Abstract
Analytical approximations to fundamental equations of continuum electrostatics on simple shapes can lead to computationally inexpensive prescriptions for calculating electrostatic properties of realistic molecules. Here, we derive a closed-form analytical approximation to the Poisson equation for an arbitrary distribution of point charges and a spherical dielectric boundary. The simple, parameter-free formula defines continuous electrostatic potential everywhere in space and is obtained from the exact infinite-series (Kirkwood) solution by an approximate summation method that avoids truncating the infinite series. We show that keeping all the terms proves critical for the accuracy of this approximation, which is fully controllable for the sphere. The accuracy is assessed by comparisons with the exact solution for two unit charges placed inside a spherical boundary separating the solute of dielectric 1 and the solvent of dielectric 80. The largest errors occur when the source charges are closest to the dielectric boundary and the test charge is closest to either of the sources. For the source charges placed within 2 Å from the boundary, and the test surface located on the boundary, the root-mean-square error of the approximate potential is less than 0.1 kcal∕mol∕∣e∣ (per unit test charge). The maximum error is 0.4 kcal∕mol∕∣e∣. These results correspond to the simplest first-order formula. A strategy for adopting the proposed method for realistic biomolecular shapes is detailed. An extensive testing and performance analysis on real molecular structures are described in Part II that immediately follows this work as a separate publication. Part II also contains an application example.
INTRODUCTION
Electrostatic interactions are often a key factor in determining properties of biomolecules,1, 2, 3, 4, 5 including their functions such as catalytic activity,6, 7 ligand binding,8, 9 complex formation,10 proton transport,11 as well as structure and stability.12, 13 In-depth studies of electrostatics-based phenomena in macromolecular systems require the ability to compute the potentials and fields efficiently and accurately on the atomic scale.2, 14 Within the framework of the so-called implicit or continuum solvent model,15, 16, 17 the Poisson–Boltzmann (PB) approach is an exact way to compute the electrostatic potential ϕ(r) produced by a molecular charge distribution ρ(r). In many practical applications its linearized form is used, in which case the following equation or its equivalent must be solved:
| (1) |
where ϵ(r) is the position-dependent dielectric constant, and the electrostatic screening effects of monovalent salt enter via the Debye–Hückel screening parameter κ.
Historically, the first quantitative approaches to computation and analysis of the electrostatic potential produced by biomolecular charge distributions relied on analytical approximations18, 19 to Eq. 1, such as the famous model due to Kirkwood.19 The use of these models led to unique insights into a number of important biophysical problems, for example, protein titration20 and protein folding.21 The limited accuracy resulting from the use of simplified shapes such as a sphere to represent the true complexity of a molecular surface was probably thought to be an inevitable drawback of these models and thus prompted the development of numerical approaches to solving the PB equation.
A prototypical numerical PB (NPB) method works by placing the molecule inside a bounding box or surface, defining a three-dimensional (3D) grid of points within it, and then solving for the ϕ(r) at every grid point through iterating a set of self-consistent equations. Currently available tools22, 23, 24, 25, 26 based on these methods produce accurate potential fields ϕ(r) for any realistic charge distribution and molecular shape. The errors of these numerical solutions can be controlled, and, in principle, made arbitrarily small (albeit at an unrealistic computational cost), by adjusting parameters of the numerical models such as the finite-difference grid resolution and the size of the bounding box.
The NPB approaches have become the de facto accuracy standard in the field.27 Despite their widespread acceptance, the methodology has several drawbacks relative to alternative analytical approaches. From the practical standpoint, the NPB methods are fundamentally more complex and generally more expensive computationally compared to closed-form analytical expressions. These differences are especially pronounced in dynamical simulation, where availability of analytical energy functions is particulary advantageous. Generally, the NPB framework does not offer as much freedom and ease in exploring parameter space of simple model systems and toy models and in making qualitative estimates. This ability may be critical for studies aimed at certain fundamental system nonspecific properties of biomolecular systems.21
The fundamental difference between NPB and analytical approaches such as the Kirkwood model is seen in the limiting case when ϕ(r) needs to be estimated at a single point in space: The NPB methodology still requires that ϕ(r) is found simultaneously at many points of a finite spacial domain, for example, at every node of a 3D cubic grid or two-dimensional (2D) surface.28, 29 The computational complexity of finding ϕ(r) combined with technical difficulties associated with computing forces due to changes in the molecular surface motivated the search for alternative methods to be used in molecular dynamics (MD) to estimate electrostatic forces within the implicit solvent framework.
While a number of promising models were proposed,30, 31, 32, 33 perhaps the most successful of these analytical alternatives is the generalized Born (GB) approximation pioneered by Still et al.34 around 1990. The model offers an analytical prescription for estimating the electrostatic part of the solvation free energy. The GB’s original formulation applies to the zero ionic strength case (the Poisson equation). Later, a heuristic prescription was introduced that successfully adapted the GB approximation to handle the nonzero salt case.35
Unlike the infinite-series Kirkwood’s solution,19 the GB expression is a mathematically simple, closed-form formula. Importantly, the GB approximation is also aimed at working for arbitrary shapes, not just spherical as in Kirkwood’s model. The algorithmic simplicity and computational efficiency of the original GB model, combined with accuracy improvements, have made it the method of choice in implicit solvent MD,15, 17, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56 although promising NPB-based alternatives have also been recently tested.26, 57
Despite the successes of the GB approximation, the model has its own serious drawbacks. First, fundamentally, the GB model does not, even in principle, permit a definition of continuous electrostatic potential everywhere in space: at best, it can only be used to define ϕ(r) at the centers of the atoms.58 This property is at odds with the very physical nature of electrostatic potential. In practice, the ability to compute the potential at any given point is critical for many applications. Second, unlike many important approximate approaches in physics, for example, the perturbation theory, or the NPB approach itself, the GB model is heuristic in nature and does not have an obvious “handle” that controls its accuracy, at least in principle. As a result, the physical origins of the observed deviations from the NPB reference are hard to trace.59
The goal of this work is to overcome these drawbacks and derive a simple analytical approximation of the Poisson equation that is closed form and controllable. Ideally, the approximation should define physically admissible electrostatic potential everywhere in space and should provide a level of accuracy acceptable in practice.
In Part I of the study presented in this paper, we derive several candidates for such an approximation and thoroughly examine their behavior and physical nature on a simple geometry (sphere) for which an exact reference solution of the Poisson problem is available. We propose a candidate approximation for realistic biomolecular shapes and show how its parameters should be redefined once the spherical symmetry is abandoned.
In Part II of this work, which is a separate paper immediately following this one, we adapt the proposed approximation to handling the screening effects of salt and thoroughly test the resulting model on a large number of realistic biomolecules. We then demonstrate how the model might be useful in a concrete problem—a search for putative RNA binding sites on the surface of a viral capsid.
Derivation of the analytical models
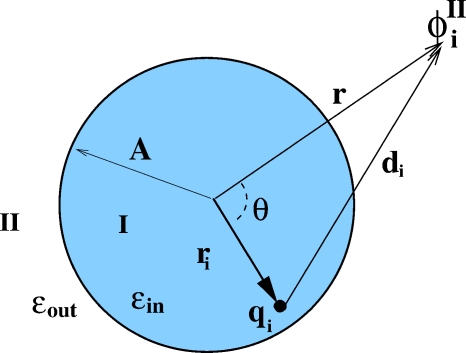
The geometric setup of the boundary value problem for the Poisson equation, Eq. 1 with κ=0, is shown in Fig. 1.
Figure 1.
The boundary value problem for Eq. 1. A spherical boundary separates the inside region I, dielectric ϵin, from the outside region II, dielectric ϵout. The point of observation is specified by its spherical coordinates (r,θ); the source charge is at (ri,0). Here A is the radius of the sphere.
We follow Kirkwood19 to obtain the exact infinite-series expressions for ϕ(r) everywhere in space. The infinite-series solutions for region I (inside) is worked out in detail in Ref. 19, with β=ϵin∕ϵout,
| (2) |
The solution for region II is worked out in detail in the appendix at the end of this paper. To summarize, we have arrived at the following solution to the Poisson equation for region II:
| (3) |
Equations 2, 3 satisfy the usual60 continuity conditions at the boundary,
| (4) |
| (5) |
The above solutions, Eqs. 2, 3, of the Poisson equation are valuable since they are exact. Unfortunately, they are not very useful in practice since each one is dependent on two infinite series that converge slowly for charge distributions relevant to biomolecules. For example, the infinite series in Eq. 3 converge slowly when (ri∕r)→1. For the potential near the molecular surface, the ratio being close to 1 is a typical case in real molecules since charged groups are rarely buried due to a high desolvation penalty. As will be discussed below, tens or even hundreds of terms might need to be kept in order to approach well-converged sums. Thus, for practical applications where speed is a factor, something different needs to be done. Also, the infinite series itself or its partial sum is not particularly helpful in illuminating the physical properties of ϕ(r). A simple closed-form approximation that retains the key physics of the Poisson equation embedded in Eqs. 2, 3 is what we are looking for. Below we present the detailed derivations for Eq. 3 and just list the end result derived from Eq. 2.
As discussed above, we need to avoid truncating the infinite series. Instead, we keep the l=0 term unchanged and approximate l∕(l+1)≈const=α for all l>0 terms in the first of the two infinite sums in Eq. 3. The approximation is both mathematically and physically motivated.
Mathematically, the approximation recasts the infinite series into a form that can be summed exactly into a closed-form simple formula. The specific algebraic form of α is motivated by a relatively small variation of l∕(l+1) for anyl>0: 1∕2≤l∕(l+1)≤1.
Physically, this approximation maintains a dependence on the constant β, which encapsulates a specific contribution of the dielectric interface to the potential. While one can easily construct other algebraically “simple” approximations that would provide equal mathematical benefit, e.g., (1+(l∕(l+1))β)≈const=α or (1+(l∕(l+1))β)−1≈const=α, these would lose the explicit dependency on β and thus were not considered.
Upon setting l∕(l+1)≈const=α for all l>0, the infinite series in Eq. 3 is approximated as
| (6) |
We now define t=(ri∕r) and use the following identity:
| (7) |
to approximate the first term in Eq. 3 as
| (8) |
Since 1∕2≤l∕(l+1)≤1 for l>0, a reasonable first guess for α is the middle of the interval, α=0.75. Applying the same identity to the second infinite sum in Eq. 3 and combining the two terms yields the following closed-form approximate expression for :
| (9) |
After algebraic manipulations, we arrive at the following analytical form for the electrostatic potential outside of the sphere, region II in Fig. 1. The corresponding expression for the inside space, region I is obtained in the same fashion. Below is the combined key result of this work,
| (10) |
| (11) |
Since only the first term, l=0, in the exact infinite sums was kept intact throughout the derivations, the above expression can be referred to as the first-order approximation, although it shall not be confused with truncating the infinite sums. To demonstrate how the accuracy of this approximation can be controlled, at least in principle, we extend Eq. 8 to include the next two terms exactly. Due to the specific symmetry of the Legendre polynomial, retaining the l=1 term exactly improves the accuracy only for antisymmetric charge distributions: ρ(θ)=−ρ(−θ) and the l=2 term improves the accuracy for symmetric charge distributions: ρ(θ)=ρ(−θ). Thus, the next order that is expected to produce overall improvements in accuracy is the third order according to the terminology just introduced,
| (12) |
After similar algebraic manipulations as before, we arrive at the following third-order expression for the outside potential:
| (13) |
An analogous third-order expression exists for the inside solution, but it will not be used in this work. An optimal α for the third-order formula must lie in the interval ; we choose the middle of the interval, α=0.875, as a reasonable initial guess.
Higher-order approximations can be defined using the approach described above. Equation 14, shown below, represents the exactly summable kth-order approximation with k∕(k+1)≤α≤1 and k≥1,
| (14) |
Properties of the analytical approximations
We now establish some basic properties of the analytical approximations we have just derived.
Relation to the Poisson equation
Each of the approximate formulas just derived satisfy the Poisson equation. For the first-order Eq. 11, this is seen immediately: The expression is the sum of two Coulomb potentials multiplied by constant prefactors. For Eq. 10 one can verify explicitly that . The statement remains true for all orders of the approximation. This is because each term in the original infinite-series solution satisfies the Poisson equation; the approximate expression contains the same terms, each multiplied by its own constant.
At first glance, the fact that the analytical approximations also satisfy the Poisson equation may seem to be at odds with the uniqueness theorem that guarantees just one solution of the Poisson problem for the specific boundary conditions. Careful examination of the behavior of our analytical approximations at the boundary resolves the apparent paradox: These analytical approximations satisfy only one of the two continuity equations at the boundary, specifically Eq. 4. The other condition, Eq. 5, is satisfied only approximately; is strictly zero only for the exact infinite-series solution making the exact solution unique. Still, the fact that our analytical approximations satisfy the Poisson equation is reassuring, since it means that these analytical approximations retain some of the key physics of the problem. Their continuity across the boundary makes this surface a natural location for simultaneously testing the accuracy of both the inside and outside solutions. For this purpose we will use defined right outside the dielectric boundary (molecular surface).
The specific form of the approximate solution of order k=1 we have just derived is peculiar: It is mathematically equivalent to the sum of scaled Coulomb potentials due to each source charge plus a scaled Coulomb potential due to the total charge of the system placed in the center of the solute sphere. The scaling factors are nontrivial, but do not depend on the geometry (size) of the solute. In contrast to the multipole expansion, the applicability domain of the approximation includes distances from the solute surface considerably smaller than the solute size A.
Accuracy
For the exact spherical geometry considered so far, the error of the analytical approximation for the potential due to a single charge inside the dielectric boundary originates solely from replacing the first infinite sum in Eq. 3 with the kth-order approximation shown in Eq. 14. A rigorous error bound for this approximation would provide useful general insights into the accuracy of the formulas we have proposed. Such an upper bound is derived in the appendix,
| (15) |
For any fixed order k of the approximation, the error decreases monotonically as the parameter t=ri∕r approaches zero, i.e., as the test charge moves away from the source. Specifically, in the limit r→∞. Perhaps more interesting is the converse statement, that is, the error bound increases monotonically as the parametert=ri∕r approaches unity. This corresponds to the point of observation approaching the source charge, Fig. 1. Obviously, the closer to the source, the larger the potential itself becomes, and so it is perhaps not so surprising that the absolute error of our approximation also increases. However, for any realistic molecular structure the error stays finite. This is because the largest value of t possible in real molecules is determined by the distance of closest approach of the center of the source and test charges to molecular surface, which is determined by the radius ρvdW of the atom carrying the charge. This physical restriction sets the “worst case” value of t to be (A−ρvdW)∕A, and thus suggests that in realistic structures the approximation be tested at a distance of 1–2 Å from the surface. For a fixed geometry of the source and test points, t=const, the error bound decreases with increasing order of the approximation k and approaches zero as k→∞.
The error bound discussed above does not describe the beneficial effects of error cancelation arising from a specific choice of α. In particular, how much of an additional benefit do higher-order approximations, k>1, provide? To investigate the accuracy of our approximations further we compare the approximate formulas directly with solutions that can be considered numerically exact.
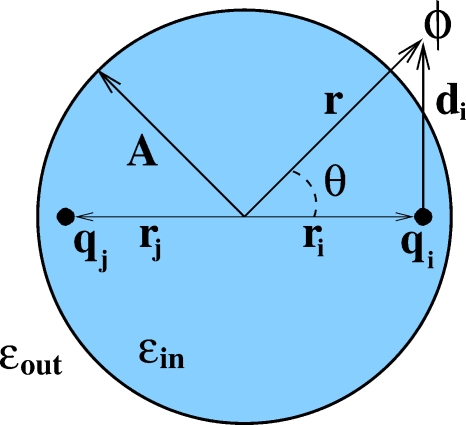
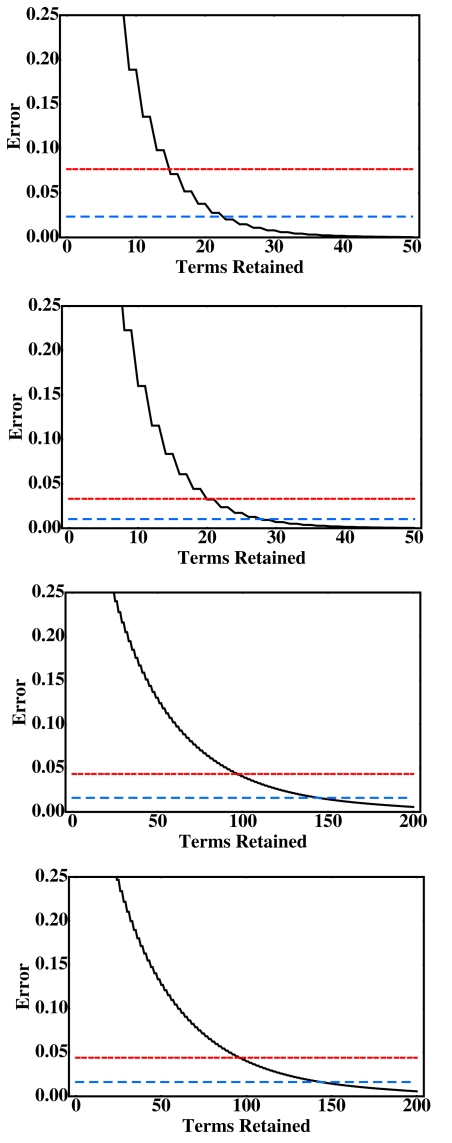
The exact solution of the Poisson equation on a sphere can be used to test the accuracy of our analytical approximations directly. In practice, we take the sum of the firstN=1000 terms in the infinite series in Eq. 3 to represent the exact solution. We use the test setup shown in Fig. 2. For a sphere of radius of 15 Å, which is the size of a typical small protein, the partial sum converges to machine precision when ∼100 terms are retained, Figs. 3a, 3b. For a larger sphere, 100 Å, which is on the order of the size of a viral capsid, all ∼1000 terms are needed for the sum to converge to machine precision, Figs. 3c, 3d. These plots demonstrate a key difference between our closed-form analytical approximations, Eqs. 11, 13, and a brute-force approach in which the first N terms in the infinite series 3 are retained to approximate ϕ(r). Depending on the size of the sphere, tens to hundreds of terms will need to be retained to achieve the same level of accuracy provided by the closed-form approximations.
Figure 2.
Setup of the test cases. Two unit charges are located on the diameter of a perfect sphere of radius A, equidistant from the center ri=rj. For the dipole case, qi=−qj, and for the dual positive case, qi=qj. The potential ϕ(r,θ) is computed at r=A for 0≤θ≤π.
Figure 3.
The root-mean-square error, in kcal∕mol per unit charge, of the various approximations to the exact solution of the Poisson equation on a sphere. The functions plotted are the error of first-order (k=1) analytical approximation, Eq. 11, with α=0.750 (double-dashed red line), the third-order (k=3) analytical approximation, Eq. 13, with α=0.875 (dashed blue line), and a partial sum solution obtained by retaining the first N terms of Eq. 3 (black curve). The potentials are computed at the surface of the sphere over the interval 0≤θ≤π; the errors are computed with respect to the exact solution, which is the converged partial sum of Eq. 3. The test geometry is shown in Fig. 2. (a) Sphere A=15 Å, dipole charge distribution, and charges located at ∣ri∣=∣rj∣=13 Å. (b) SphereA=15 Å, dual positive charge distribution, and charges at ∣ri∣=∣rj∣=13 Å. (c) Sphere A=100 Å, a dipole charge distribution, and ∣ri∣=∣rj∣=98 Å. (d) Sphere with A=100 Å, a dual positive charge distribution, and ∣ri∣=∣rj∣=98 Å.
It should be stressed that the “controllability” of the approximations just derived strictly applies only in the case of a perfectly spherical dielectric boundary. In particular, one cannot a priori expect that limk→∞∣ϕapprox(k)−ϕexact∣=0 for realistic biomolecular structures. We speculate that one may use higher orders k>1 of the approximation to explore the limits of the sphere-based approach on different classes of realistic biomolecular shapes. Namely, for some shapes and∕or regions of space one may observe systematic improvement in the accuracy with increasing k. For these shapes, one may consider the use of k>1 formulas. However, our first priority will be to adapt and test the basick=1 approximation on realistic biomolecular shapes. This is because the error analysis presented above for the spherical shape shows that the bulk of the agreement between the analytical approximations and the exact solution is already achieved within just the first-order approximation, Fig. 3. The next step, the third-order approximation given by Eq. 13, only marginally improves the agreement with the exact solution while substantially increasing the approximation’s complexity. This additional increase in complexity may not be justified, especially if one aims at using the formulas in applications where speed and stability of the algorithms are critical.
Setting parameters of the model
Later in this work we will present additional arguments for using the simpler Eqs. 10, 11 for real biomolecules. At this point we need to decide what value of the parameter α in Eqs. 10, 11 is best. While we could simply take the ad hoc value of α=0.75 that was used in Fig. 3 above, we prefer to derive the optimal α based on more rigorous grounds. A physically justified choice of α can come from the requirement that it minimizes the error between the approximate and exact ϕ(r). There are many reasonable ways to compare two scalar fields defined in 3D space (or 2D if one limits comparison to some Gaussian surface around the charge distribution, for example, the molecular surface). Here, we will use the following approach to set the value of α: Require that the best α minimizes the error in the solvation energy of a random charge distribution inside a sphere. We chose this strategy because comparing two real numbers is more straightforward than comparing two scalar fields. This comparison also allows us to make a connection between the current model and the previous ones such as the GB. To this end, we consider an arbitrary charge distribution and define the reaction field potential Φ inside the sphere. The Φ is given by the inside part of the analytical approximation, Eq. 10, less the Coulomb field: . The electrostatic part of the solvation energy is then
| (16) |
with .
A closer look at the above expression reveals that it is equivalent to Eq. (3) of Ref. 61 which is the analytic linearized PB (ALPB) model developed in Refs. 33, 61. Thus, the ALPB model with the above fij can be considered a special “discrete” case of the current first-order approximation, Eqs. 10, 11, for ϕ(ri) defined only at the location of the point charges qi. This connection allows us to use the optimal value of α=32(3 ln 2−2)∕(3π2−28)−1≈0.580127 which was rigorously derived for the ALPB model.33 This value of α should be appropriate for a random charge distribution inside the sphere. One can also check explicitly that the GB model (on a sphere) is also just a particular case of the current theory in the limits ϵout→∞ or α→0. In the ϵout→∞ limit, the analytical approximations, Eqs. 10, 11, 13, all become exact solutions of the Poisson equation on a sphere.
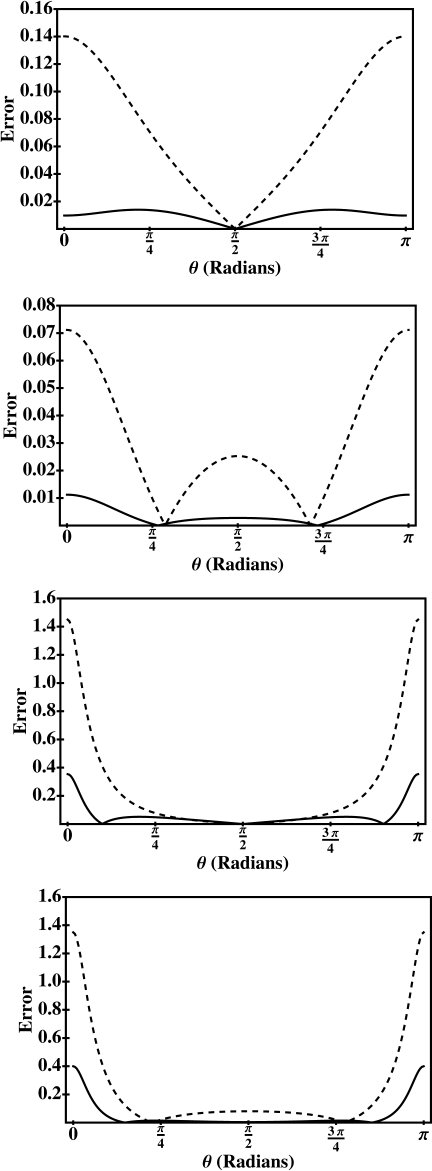
With the rigorously justified choice of an optimal value for α, our approximations, Eqs. 10, 11, become parameter-free. Their performance for the entire range 0≤θ≤π is compared to the exact solution on the surface of a sphere, Fig. 4. For comparison, the “Null model”—screened Coulomb potential 1∕ϵout∑i(qi∕di) due to the same set of charges qi—is also shown.
Figure 4.
Absolute error, in kcal∕mol per unit charge, of the first-order analytical approximation, Eq. 11, with α=0.580 127 (solid lines). The error is computed as the absolute difference between the analytical approximation and the exact solution (converged partial sum). For comparison, the absolute error of the screened Coulomb potential produced by the same charge distribution is also shown (dashed lines). The geometric setup is shown in Fig. 2. (a) Sphere A=15 Å, dipole charge distribution, and unit charges located at ∣ri∣=∣rj∣=6 Å. (b) Sphere A=15 Å, dual positive charge distribution, and unit charges at ∣r∣=∣rj∣=6 Å. (c) Sphere A=15 Å, dipole charge distribution, and unit charges located at ∣ri∣=∣rj∣=13 Å. (d) SphereA=15 Å, dual positive charge distribution, and charges at ∣ri∣=∣rj∣=13 Å.
In agreement with the considerations presented above for the error bound, the largest errors of the approximation occur when the source charges are closest to the boundary and the test charge is closest to one of the sources. For the geometry used to produce the error curves in Fig. 4, these maximal errors for k=1 approximation are ∼0.4 kcal∕mol∕∣e∣ or ∼10% of the corresponding exact value. These are of the same order of what one may expect from a “typical” numerical solution of the PB equation for a similar test charge geometry. Namely, in an earlier study,62 a geometric setup similar to ours and the same reference—numerically converged partial sum of the exact series solution for a sphere—was used to assess the accuracy of a finite-difference algorithm that was at the time implemented in the popular package DELPHI. The largest error reported in that study was ∼15% of the exact reference, for the source charge located 1 Å deep inside the dielectric boundary, and the test charges being 3 Å away from the source. One should be careful, however, not to overinterpret such comparisons between two fundamentally different approaches: The accuracy of both can be increased, albeit at additional computational expense. In the case of our analytical approximation this can be achieved by using its higher ordersk>1, while the accuracy of the NPB solutions can be improved through a variety of techniques that include focusing62 or multigrid methods.24
The errors of the approximate electrostatic solvation energies ΔGel computed via Eq. 16 for our test geometries are appreciably smaller than the errors (per unit charge) in the potential itself. Namely, for the two source charge geometries described in Fig. 4 the maximum error in ΔGel is ∼0.13 kcal∕mol or only 0.1% of the corresponding exact value. We therefore conclude that direct comparisons between approximate and exact potentials over the entire dielectric boundary is a more sensitive test of the accuracy of the type of approximation considered here. Although quite tedious, these comparisons may thus be preferred to “global metrics” such as ΔGel.
Adaptation to nonspherical shapes
The key question now is how well our analytical approximation for the solution of the Poisson equation on a sphere will perform on shapes that are not exactly spherical. The extensive testing on realistic biomolecular shapes will be presented in Part II of this work that immediately follows this paper. Here, we conclude by showing how our model can be adapted to the nonspherical case.
The first step is to decide what order k of the analytical expressions derived above is appropriate for realistic biomolecular shapes. We have already argued that since the first-order Eqs. 10, 11 and the third-order Eq. 13 perform similarly against the exact solution, Fig. 3, the extra computational complexity of introducing dependencies on Legendre polynomials might be unwarranted. Therefore, we propose that the adaptation of our approximations for realistic molecular shapes begins with the k=1, Eqs. 10, 11.
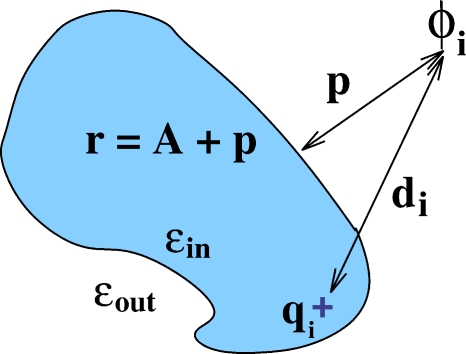
Next, we need to define all the geometrical parameters that enter Eqs. 11, 10 for the nonspherical case. The distance from the point charge to the point of observation di does not present a problem as it translates directly to the nonspherical case. The distance from the center of the sphere to the observation point r is less straightforward. Fortunately, we do have a physical parameter that characterizes the global shape of the structure and replaces the radius of the sphere in the general case—the so-called effective electrostatic radius that was introduced earlier.33 Once this parameter is computed, which can be done analytically,61 the r distance can be defined as electrostatic radius plus (or minus, if the point of observation is inside the structure) the distance p to molecular surface, see Fig. 5.
Figure 5.
Definition of the geometric parameters that enter the analytical formulas 10, 11 and can be used to compute the electrostatic potential ϕi due to a single charge located inside an arbitrary biomolecule (in the absence of mobile ions). Here di is the distance from the point of observation where ϕi needs to be computed, to the source charge qi. The distance from the point of observation to the molecular surface is p (p<0 for points inside the boundary). The so-called effective electrostatic size of the molecule, A, characterizes its global shape and is computed analytically as described in Ref. 61. The distance from the point of observation to the “center” of the molecule is then defined as r=A+p. Likewise the position of the charge ri is defined as A minus the distance of the charge to surface (not shown).
The above definition of the geometric parameters that enter formulas 10, 11 for nonspherical geometries is attractive because it treats all regions of space on the same footing. This is why it will be used throughout this work, particularly in Part II. However, depending on specific application, one may find some more restrictive alternatives useful. We note in this respect that the accuracy of the outside solution, Eq. 11, is rather insensitive to the precise definition of r. This is because the maximum error of the approximation occurs closest to the source on the dielectric boundary, and at this region the 1∕di terms dominate. To be specific, consider the following example. Suppose the goal is to get a quick estimate of just (solvent space), then one can proceed by determining a meaningful geometric center of the structure, and then define r simply as the distance to it. Since, according to the main definition in Fig. 5, r cannot be less than A for points outside the structure, one should set r=A for all r≤A. For an overall neutral molecule, ∑iqi=0, and the computation simplifies even further as the explicit dependence on r cancels from the total potential obtained via Eq. 11.
CONCLUSIONS
In this study we have shown how the exact infinite-series solution of the Poisson equation for an arbitrary charge distribution inside a spherical dielectric boundary can be approximated by a simple analytical formula. We have derived such expressions for the potentials both inside and outside the dielectric boundary, for arbitrary internal and external dielectrics. Unlike the GB model, our model defines electrostatic potential everywhere in 3D space; this parameter-free approximate expression is itself a solution of the Poisson equation, which means that it retains some of the key physics of the problem. We show how an apparent contradiction with the uniqueness theorem of electrostatics is resolved. We have extensively tested the accuracy of the approximation against the exact infinite-series solution represented by its numerically converged partial sum. The errors are assessed for two source charges placed inside the spherical boundary separating the solute of dielectric 1 and the solvent of dielectric 80. We analyzed the errors resulting from several locations of the source charges on the opposite sides of the diameter of the sphere. For unit source charges placed within 2 Å from the boundary, and the test surface located on the boundary, we find the root-mean-square error of the approximate potential to be less than 0.1 kcal∕mol∕∣e∣ (per unit test charge). In agreement with the predictions based on a rigorously derived error bound, the largest errors in the approximate potential arise from configurations in which the source charge is closest to the dielectric boundary and the test charge is closest to the source. This maximum error of 0.4 kcal∕mol∕∣e∣ or ∼10% of the exact value corresponds to the source charges being 2 Å apart in our test geometry, that is, less than a typical salt-bridge distance. The errors of the approximate electrostatic solvation energies computed via the approximation are noticeably smaller than the corresponding errors in the potential itself. Thus, direct comparisons between approximate and exact potential over the entire dielectric boundary, although tedious, appear to be a more sensitive test of the accuracy of the type of approximation considered here than comparisons based on solvation energy.
Just like the perturbation theory, our approximation is fully controllable, at least in the perfect spherical case considered in this work: it is rigorously shown that the error approaches zero with the increasing order of the approximation. However, unlike the perturbation theory, the approximation is not equivalent to a sum of the first few terms of the infinite-series solution: it effectively retains all of the terms, albeit approximately. To achieve the equivalent accuracy by a straightforward summation of the exact infinite-series solution, tens or even hundreds of terms would have to be retained for realistic charge distributions. While we cannot claim full controllability for realistic biomolecular shapes, we speculate that for some shapes and∕or regions of space one may observe systematic improvement in the accuracy with increasing order of the approximation. These improvements are likely to be small though: for the perfectly spherical shape the bulk of the agreement between the analytical approximations and the exact solution is already achieved within just the first-order approximation. Thus, testing the first-order formulas on realistic molecular structures should be the first priority. These tests are performed in Part II of this study that immediately follows.
ACKNOWLEDGMENTS
The authors thank Grigori Sigalov for reading the manuscript and providing valuable feedback. This work was supported by NIH Grant No. GM076121 and ASPIRES seed grant from Virginia Tech. A.T.F. acknowledges support from NSF IGERT Grant No. DGE-0504196.
APPENDIX: DERIVATION DETAILS
Boundary value problem
The derivation refers to the setup shown in Fig. 1. The fixed charges exist only in region I, and so the corresponding Poisson equation is
| (A1) |
where the point charge density is placed on the z-axis at position ri.
In region II,
| (A2) |
These two regions in the spherically symmetric case are 0≤r≤A and A≤r<∞, with the charge located on the z-axis, a distance ri from the origin. The solution of the Poisson equation for region I, Eq. A1, is the sum of Coulomb’s potential due to the point charge qi and the reaction field part. Due to azimuthal symmetry, the solution depends only on the angle θ through Legendre polynomials Pl(cos θ),
| (A3) |
Using the following definitions:
| (A4) |
and the well-known identity,60
| (A5) |
the solution for region I is
| (A6) |
No fixed charges are present in region II, which gives
| (A7) |
where B and C are constants determined by the continuity conditions at the boundary r=A: and . For the remaining boundary condition, the continuity of the tangential components of the electric field ∂ϕi∕∂θ will be satisfied automatically for the unique exact solution of the Poisson equation.
The first boundary condition gives
| (A8) |
Because of the orthogonality of the Legendre polynomials, the equality simplifies to a relation between Bl and Cl,
| (A9) |
or, after integration,
| (A10) |
The second boundary condition equates the normal components of the electric displacement fields of the two regions,
| (A11) |
The orthogonality relation between the Legendre polynomials is used again to simplify Eq. A11, thus providing the second relationship between Bl and Cl,
| (A12) |
Equations A10, A12 are solved simultaneously to give independent expressions for Bl and Cl,
| (A13) |
| (A14) |
Recall that the equation for region I is
| (A15) |
Let t=r<∕r>, then the equation for region I becomes
| (A16) |
After summing up the first infinite series, Eq. A16 becomes
| (A17) |
Figure 1 represents the geometry definition and defines . By replacing cos θ with this identity and simplifying the potential in region I, I becomes
| (A18) |
To simplify the equation, define the dimensionless distance parameter t=(rir∕A2). Then
| (A19) |
For region II, the dimensionless distance parameter ist=ri∕r; substituting the result for Cl into Eq. A7 yields the potential in region II,
| (A20) |
Error bound
The error of the approximate analytic solution for the potential in region II for a single charge in a sphere originates from replacing the first infinite sum in Eq. 3 with the kth-order approximation shown in Eq. 14. Since the terms with l<k in this approximation are exact, the error is
| (A21) |
A relatively simple upper bound for the above infinite sum is available, which depends on the value of k chosen for the order of the approximation. First, notice that since ∣∑ab∣≤∑∣a∣∣b∣, the above error is largest when all tlPl(cos θ) are largest and of the same sign, which occurs at cos θ=0 when Pl(cos θ)=1 (t≥0 by definition). Then, sincek∕(k+1)<α<1, l∕(l+1)<1, and l≥k in Eq. A21, one can check that ∣[1∕(1+αβ)−1∕(1+(l∕(l+1))β)]∣≤[1∕(1+(k∕(k+1))β)−1∕(1+β)]. This yields the following expression for the upper bound on :
| (A22) |
After performing the summation of the geometric series in the above equation along with some algebraic manipulation, we arrive at
| (A23) |
In reality, β is always positive, which allows us to also write
| (A24) |
In the important case of aqueous solvation, β⪡1, this somewhat simpler expression has essentially the same numerical value as the one above it.
References
- Perutz M., Science 10.1126/science.694508 201, 1187 (1978). [DOI] [PubMed] [Google Scholar]
- Honig B. and Nicholls A., Science 10.1126/science.7761829 268, 1144 (1995). [DOI] [PubMed] [Google Scholar]
- Davis M. E. and McCammon J. A., Chem. Rev. (Washington, D.C.) 10.1021/cr00101a005 90, 509 (1990). [DOI] [Google Scholar]
- Baker N. A. and McCammon J. A., Structural Bioinformatics (Wiley, New York, 2002). [Google Scholar]
- Warshel A. and Åqvist J., Annu. Rev. Biophys. Biophys. Chem. 10.1146/annurev.bb.20.060191.001411 20, 267 (1991). [DOI] [PubMed] [Google Scholar]
- Warshel A., Biochemistry 10.1021/bi00514a028 20, 3167 (1981). [DOI] [PubMed] [Google Scholar]
- Fersht A., Shi J., Knill-Jones J., Lowe D., Wilkinson A., Blow D., Brick P., Carter P., Waye M., and Winter G., Nature (London) 10.1038/314235a0 314, 235 (1985). [DOI] [PubMed] [Google Scholar]
- Szabo G., Eisenman G., McLaughlin S., and Krasne S., Ann. N.Y. Acad. Sci. 195, 273 (1972). [PubMed] [Google Scholar]
- Douglas T. and Ripoll D. R., Protein Sci. 7, 1083 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman F. B., Norel R., and Honig B., Curr. Opin. Struct. Biol. 10.1016/S0959-440X(00)00065-8 10, 153 (2000). [DOI] [PubMed] [Google Scholar]
- Onufriev A., Smondyrev A., and Bashford D., J. Mol. Biol. 10.1016/S0022-2836(03)00903-3 332, 1183 (2003). [DOI] [PubMed] [Google Scholar]
- Yang A. -S. and Honig B., Curr. Opin. Struct. Biol. 10.1016/0959-440X(92)90174-6 2, 40 (1992). [DOI] [Google Scholar]
- Whitten S. and Garcia-Moreno B., Biochemistry 10.1021/bi001015c 39, 14292 (2000). [DOI] [PubMed] [Google Scholar]
- Chin K., Sharp K. A., Honig B., and Pyle A. M., Nat. Struct. Biol. 10.1038/14940 6, 1055 (1999). [DOI] [PubMed] [Google Scholar]
- Cramer C. and Truhlar D., Chem. Rev. (Washington, D.C.) 10.1021/cr960149m 99, 2161 (1999). [DOI] [PubMed] [Google Scholar]
- Roux B. and Simonson T., Biophys. Chem. 10.1016/S0301-4622(98)00226-9 78, 1 (1999). [DOI] [PubMed] [Google Scholar]
- Gallicchio E. and Levy R., J. Comput. Chem. 10.1002/jcc.10400 25, 479 (2004). [DOI] [PubMed] [Google Scholar]
- Linderström-Lang K., C. R. Trav. Lab. Carlsberg 15, 1 (1924). [Google Scholar]
- Kirkwood J. G., J. Chem. Phys. 10.1063/1.1749489 2, 351 (1934). [DOI] [Google Scholar]
- Tanford C. and Roxby R., Biochemistry 10.1021/bi00761a029 11, 2192 (1972). [DOI] [PubMed] [Google Scholar]
- Stigter D., Alonso D. O., and Dill K. A., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.88.10.4176 88, 4176 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura J. D., Davis M. E., Gilson M. K., Wade R. C., Luty B. A., and McCammon J. A., Rev. Comput. Chem. 10.1002/9780470125823.ch4 5, 229 (1994). [DOI] [Google Scholar]
- Bashford D., in Scientific Computing in Object-Oriented Parallel Environments, Lecture Notes in Computer Science, ISCOPE97 Vol. 1343, edited by Ishikawa Y., Oldehoeft R. R., Reynders J. V. W., and Tholburn M. (Springer, Berlin, 1997), pp. 233–240. [Google Scholar]
- Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.181342398 98, 10037 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchia W., Sridharan S., Nicholls A., Alexov E., Chiabrera A., and Honig B., J. Comput. Chem. 10.1002/jcc.1161 23, 128 (2002). [DOI] [PubMed] [Google Scholar]
- Luo R., David L., and Gilson M., J. Comput. Chem. 10.1002/jcc.10120 23, 1244 (2002). [DOI] [PubMed] [Google Scholar]
- Baker N. A., Curr. Opin. Struct. Biol. 10.1016/j.sbi.2005.02.001 15, 137 (2005). [DOI] [PubMed] [Google Scholar]
- Totrov M. and Abagyan R., Biopolymers 60, 124 (2001). [DOI] [PubMed] [Google Scholar]
- Lu B., Cheng X., Huang J., and McCammon J. A., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0605166103 103, 19314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abagyan R. and Totrov M., J. Mol. Biol. 10.1006/jmbi.1994.1052 235, 983 (1994). [DOI] [PubMed] [Google Scholar]
- Havranek J. J. and Harbury P. B., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.96.20.11145 96, 11145 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Deng S., and Jacobs D., J. Comput. Phys. 223, 846 (2006). [Google Scholar]
- Sigalov G., Scheffel P., and Onufriev A., J. Chem. Phys. 10.1063/1.1857811 122, 094511 (2005). [DOI] [PubMed] [Google Scholar]
- Still W. C., Tempczyk A., Hawley R. C., and Hendrickson T., J. Am. Chem. Soc. 10.1021/ja00172a038 112, 6127 (1990). [DOI] [Google Scholar]
- Srinivasan J., Trevathan M., Beroza P., and Case D., Theor. Chem. Acc. 10.1007/s002140050460 101, 426 (1999). [DOI] [Google Scholar]
- Hawkins G., Cramer C., and Truhlar D., Chem. Phys. Lett. 10.1016/0009-2614(95)01082-K 246, 122 (1995). [DOI] [Google Scholar]
- Hawkins G., Cramer C., and Truhlar D., J. Phys. Chem. 10.1021/jp961710n 100, 19824 (1996). [DOI] [Google Scholar]
- Schaefer M. and Karplus M., J. Phys. Chem. 10.1021/jp9521621 100, 1578 (1996). [DOI] [Google Scholar]
- Qiu D., Shenkin P., Hollinger F., and Still W., J. Phys. Chem. A 10.1021/jp961992r 101, 3005 (1997). [DOI] [Google Scholar]
- Edinger S., Cortis C., Shenkin P., and Friesner R., J. Phys. Chem. B 10.1021/jp962156k 101, 1190 (1997). [DOI] [Google Scholar]
- Jayaram B., Liu Y., and Beveridge D., J. Chem. Phys. 10.1063/1.476697 109, 1465 (1998). [DOI] [Google Scholar]
- Ghosh A., Rapp C., and Friesner R., J. Phys. Chem. B 10.1021/jp982533o 102, 10983 (1998). [DOI] [Google Scholar]
- Bashford D. and Case D., Annu. Rev. Phys. Chem. 10.1146/annurev.physchem.51.1.129 51, 129 (2000). [DOI] [PubMed] [Google Scholar]
- Lee M., F.Salsbury, Jr., and C.BrooksIII, J. Chem. Phys. 10.1063/1.1480013 116, 10606 (2002). [DOI] [Google Scholar]
- Felts A., Harano Y., Gallicchio E., and Levy R., Proteins 10.1002/prot.20104 56, 310 (2004). [DOI] [PubMed] [Google Scholar]
- Dominy B. and Brooks C., J. Phys. Chem. B 10.1021/jp984440c 103, 3765 (1999). [DOI] [Google Scholar]
- David L., Luo R., and Gilson M., J. Comput. Chem. 21, 295 (2000). [DOI] [PubMed] [Google Scholar]
- Spassov V., Yan L., and Szalma S., J. Phys. Chem. B 10.1021/jp020674r 106, 8726 (2002). [DOI] [Google Scholar]
- Calimet N., Schaefer M., and Simonson T., Proteins 10.1002/prot.1134 45, 144 (2001). [DOI] [PubMed] [Google Scholar]
- Tsui V. and Case D., J. Am. Chem. Soc. 10.1021/ja9939385 122, 2489 (2000). [DOI] [Google Scholar]
- Wang T. and Wade R., Proteins 10.1002/prot.10248 50, 158 (2003). [DOI] [PubMed] [Google Scholar]
- Onufriev A., Bashford D., and Case D., Proteins 10.1002/prot.20033 55, 383 (2004). [DOI] [PubMed] [Google Scholar]
- Simmerling C., Strockbine B., and Roitberg A., J. Am. Chem. Soc. 10.1021/ja0273851 124, 11258 (2002). [DOI] [PubMed] [Google Scholar]
- Nymeyer H. and Garcia A., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2232868100 100, 13934 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. and Duan Y., Proteins 10.1002/prot.10470 55, 620 (2004). [DOI] [PubMed] [Google Scholar]
- Case D. A., Cheatham T. E., Darden T., Gohlke H., Luo R., Merz K. M., Onufriev A., Simmerling C., Wang B., and Woods R. J., J. Comput. Chem. 10.1002/jcc.20290 26, 1668 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu N. V., Zhu P., and Sharp K. A., J. Comput. Chem. 10.1002/jcc.20138 25, 2049 (2004). [DOI] [PubMed] [Google Scholar]
- Onufriev A., Bashford D., and Case D., J. Phys. Chem. B 10.1021/jp994072s 104, 3712 (2000). [DOI] [Google Scholar]
- Roe D. R., Okur A., Wickstrom L., Hornak V., and Simmerling C., J. Phys. Chem. B 10.1021/jp066831u 111, 1846 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J., Classical Electrodynamics, 3rd ed. (Wiley, New York, 1999). [Google Scholar]
- Sigalov G., Fenley A., and Onufriev A., J. Chem. Phys. 10.1063/1.2177251 124, 124902 (2006). [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Sharp K. A., and Honig B. H., J. Comput. Chem. 10.1002/jcc.540090407 9, 327 (1988). [DOI] [Google Scholar]