Abstract
Annotating the tremendous amount of sequence information being generated requires accurate automated methods for recognizing homology. Although sequence similarity is only one of many indicators of evolutionary homology, it is often the only one used. Here we find that supplementing sequence similarity with information from biomedical literature is successful in increasing the accuracy of homology search results. We modified the PSI-BLAST algorithm to use literature similarity in each iteration of its database search. The modified algorithm is evaluated and compared to standard PSI-BLAST in searching for homologous proteins. The performance of the modified algorithm achieved 32% recall with 95% precision, while the original one achieved 33% recall with 84% precision; the literature similarity requirement preserved the sensitive characteristic of the PSI-BLAST algorithm while improving the precision.
1. Introduction
The sequence information generated by genome sequencing projects offers opportunities for understanding biology at an unprecedented fine level of detail. At the same time, the biomedical literature provides a record of high level biological phenomena as observed and reported over many decades. There is an opportunity to combine the power of the genome sequence information with the published biological record to accelerate progress and gain insight. Here we show that including literature to tailor homology searches against sequence databases can improve performance.
The concept of homology between two protein or nucleotide sequences is often used to infer that two genes or their protein products are related by evolution. Divergence between the two entities may have occurred when two species evolved from a single ancestor (orthologs) or when gene duplication occurs within a species (paralogs). We usually expect that homologous sequences have common functional roles in enzymatic activity, cellular functions, or overall cellular processes, and may have common structural features, such as in their protein tertiary structure or active site mechanisms. Since attributing structure, function, or process to a protein sequence experimentally can be expensive in time and effort, biologists look to other sequences that share similarity to predict homology and then infer these features. This approach has been used widely for structure prediction, function prediction, and genome annotation [1–7].
Well-known approaches to assess sequence similarity include dynamic programming [8,9] and BLAST [10]. The dynamic programming approaches find the alignment between any two sequences that generates the most optimal score based on user-specified parameters. The BLAST approach is an approximation of an optimal algorithm and was designed to search databases rapidly for sequences that align significantly well to a query sequence. Thus, it is often used for applications that require high performance, such as genome annotation.
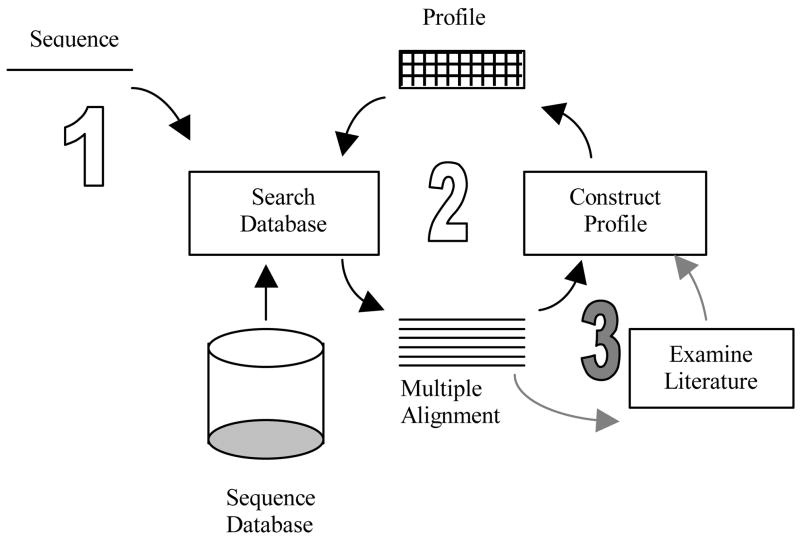
PSI-BLAST (Position Specific Iterated BLAST) is an iterative version of BLAST designed to increase the sensitivity of searches [11]. In the first iteration, a BLAST search obtains significantly similar sequences that are used to create a probabilistic sequence profile. In subsequent iterations that profile is used to search the database and to update the significant sequences (see Figure 1). By including more diverse sequences into the query, sensitivity is improved. PSI-BLAST approaches the problem of homology searching by assuming that the query sequence is part of a larger family of sequences; the aim of iterative profile refinement is to ascertain the underlying common structure of the unknown family and discover its members.
Figure 1.
An illustration of PSI-BLAST and our modification. A sequence is used in the initial query to BLAST search the database for similar sequences (1), a multiple alignment is then used to construct a profile to search the database again (2). Our modification (3) involves screening the sequences that constitute the multiple alignment for literature similarity; the sequences for which the associated literature is least concordant with that of the original sequence used in (1) are eliminated from the profile.
As PSI-BLAST iterates, it includes a more diverse array of sequences, and the possibility of including a sequence that is not properly considered a homolog of the original query sequence increases. Thus, any errors introduced into the profile can be magnified, eventually diluting the signal from the original sequence; this situation has been called “profile drift”. In these situations the algorithm fails to converge or converges to an imperfect solution.
PSI-BLAST considers only sequence similarity and no other biological knowledge, such as the scientific literature associated with the sequences. For example, if a query sequence is similar to many cell cycle proteins, a reasonable refinement may be to consider only those proteins involved in the cell cycle. Including more information may result in a search that is relatively resistant to contamination. Our adaptation of PSI-BLAST removes sequences that lack sufficient literature similarity in each of the iterations. Evidence that literature scores are useful for protein structure and functional analysis has previously been presented in [5,12,13].
2. Method
The code for the modified PSI-BLAST algorithm was implemented in the Python programming language [14] using the Biopython toolkit (www.biopython.org) on a Sun E450 platform. All experiments were performed on protein sequences obtained from SWISS-PROT Release 39 (May 2000) [15]. SWISS-PROT 39 is a human-curated database of 86,593 protein sequences and contains cross-references to databases including Protein Data Bank (PDB) [16] and MEDLINE [17].
To validate our approach we created a database of sequences that are associated with at least a minimal amount of biological literature. Next, we defined a gold standard of homologous families of sequences. Finally, to assess performance we ran sequence homology searches with PSI-BLAST, varying the parameters used for profile construction.
Modified PSI-BLAST approach
For each homology search, PSI-BLAST was run against the SWISS-PROT database for a maximum of ten iterations with the profile inclusion criteria that the e-value significance of a hit must be at least 0.001. To prevent trivial sequence similarities, we filtered both the query and database sequences for low-complexity regions with SEG using the recommended parameters (12 1.8 2.0) [18].
Our modification to PSI-BLAST involved throwing out sequences that have poor literature similarity to the query sequence. After each iteration of the search, we ranked the significant hits according to a literature similarity score and discarded the lowest scoring fraction, thereby excluding them from the profile (Figure 1).
Collecting Sequence Information and Literature
To obtain literature pertaining to a sequence, we used the information indexed from its SWISS-PROT record. First, we collected the description, comments, and keywords in the record. Next, we retrieved the record’s MEDLINE cross-references and downloaded the citation and its MeSH headings, subheadings, and abstracts. We defined the literature of a sequence as the concatenation of these unstructured texts.
Once we collected literature for each sequence, we created a list of domain specific stop words. These are words that contain little information for distinguishing the sequences. We defined stop words as those words that appear with less than 3 sequences or more than 85,000 sequences. We found 80,479 stop words out of a total of 147,639 words in the corpus. This simple method for identifying stop words has previously been shown to be effective in similar tasks [19].
Calculating Document Similarity
The similarity between the literature of two sequences was calculated using a vector cosine measure [20]. In this model documents are represented as a vector in which each dimension represents the number of times a word appeared in a document. Documents were tokenized using all non-alphanumeric characters as delimiters. Words are then any lowercased token that is not a stop word.
The similarity between two documents is the cosine of the angle between their word vectors:
| (1) |
where A and B are the word vectors of two documents. Documents with similar word content yield scores close to 1, while those with different words yield scores close to 0. The lengths of the documents are not relevant to the similarity, as the cosine measure normalizes the vectors.
Defining a Gold Standard for Validation
To validate our approach we created families of homologous protein sequences to use as a gold standard. Homology families should contain sequences that are related by evolution, rather than just by sequence similarity. Since this is difficult to define, we choose a definition based on the Structural Classification of Proteins Database (SCOP), release 1.50 (February 2000) [21]. SCOP is a manually constructed hierarchical categorization of proteins based on structure and function. Since biological relatedness is implied at the superfamily level, we defined a homology family as the set of SWISS-PROT sequences that reference structures in the same SCOP superfamily. All SWISS-PROT sequences that map into a single SCOP superfamily via PDB were selected for the gold standard.
Choice of Test Set
Our test set consisted of one query sequence per family. Candidate sequences were selected from the gold standard based on two criteria: 1) they must contain at least four MEDLINE references with abstracts and 2) they must be in families with at least five members. For each family we selected the most divergent candidate sequence to be in our test set. We identified this sequence as the one that detects the least number of homologous sequences in a BLAST search. If multiple sequences are equally divergent, one was chosen randomly.
Validation
We conducted four homology searches for each test sequence. One search used the standard PSI-BLAST. Three searches used a PSI-BLAST modified to account for literature similarity with various degrees of stringency; we dropped sequences with the lowest 5%, 10%, and 20% of literature similarity per iteration.
3. Results
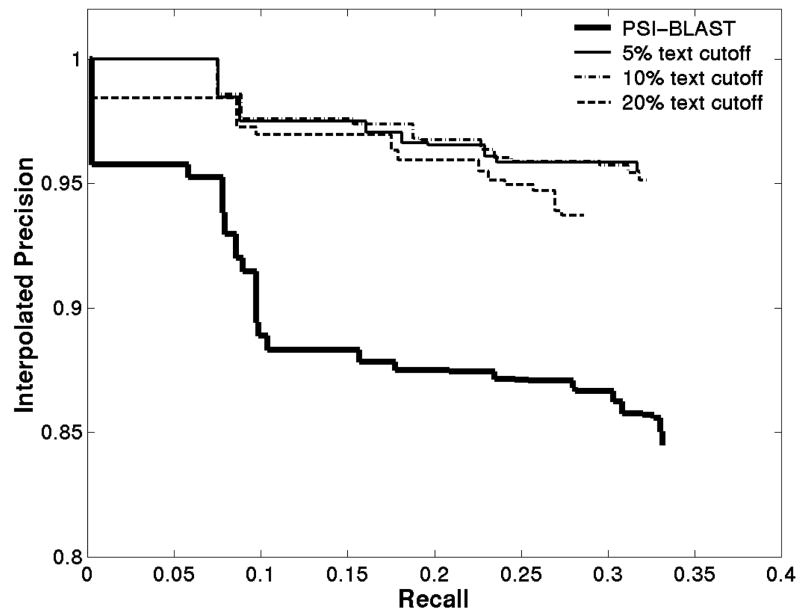
Figure 2 shows a comparison of the performance of PSI-BLAST to the various modified PSI-BLAST approaches. Recall is the number of homologous sequences surpassing a fixed e-value cutoff divided by the total number of homologous sequences. At a fixed recall, precision is the number of homologous sequences detected divided by the total number of sequences detected. The modified PSI-BLAST was more precise than the original at any recall. In addition, the precision did not decay as rapidly as recall was increased.
Figure 2.
Using Text Comparison Improves Homology Search Results. Results of homology search for 54 training sequences from different families. Precision is interpolated to insure that the curves are monotonic. The solid bold line represents the unmodified PSI-BLAST algorithm; other lines represent modified PSI-BLAST algorithm that drops the sequences with the lowest 5%, 10%, and 20% of literature similarity.
Table 1 lists all the families for which the outcome of PSI-BLAST was altered by inclusion of our literature criteria; these are the families that account for the differences in Figure 2. For 46 of the 54 families that were tested, the outcome was identical for the modified and the unmodified PSI-BLAST. Out of the eight queries remaining, five differed in convergence, while three differed in performance. These eight families fall into three categories. The first two families in Table 1 converged to poor solutions with standard PSI-BLAST and failed to converge for the modified PSI-BLAST. The next three failed to converge for PSI-BLAST, but converged to reasonably good solutions for modified PSI-BLAST. The final three converged for both modified and standard PSI-BLAST; the solutions are slightly better for the standard one.
Table 1.
Homologous families for which performance differed between standard PSI-BLAST and modified PSI-BLAST (drop 10% of sequences with poorest literature similarity). Most of the 54 families have identical performance for both algorithms and are not shown. Superfamily is a SCOP homology family and Query Sequence is its test sequence. Words is the number of document words associated with the query sequence. # Seqs is the number of sequences in the family. The final six columns describe the results of a search with the query sequence. Here, precision and recall were calculated for each individual family using all the results from the homology search.
| Convergence | Precision | Recall | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Superfamily | Query Sequence | Words | # Seqs | PSI-BLAST | Text 10% | PSI-BLAST | Text 10% | PSI-BLAST | Text 10% |
|
| |||||||||
| EGF/Laminin | C1R_HUMAN | 1661 | 5 | yes | no | 0.11 | N/A | 0.8 | N/A |
| Acid proteases | POL_HV2RO | 1271 | 22 | yes | no | 0.6 | N/A | 0.27 | N/A |
| PLP-dependent transferases | GLYC_RABIT | 1052 | 21 | no | yes | N/A | 1 | N/A | 0.1 |
| Thioredoxin-like | CAQS_RABIT | 1516 | 13 | no | yes | N/A | 1 | N/A | 0.38 |
| Glucocorticoid receptor-like(DNA-binding domain) | CYSR_CHICK | 1738 | 10 | no | yes | N/A | 0.8 | N/A | 0.4 |
| EF-hand | SCP_NERDI | 963 | 31 | yes | yes | 0.92 | 0.92 | 0.74 | 0.71 |
| Glycosyl-transferases | CHLY_HEVBR | 1007 | 20 | yes | yes | 1 | 1 | 0.2 | 0.15 |
| Snake toxin-like | CD59_HUMAN | 2435 | 23 | yes | yes | 1 | 1 | 0.13 | 0.09 |
4. Discussion
The figures demonstrate the major strength of our approach. Inclusion of biomedical literature into homology searching in some cases improved performance and otherwise did not deteriorate it. As greater precision was achieved, recall was not as dramatically reduced as it was for the standard PSI-BLAST.
For the protein family “Thioredoxin-like”, the PSI-BLAST homology search with the “CAQS-RABIT” test sequence failed to converge. The modified PSI-BLAST that accounted for literature similarity did converge on a precise solution; it correctly detected 5 sequences. In this case, removing sequences with low literature similarity prevented profile drift and allowed the search to converge on a correct solution.
Alternatively, for the “EGF/Laminin” and “Acid proteases” families the standard PSI-BLAST converged upon incorrect answers, indicating that drift occurred. In the modified PSI-BLAST, removing sequences with unrelated literature slowed the drift, preventing it from converging in 10 iterations. These families suffered because non-homologous sequences had high similarity to family sequences. Although excluding these sequences did not prevent them from being detected in the next round, it did prevent further drift in the profile. Literature similarity checking added an additional constraint against including erroneous sequences.
However, the literature similarity constraint made no difference in the performance of PSI-BLAST in the majority of the families. Out of the 54 families, only 5 of the searches benefited from the additional constraint, and only 2 of those resulted in major improvements. In the 3 cases in which the performances were worse, they resulted in slightly lower recalls that can be attributed to a single missed sequence in each family.
A limitation of any natural language processing approach to biological problems is that areas for which the appropriate quantity of text is unavailable may be difficult to study. In the context of this work, for example, annotation of newly discovered sequences are unlikely to benefit from the literature if the literature simply does not provide any information about the related sequences. In the algorithm we tested, the literature of the original document is used to screen additional sequences. Instead, an adaptive method where the literature of the original document is supplemented with the literature of the queried sequences may be appropriate. This would correspond to an assumption that the literature gathered in subsequent iterations was sufficiently representative of the original sequence, to allow it to be used to create a “literature profile.” However, such an approach might be subject to the same drift phenomena that limit PSI-BLAST!
Aside from homology searching, combining literature similarity with sequence similarity has applications in any area in which sequence differences can be supplemented with expert knowledge. For example, single nucleotide polymorphisms and other sequence level differences between individuals are now being characterized and may soon be relevant clinically [22,23]. In exploring these polymorphisms in the context of clinical data, it may be useful to look not only for similarities at the genomic level, but also at the level of the patient record. In this setting, biomedical literature is replaced with an electronic medical record, and comparisons are made between individual patient genomes. For example, the genomic sequence of a presenting patient can be queried against a patient database of sequences and records. Similar patients can be examined and studied to understand the history of diagnosis and treatment, and to correlate these with genomic variations.
In conclusion, we have shown that the biological literature can be used to improve the detection of sequence homology. Simple natural language processing techniques capture enough information from free text to improve the accuracy of homology searches.
Acknowledgments
We acknowledge the support of Stanford Graduate Fellowship (JTC), NIH GM-07365 (SR), NIH-1U01-GM61374-01, NSF-DBI-9600367, and the Burroughs Wellcome Foundation. The authors would like to thank Steven Brenner for insightful conversations and Peter Cooper for technical help with the NCBI toolkit.
References
- 1.Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297:233–49. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- 2.Dunbrack RL., Jr Comparative modeling of CASP3 targets using PSI-BLAST and SCWRL. Proteins. 1999;(Suppl):81–7. doi: 10.1002/(sici)1097-0134(1999)37:3+<81::aid-prot12>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 3.Huynen M, et al. Homology-based fold predictions for Mycoplasma genitalium proteins. J Mol Biol. 1998;280:323–6. doi: 10.1006/jmbi.1998.1884. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl E, Elofsson A. Identification of related proteins on family, superfamily and fold level. J Mol Biol. 2000;295:613–25. doi: 10.1006/jmbi.1999.3377. [DOI] [PubMed] [Google Scholar]
- 5.MacCallum RM, Kelley LA, Sternberg MJ. SAWTED: structure assignment with text description--enhanced detection of remote homologues with automated SWISS-PROT annotation comparisons. Bioinformatics. 2000;16:125–9. doi: 10.1093/bioinformatics/16.2.125. [DOI] [PubMed] [Google Scholar]
- 6.Sauder JM, Arthur JW, Dunbrack RL., Jr Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins. 2000;40:6–22. doi: 10.1002/(sici)1097-0134(20000701)40:1<6::aid-prot30>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 7.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 8.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 9.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–7. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrade MA. Position-specific annotation of protein function based on multiple homologs. ISMB. 1999:28–33. [PubMed] [Google Scholar]
- 13.Andrade MA, Valencia A. Automatic annotation for biological sequences by extraction of keywords from MEDLINE abstracts. Development of a prototype system. ISMB. 1997:25–32. [PubMed] [Google Scholar]
- 14.Lutz M, Ascher D, Willison F. Learning Python. O’Reilly; Sebastopol, CA: 1999. [Google Scholar]
- 15.Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991;19 (Suppl):2247–9. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein FC, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–42. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 17.Hutchinson D. Medline for health professionals: how to search PubMed on the Internet. New Wind; Sacramento: 1998. [Google Scholar]
- 18.Wootton JC, Federhen S. Statisitics of local complexity in amino acid sequences and sequence databases. Computers and Chemistry. 1993;17:149–163. [Google Scholar]
- 19.Yang Y, Pedersen JP. A Comparative Study on Feature Selection in Text Categorization Proceedings of the Fourteenth International Conference on Machine Learning. International Conference on Machine Learning. 1997 [Google Scholar]
- 20.Wilbur WJ, Yang Y. An analysis of statistical term strength and its use in the indexing and retrieval of molecular biology texts. Comput Biol Med. 1996;26:209–22. doi: 10.1016/0010-4825(95)00055-0. [DOI] [PubMed] [Google Scholar]
- 21.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 22.Cargill M, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–8. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 23.Halushka MK, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–47. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]