg per mg crude protein, respectively. Therefore, the translational efficiency of the selenoprotein MsrA was low, estimated to be 0.15% of that of its Cys variant. The activities of the wild-type selenoprotein and Cys mutant forms in the crude extracts as shown in Figure 5C agreed well with those of the purified forms, respectively (Table 2).
Figure 5.
Expression of Clostridium selenoprotein MsrA in E. coli. (A) 75Se metabolic labeling. BL21(DE3) cells transformed with an empty vector (lane 1) or pET-CLOS-MsrA (lane 2) were metabolically labeled with 75Se. Proteins were separated by SDS-PAGE, transferred onto a PVDF membrane, and visualized with a PhosphorImager. (B) Western blot analysis. Purified Cys mutant (U16C) and crude extracts from cells expressing Clostridium selenoprotein MsrA were analyzed using anti-His antibodies. Lane 1, purified U16C; lane 2, wild-type selenoprotein MsrA from cell extracts. (C) Enzyme assays. Crude extracts from cells containing an empty vector, pET-CLOS-MsrA (wild-type), or pET-CLOS-MsrA/U16C (U16C mutant) were prepared, respectively. A total of 200 μg crude protein from control and CLOS-MsrA extracts and 2 μg crude protein of the CLOS-MsrA/U16C extract were used in a DTT-dependent assay in the presence of 200 μM DABSyl-methionine-S-sulfoxide.
Table 2.
Specific activity and kinetic constants of Clostridium MsrA forms
| MsrA forms | Specific activity (nmol/min/mg protein) |
Km (mM) |
kcat (s−1) |
kcat/ Km (M−1 S−1) |
|---|---|---|---|---|
| WT | 4,185 ± 46 | 10.2 ± 1.2 | 84 ± 5 | 8,235 |
| U16C | 464 ± 14 | 4.0 ± 0.2 | 3.8 ± 0.3 | 950 |
Purified proteins (90 ng WT and 1 μg U16C) were assayed in the DTT-dependent reaction. The substrate (DABSyl-methionine-S-sulfoxide) concentration used was 200 μM for determination of specific activity. Km and kcat values were determined from Lineweaver-Burk plots using 50-400 μM substrate in the presence of DTT. WT, wild-type selenoprotein; U16C, Sec to Cys mutant.
There have been several reports on heterologous expression of eukaryotic selenoproteins in E. coli6,26,44,45. Either the entire SECIS element of E. coli fdhH (as in most studies) or a minimal SECIS stem-loop had to be introduced downstream of UGA codon to express eukaryotic selenoproteins. In E. coli, heterologous expression of bacterial selenoproteins that contain their own SECIS elements often fails due to SECIS structure barriers 46. However, a few successful cases have been previously reported 47,48. In our present study, irrespective of discrepancies in Clostridium msrA and E. coli SECIS elements, the Clostridium selenoprotein msrA gene could be expressed in E. coli. However, as discussed above, the translational efficiency was low (less than 0.2% compared to its Cys variant) even when the cells were co-expressed with selA, selB, and selC genes encoding selenocysteine synthase, SelB and tRNASec, respectively. These results suggest a low functionality of the SECIS element of Clostridium msrA in the E. coli Sec insertion system.
Characterization of Clostridium selenoprotein MsrA
To investigate catalytic properties of Clostridium MsrA, we purified this protein (tagged at the C-terminus with His) from E. coli cells. As shown in Table 2, the specific activity of Clostridium selenoprotein MsrA was found to be 4,185 nmol/min/mg protein in a DTT-dependent reaction. This value was 9-fold higher than that of its Cys mutant (U16C). The kcat value of the selenoprotein MsrA was 22-fold higher than that of its Cys mutant and the Km value of the selenoprotein was 2.5-fold higher. Overall, these data show that selenium provides catalytic advantage to Clostridium MsrA. Importantly, this enzyme appears to be a highly efficient catalyst with kinetic properties comparable with those of Chlamydomonas MsrA characterized previously 31.
Trx is typically a natural reductant for regeneration of MsrA enzymes. To test if Trx can function as a reductant for Clostridium MsrA, we measured Trx-dependent activities of its natural Sec-containing and mutant Cys-containing forms using human Trx1 and Trx reductase 1, and compared the data with DTT-dependent activities of these MsrA forms. In addition, mouse MsrA served as control because this protein is known as a target of Trx. As shown in Table 3, the ratio of Trx- to DTT-dependent activities of mouse MsrA was 0.73, indicating that human Trx1 as well as DTT could efficiently reduce this protein. However, for Clostridium selenoprotein MsrA, this ratio was less than 0.01, suggesting that Trx1 was a poor reductant for this selenoprotein. Similarly, the Cys mutant could not be efficiently reduced by Trx1 (the ratio of Trx- to DTT-dependent activity was 0.03). To examine the possibility that human Trx couples poorly to the bacterial selenoprotein MsrA, we carried out the enzyme assay in the presence of bacterial (E. coli) Trx. The ratios of Trx- to DTT-dependent activities were consistent with the results for human Trx1; 0.93 for mouse MsrA, 0.016 for Clostridium selenoprotein MsrA, and 0.05 for its Cys mutant. Therefore, our data suggested that Trx is not a reducing agent in vivo for Clostridium MsrA and that unknown physiological reductant(s) must function in the reduction of this enzyme.
Table 3.
Ratios of Trx- to DTT-dependent activities
| MsrA forms | Specific activity (nmol/min/mg protein) |
Ratio of Trx/DTT activity |
|||
|---|---|---|---|---|---|
| DTT-dependent reaction |
Trx-dependent reaction | ||||
| hTrx1 | eTrx | hTrx1 | eTrx | ||
| WT | 4,185 ± 46 | 34 ± 5 | 70 ± 6 | 0.008 | 0.017 |
| U16C | 464 ± 14 | 14 ± 2 | 23 ± 2 | 0.03 | 0.05 |
| Mouse MsrA | 256 ± 24 | 188 ± 9 | 238 ± 8 | 0.73 | 0.93 |
Purified proteins (90 ng WT, 1 μg U16C, and 1 μg mouse MsrA) were used in the DTT- and Trx-dependent assays containing 200 μM DABSyl-methionine-S-sulfoxide. hTrx1, human cytosolic Trx; eTrx, E. coli Trx; WT, Clostridium selenoprotein MsrA; U16C, its Cys mutant.
Recent studies revealed that thionein (a low-molecular-mass Cys-rich protein) can function as a reducing agent for MsrA and MsrB 49, and that selenium compounds such as selenocysteamine could reduce mammalian MsrBs 50. Our previous study on Chlamydomonas selenoprotein MsrA and its Cys mutant revealed that the Trx-dependent activities of these enzyme forms were more than 20-fold lower than their DTT-dependent activities 31. Taken together and combined with the observation that known selenoprotein MsrAs lack conserved resolving Cys residues (or do not have any Cys as in Clostridium MsrA) 31, our data on Chlamydomonas and Clostridium MsrAs suggest that Trx is not a natural reducing agent for the selenenic acid intermediates of selenoprotein MsrAs.
Conclusion
This study shows that Clostridium sp. OhILAs, a gram-positive anaerobic bacterium, is predicted to be a selenoprotein-rich organism that may contain 13 selenoproteins, including 5 that have not been previously described. Interestingly, one of the detected selenoproteins was MsrA, which serves an antioxidant function by catalyzing the reduction of methionine-S-sulfoxide. Little is known about anaerobic bacterial MsrAs. We cloned and expressed this Clostridium selenoprotein in E. coli. Although its SECIS element did not resemble those in E. coli, we could verify the recombinant protein expression by 75Se labeling, western blots, and activity assays. Thus, the SECIS element of Clostridium selenoprotein msrA is functional and recognized by the E. coli Sec insertion machinery, although the translational efficiency was found to be low. The enzyme assays and kinetic analyses revealed that the selenoprotein MsrA is much more active than its Cys mutant form, indicating a key contribution of selenium to the catalytic efficiency of this selenoenzyme. Our data also suggest that Trx, a general in vivo reducing agent for MsrA, is not a reductant for Clostridium selenoprotein MsrA. Further studies will be needed to characterize the detailed reductase steps including determination of the natural reducing agent for this selenoprotein MsrA.
Supplementary Material
Acknowledgments
We thank Dr. John Stolz (Duquesne University, USA) for providing Clostridium sp. OhILAs genomic DNA and Dr. Dmitri Fomenko (University of Nebraska, USA) for help with sequence information. This work was supported by the KOSEF (Korea Science and Engineering Foundation) grant R13-2005-005-01004-0 (to HYK), the Yeungnam University research grant in 2006 (to HYK), and the NIH (National Institutes of Health) grant AG021518 (to VNG).
Abbreviations
- DTT
dithiothreitol
- Msr
methionine sulfoxide reductase
- Sec
selenocysteine
- SECIS
selenocysteine insertion sequence
- Trx
thioredoxin
References
- 1.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 2.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 4.Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci U S A. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci U S A. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner DC, Stadtman TC. Purification of protein components of the clostridial glycine reductase system and characterization of protein A as a selenoprotein. Arch Biochem Biophys. 1973;154:366–381. doi: 10.1016/0003-9861(73)90069-6. [DOI] [PubMed] [Google Scholar]
- 8.Cone JE, Del Rio RM, Davis JN, Stadtman TC. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci U S A. 1976;73:2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rother M, Resch A, Wilting R, Bock A. Selenoprotein synthesis in archaea. Biofactors. 2001;14:75–83. doi: 10.1002/biof.5520140111. [DOI] [PubMed] [Google Scholar]
- 10.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 12.Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 13.Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigo R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Gladyshev VN. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- 16.Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc Natl Acad Sci U S A. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaye L, Becerra A, Orgel L, Lazcano A. Molecular evolution of peptide methionine sulfoxide reductases (MsrA and MsrB): on the early development of a mechanism that protects against oxidative damage. J Mol Evol. 2007;64:15–32. doi: 10.1007/s00239-005-0281-2. [DOI] [PubMed] [Google Scholar]
- 19.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews B, John G, Nathan C, Brot N. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- 21.Moskovitz J. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim Biophys Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 23.Schoneich C. Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer's disease. Biochim Biophys Acta. 2005;1703:111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci U S A. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 27.Turanov AA, Su D, Gladyshev VN. Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J Biol Chem. 2006;281:22953–22963. doi: 10.1074/jbc.M604326200. [DOI] [PubMed] [Google Scholar]
- 28.Kumar RA, Koc A, Cerny RL, Gladyshev VN. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J Biol Chem. 2002;277:37527–37535. doi: 10.1074/jbc.M203496200. [DOI] [PubMed] [Google Scholar]
- 29.Kim HY, Gladyshev VN. Role of structural and functional elements of mouse methionine-S-sulfoxide reductase in its subcellular distribution. Biochemistry. 2005;44:8059–8067. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. Embo J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 34.Antoine M, Boschi-Muller S, Branlant G. Kinetic characterization of the chemical steps involved in the catalytic mechanism of methionine sulfoxide reductase A from Neisseria meningitidis. J Biol Chem. 2003;278:45352–45357. doi: 10.1074/jbc.M307471200. [DOI] [PubMed] [Google Scholar]
- 35.Lowther WT, Brot N, Weissbach H, Matthews BW. Structure and mechanism of peptide methionine sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry. 2000;39:13307–13312. doi: 10.1021/bi0020269. [DOI] [PubMed] [Google Scholar]
- 36.Boschi-Muller S, Azza S, Sanglier-Cianferani S, Talfournier F, Van Dorsselear A, Branlant G. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J Biol Chem. 2000;275:35908–35913. doi: 10.1074/jbc.M006137200. [DOI] [PubMed] [Google Scholar]
- 37.Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 38.Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol. 2005;12:198–203. doi: 10.1038/nsmb890. [DOI] [PubMed] [Google Scholar]
- 39.Ose T, Soler N, Rasubala L, Kuroki K, Kohda D, Fourmy D, Yoshizawa S, Maenaka K. Structural basis for dynamic interdomain movement and RNA recognition of the selenocysteine-specific elongation factor SelB. Structure. 2007;15:577–586. doi: 10.1016/j.str.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Fourmy D, Guittet E, Yoshizawa S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J Mol Biol. 2002;324:137–150. doi: 10.1016/s0022-2836(02)01030-6. [DOI] [PubMed] [Google Scholar]
- 41.Sandman KE, Tardiff DF, Neely LA, Noren CJ. Revised Escherichia coli selenocysteine insertion requirements determined by in vivo screening of combinatorial libraries of SECIS variants. Nucleic Acids Res. 2003;31:2234–2241. doi: 10.1093/nar/gkg304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Reches M, Groisman I, Engelberg-Kulka H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tormay P, Bock A. Barriers to heterologous expression of a selenoprotein gene in bacteria. J Bacteriol. 1997;179:576–582. doi: 10.1128/jb.179.3.576-582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bar-Noy S, Moskovitz J. Mouse methionine sulfoxide reductase B: effect of selenocysteine incorporation on its activity and expression of the seleno-containing enzyme in bacterial and mammalian cells. Biochem Biophys Res Commun. 2002;297:956–961. doi: 10.1016/s0006-291x(02)02314-8. [DOI] [PubMed] [Google Scholar]
- 45.Sun QA, Su D, Novoselov SV, Carlson BA, Hatfield DL, Gladyshev VN. Reaction mechanism and regulation of mammalian thioredoxin/glutathione reductase. Biochemistry. 2005;44:14528–14537. doi: 10.1021/bi051321w. [DOI] [PubMed] [Google Scholar]
- 46.Söhling B, Parther T, Rucknagel KP, Wagner MA, Andreesen JR. A selenocysteine-containing peroxiredoxin from the strictly anaerobic organism Eubacterium acidaminophilum. Biol Chem. 2001;382:979–986. doi: 10.1515/BC.2001.123. [DOI] [PubMed] [Google Scholar]
- 47.Wilting R, Vamvakidou K, Bock A. Functional expression in Escherichia coli of the Haemophilus influenzae gene coding for selenocysteine-containing selenophosphate synthetase. Arch Microbiol. 1998;169:71–75. doi: 10.1007/s002030050542. [DOI] [PubMed] [Google Scholar]
- 48.Gursinsky T, Grobe D, Schierhorn A, Jager J, Andreesen JR, Söhling B. Factors and selenocysteine insertion sequence requirements for the synthesis of selenoproteins from a gram-positive anaerobe in Escherichia coli. Appl Environ Microbiol. 2008;74:1385–1393. doi: 10.1128/AEM.02238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sagher D, Brunell D, Hejtmancik JF, Kantorow M, Brot N, Weissbach H. Thionein can serve as a reducing agent for the methionine sulfoxide reductases. Proc Natl Acad Sci U S A. 2006;103:8656–8661. doi: 10.1073/pnas.0602826103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sagher D, Brunell D, Brot N, Vallee BL, Weissbach H. Selenocompounds can serve as oxidoreductants with the methionine sulfoxide reductase enzymes. J Biol Chem. 2006;281:31184–31187. doi: 10.1074/jbc.M606962200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.