Abstract
Although quantitative traits loci (QTL) analysis has been widely performed to isolate agronomically important genes, it has been difficult to obtain molecular markers between individuals with similar phenotypes (assortative mating). Recently, the miniature inverted-repeat transposable element mPing was shown to be active in the japonica strain Gimbozu EG4 where it had accumulated more than 1000 copies. In contrast, most other japonicas, including Nipponbare, have 50 or fewer mPing insertions in their genome. In this study we have exploited the polymorphism of mPing insertion sites to generate 150 PCR markers in a cross between the closely related japonicas, Nipponbare × Gimbozu (EG4). These new markers were distributed in genic regions of the whole genome and showed significantly higher polymorphism (150 of 183) than all other molecular markers tested including short sequence repeat markers (46 of 661). In addition, we performed QTL analysis with these markers using recombinant inbred lines derived from Nipponbare × Gimbozu EG4, and successfully mapped a locus involved in heading date on the short arm of chromosome 6. Moreover, we could easily map two novel loci involved in the culm length on the short arms of chromosomes 3 and 10.
Key words: Linkage mapping, Transposon, japonica, Oryza sativa L., QTL analysis
1. Introduction
The development of DNA markers has made it possible to study the naturally occurring allelic variation controlling quantitative traits loci (QTL).1,2 QTL analysis based on high-resolution linkage maps is now routinely performed in various plant species and has resulted in the mapping loci of many genes involved in agronomically important traits.3–9
A high-resolution, high-density linkage map of rice has been constructed10–12 and used for QTL analyses that identified loci and genes involved in heading date13–16 and culm length.17 Like most other QTL analyses in rice, this was performed using crosses between the subspecies indica and japonica where it is relatively easy to obtain molecular markers and construct saturated linkage maps. However, the use of such distant cross combinations can disturb efficient identification of QTLs because of the prevalence of hybrid seed sterility and the simultaneous segregations of many other loci. As such, it is difficult to identify loci or alleles of minor effect. To do so usually requires the generation of additional genetic resources including near isogenic lines or chromosome segment substitution lines which need repeated backcrosses.
In contrast, japonica × japonica cross combinations have an apparent advantage in analyzing minor QTL and identifying alleles responsible for local variation, because of the segregation of only a few loci.18 Thus, to breed japonica cultivars, DNA markers that can be used among closely related lines need to be developed. However, the difficulty in obtaining a sufficient number of evenly distributed molecular markers severely restricts the widespread applicability of linkage analysis using cross combination within temperate japonica cultivars. Although several projects to obtain more markers are currently proceeding, the recent reports reaffirmed that it is costly and time consuming to obtain suitable DNA markers.19–21 For example, Shirasawa et al.21 indicated that the average frequencies of genes having single nucleotide polymorphisms (SNP) in the whole genome within japonica cultivars are 9.2% (2944/32 000), while those between japonica and indica cultivars are 94.8% (30 336/32 000). Although some QTL analyses using short sequence repeat (SSR), SNP and restriction fragment length polymorphism (RFLP) markers on temperate japonica × temperate japonica populations have been conducted,18,22–27 the ratio of polymorphic markers among these varieties were about 10% (7.3–15.7%) (Table 1). That means, in order to obtain 100 DNA markers, screening ∼1000 markers is required.
Table 1.
Summary of QTL analysis among temperate japonica × japonica cross combinations
| Cross cultivars | Population structure | Markers used | No. of polymorphic markers/no. of screening markers (polymorphism frequency, %) | Total length, cM | Reference |
|---|---|---|---|---|---|
| Akihikari | DHLs | RFLP | 127/1252 (10.1) | No data | 23 |
| Koshihikari | RAPD | 28/862 (3.0) | |||
| Chiyonishiki | RILs | SSR | 199/1663 (11.9) | 891.1 | 25 |
| Koshijiwase | |||||
| Hana-echizen | F2, F3 | SSR | 64/407 (15.7) | No data | 19 |
| Nigatawase | |||||
| Koshihikari | BILs | SSR | No data (11.8) | 956.1, 973.1 | 26 |
| Nipponbare | SNP | No data (?) | 50 | ||
| Sakihikari | RILs | SSR | No data (?) | No data | 27 |
| Nipponbare | |||||
| Moritawase | RILs | SSR | 130/1780 (7.3) | 1060 | 24 |
| Koshihikari | |||||
| Suweon365 | RILs | SSR | 121/621 (19.5) | 2227 | 27 |
| Chucheongbyeo | AFLPs | 118/908 (12.9) | |||
| MITEs | 52/71 (42.3) | ||||
| Gimbozu | RILs | mPing-SCAR | 150/183 (82.3) | 1771 | This study |
| Nipponbare | SSR | 46/661 (7.0) |
Transposable elements (TEs) have also been exploited in the development of molecular markers.28–33 In this regard, the most valuable TEs are those that are actively transposing and generating insertion site polymorphisms. Unfortunately, the rice genome is relatively stable with very few actively transposing elements. So far, only two TEs have been employed as markers among temperate japonica varieties.27 Although TE insertion polymorphism was shown to be more frequent (42.3%) than other DNA markers, the total number of applicable TE markers was only 52.27
Here we report the successful generation of molecular markers using the newly characterized mPing element. This element was independently discovered by three labs as the first active miniature inverted-repeat transposable element as well as the first active DNA transposon in rice.34–36 A subsequent study revealed that mPing copy number was generally less than 50 in most cultivars, but that it had amplified to over 1000 copies in one strain, Gimbozu EG4 (EG4, hereafter). Thus, there are more than 1000 insertion site polymorphisms of mPing when EG4 is compared with other cultivars including all characterized strains of the japonica subspecies. Of these 1000 insertions, most of them (90%) were into the single copy regions, and more than 70% were located within 5 kb of transcribed DNA.37
In this study, we have designed 150 sequence characterized amplified region (SCAR) markers based on the sequence information of mPing insertion sites in EG4. We will discuss the advantage of this novel marker system, which can be easily applicable for genetic analyses between closely related genomes.
2. Materials and methods
2.1. Plant materials and genomic DNA extraction
In 2005, a total of 190 F5 recombinant inbred lines (RILs) (12 plants per line) of EG4 × Nipponbare were grown in the paddy field of Kyoto University (35°01′N) under natural day length conditions. Seeds were sown on 17 May and transplanted on 7 June in an irrigated rice field. The heading date of F5 plants (190 lines × × 12 plants/line = 2280) and parental plants was determined by calculating the average value of each line, monitoring for the appearance of the first panicle. Leaf material for DNA extraction was collected after monitoring heading date to minimize damage to F5 plants.
In 2007, a total of 96 F7 RILs (12 plants/line) were grown under the same conditions as described above. Seeds were sown on 1 May and transplanted on 24 May in an irrigated rice field. The culm length of those plants was measured to the top internodes. The heading date of those plants was also determined as described above. Genomic DNA of all plant materials was extracted by the cetyl tri-methylammonium bromide (CTAB) method.38
2.2. SSR markers
PCR for SSR markers was performed in 5 µL containing 0.5 µL 10× Ex-Taq buffer, 0.5 µL dNTPs (2 mM each), 0.25 µL DMSO, 0.02 µL Ex taq, l00 ng genomic DNA and 5 pmol of each primer. PCR conditions were as follows: 94°C for 10 min; 35 cycles of 94°C for 30 s, 50°C for 1 min and 72°C for 30 s; 72°C for 7 min.
Reaction products were loaded on 12% polyacrylamide gels, run at 500 v for 2 h, stained with ethidium bromide and gels were scanned with molecular imager FX (Nippon Bio-Rad Laboratories, Tokyo).
2.3. mPing-SCAR markers
Primers flanking mPing insertion sites in EG4 were designed based on sequence information from a previous study.37 PCR for mPing-SCAR markers was performed in 5 µL containing 0.5 µL 10× Ex-Taq buffer, 0.5 µL dNTP mix (2 mM each), 0.25 µL DMSO, 0.02 µL Ex-Taq, l00 ng genomic DNA and 8 pmol of each primer. The cycling parameters were as follows: 94°C for 3 min; 30 cycles of 95°C for 30 s, 58°C for 45 s and 72°C for 3 min; 72°C for 3 min.
Amplicons of mPing-SCAR markers were resolved in 1.0% agarose gels, stained with ethidium bromide and visualized under UV light.
2.4. Determining mPing excision frequency
mPing excision was assessed by PCR. Primers were designed flanking six mPing insertion sites that are shared by Nipponbare and EG4. Their sequences are: AC107315_U1: TGAAATAACATAGCCATACCAG, AC107315_L1: AGGGTTTTTGGATACGAATGA, AC083492_U1: GATGTTGATGGTAGTGTGAGAG, AC083492_L1: TGCTGTAATAGTTTGGGGGTAG, AC121491_U1: GGTAACTGTAGTAGCGTAGTG, AC121491_L1: GAGAGCATCCACAACGAATAAT, AP003542_U1: TTGGGTAGGGTTGTGGGGATTT, AP003542_L1: GAGATGGGACTTGAGGAGAGAC, AP003634_U1: AAAAGAAAACAGAAAACAGTCG, AP003634_L1: CGCTCAACATAAACCAAAAACC, BX000500_U1: AGTTGATACGATAATGCTTCTA, and BX000500_L1: CTTCTTTTCTCTCCTCCTTTTC. For EG4-specific insertion sites, 20 primer pairs of mPing-SCAR markers (MK1_1, MK1_10, MK1_12, MK1_13, MK1_22, MK1_23, MK1_28, MK1_29, MK1_30, MK1_32, MK2_1, MK2_2, MK2_4, MK2_5, MK2_8, MK2_23, MK3_2, MK3_9, MK3_13 and MK5_2) were chosen to analyze excision. PCR conditions were the same as described above.
2.5. Mapping of mPing-insertion sites
Chromosomal distribution of the mPing-SCAR markers and SSR markers were analyzed with 190 F5 RILs derived from a cross between EG4 and Nipponbare. Linkage analysis was performed with Mapmaker version 3.0.39
2.6. QTL analysis of heading date and culm length
Composite interval mapping,40 which combines interval mapping with multiple regression, was done using Windows QTL Cartographer 2.0 with forward and backward regression.41 The experiment-wise LOD threshold significance level was determined by computing 1000 permutations,42 as implemented by the QTL cartographer.
3. Results and discussion
3.1. Screening for SSR markers
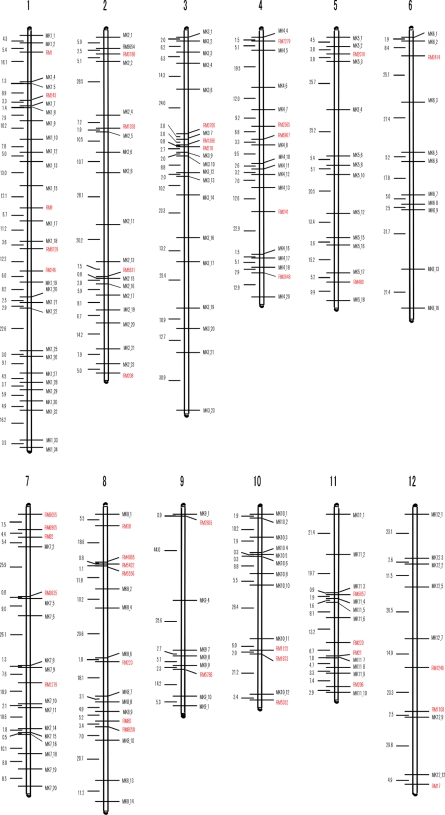
SSR markers were first used for genotyping Nipponbare/EG4 RILs. However, only 46 of 661 markers tested (7.0%) were polymorphic. In addition, the distribution of these markers was uneven. Only a few markers were found polymorphic on chromosomes 3, 5, 6 and 9–12. In addition, there were long stretches of the genome without any markers on each of the 12 chromosomes of rice (see Fig. 1 for details). These results illustrate the difficulty of using conventional molecular markers for linkage analysis of japonica × japonica cross.
Figure 1.
The genetic map based on the mPing-SCAR markers and SSR markers. mPing-SCAR markers are shown in black whereas SSR markers are shown in red (see text for details).
3.2. Designing mPing-SCAR markers
In a prior study it was determined that there were over 1000 insertion site polymorphisms of mPing between EG4 and Nipponbare.37 To exploit these polymorphisms in the development of molecular markers, we designed primer pairs based on genomic sequences flanking carefully selected mPing insertions (Fig. 2A). Insertion site polymorphisms were easily detected in both homozygous and in heterozygous genotypes, indicating that these primer sets could serve as co-dominant markers (Fig. 2B). The genotypes could be easily determined by 15-min electrophoresis in 1.0% agarose gels (Fig. 2B). Of 219 primer sets tested, 183 were successfully amplified, and 150 were found to be polymorphic between EG4 and Nipponbare.
Figure 2.
The use of mPing-SCAR markers. (A) Schematic of a polymorphic locus. Grey triangles represent the inverted-repeat of mPing and arrows indicate the positions of locus specific primers. (B) An example of a mPing-SCAR marker. 100 bp-ladder (M) is followed by EG4 (G), Nipponbare (N) and the RILs. The gel image was visualized after 15 min running at 100 V.
3.3. Stability of mPing-SCAR markers
Transposition usually involves two events, element excision and element reinsertion elsewhere in the genome. In the previous finding, there are approximately 50 new mPing insertions per plant per generation in EG4.37 If mPing excises frequently, it has no value as a molecular markers. Thus, the excision frequency of six insertion sites was checked in 400 Nipponbare and 399 EG4 plants by PCR with flanking primers. Of all insertion sites tested, not a single empty site was detected (data not shown). In addition, 20 EG4-specific insertion sites in 96 EG4 plants were also tested for excision events by PCR using flanking primers. Only one plant showed a single excision at MK1_22 on chromosome 1 (see Materials and methods for details and Fig. 1). No empty sites were detected in any other plants (data not shown).
Based on the haploid copy number of EG4 (1163)37 and the average number of new insertions (50)37 per plant per generation, the probability of excision for any single copy of mPing should be 0.0215 (=50/(1163 × 2)). However, the observed excision frequency in the 96 EG4 plants was 80-fold lower than the expected value (only 0.00 026 (=1/(20 × 96 × 2))). How can there be so many new insertions without any apparent excision of mPing? This contradiction can be explained if mPing moves by the gap repair mechanism where double-strand breaks generated by mPing excisions are repaired by utilizing a copy of mPing from either the sister chromatid or from the homologous chromosome.43,44 In total, mPing excisions are very rare events and do not disturb linkage analysis.
On the other hand, segregation distortion has always been observed in other linkage analysis. For example, in the F2 population of Nipponbare (japonica) × Kasalath (indica), 11 loci were detected to deviate from the expected 1:2:1 segregation.45 Kwon et al.31 also reported that 40–60% of the markers using insertion polymorphisms of TEs showed distorted segregation in the RILs of Milyang23 (indica) × Gihobyeo (japonica). In contrast, when 190 F5 RILs were surveyed with the 150 SCARs, the segregation ratio of genotypes (EG4:Nipponbare) was 1:1.01 (16 356:16 502), indicating that segregation was not significantly distorted. While this can be attributed to the close relationship between EG4 and Nipponbare, it also demonstrates the reliability of mPing-SCAR markers. However, it should be noted that the frequency of genotypes of the 150 mPing-SCAR marker-loci (EG4:Nipponbare=16 356:16 502) was slightly lower than that of the 46 SSR marker loci (3943:3894). This may reflect mPing activity in the EG4 background, that is, some of the mPing elements may have excised.
3.4. Genetic mapping of mPing-SCAR and SSR markers
We constructed a genetic map (with Mapmaker version 3.0) that included 150 mPing-SCAR markers and 46 SSR markers in all 12 linkage groups (Fig. 1; Table 2 and see Supplementary Table S1). The total length of the genetic map was 1771 cM with an average genetic distance between markers of 9.0 cM. The number of markers per chromosome ranged from 9 on chromosome 9 to 31 on chromosome 1, and the length of genetic region per chromosome was from 92.9 cM (chromosome 11) to 223 cM (chromosome 1) (Table 2). This result is consistent with the lengths of these chromosomes as determined by the International Rice Genome Sequencing Project (2005): chromosomes 1 through 3 are larger than chromosomes 9 through 11.46 Moreover, by comparing the genetic distances of mPing-SCAR markers with their physical locations (according to the annotated rice genome) (see Supplementary Table S1), we find that the genetic distances strongly correlate with physical distances, thus providing another indication of the accuracy of mPing-SCAR markers (Table 2; see Supplementary Table S1).
Table 2.
The number of mapped markers and genetical distances on each chromosome in the RIL mapping populations
| Molecular marker | Number of markers on chromosome |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Total | |
| mPing-SCAR markers | 26 | 15 | 17 | 13 | 12 | 10 | 13 | 10 | 7 | 10 | 10 | 7 | 150 |
| SSR markers | 5 | 5 | 3 | 5 | 2 | 1 | 6 | 7 | 2 | 3 | 4 | 3 | 46 |
| Total | 31 | 20 | 20 | 18 | 14 | 11 | 19 | 17 | 9 | 13 | 14 | 10 | 196 |
| Chromosome length (cM) | 223 | 179 | 202 | 137 | 141 | 146 | 162 | 153 | 100 | 96 | 93 | 139 | 1,771 |
| Average intervals (cM) | 7.2 | 9.0 | 10.1 | 7.6 | 10.1 | 13.3 | 8.5 | 9.0 | 11.1 | 7.4 | 6.6 | 13.9 | 9.0 |
As can be seen in Fig. 1, there are still chromosomal regions lacking markers (the short arm of chromosome 4), or with few markers (only two markers on the short arm of chromosome 9) (Fig. 1; see Supplementary Table S1). However, the upper half of chromosomes 4 and 9 including centromeres is known to be heterochromatic with low gene density (http://www.tigr.org/tdb/e2k1/osa1/).47–49 In addition, no SCAR markers were in centromeric, pericentromeric or telomeric regions, according to their physical positions in the annotated rice genome (see Supplementary Table S1). These facts strongly indicate that mPing-SCAR markers are enriched in euchromatic, gene-rich regions. As such, the lack of mPing markers in heterochromatic regions may not be a serious problem if gene discovery is the objective. However, a total of 926 insertion sites were already identified and some of them are located on such heterochromatic regions (Naito et al., unpublished data). Designing more primer sets to cover the whole genomic region is now underway.
3.5. QTL analysis of the heading date
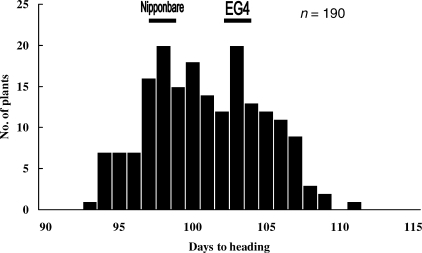
To perform QTL analysis, days to heading of Nipponbare, EG4 and RIL populations were investigated in 2005 and 2007. The difference in days to heading was very small between these parental lines, with mean values for Nipponbare and EG4 of 98 and 103 days in 2005, and 102 and 104 days in 2007. However, the variation for days to heading of RILs showed transgressive segregation (range 93–111 days in 2005 and 99–110 days in 2007) and was continuous, suggesting a bimodal distribution (Fig. 3 and Supplementary Fig. S1). These results indicated that more than two loci with opposing effects were involved in the segregation of heading date in this cross. Both QTL analyses performed in 2005 and 2007 (with QTL chartographer) detected a single QTL peak (LOD score >8.5) on the short arm of chromosome 6. Significant threshold LOD values (P < 0.05) in the F5 populations (2005) and in the F7 populations (2007) were 2.9 and 3.71, respectively. This peak was located within a 847 kb region between MK6_2 (2 034 336 nt) and RM3414 (2 881 883 nt) (Fig. 4). The additive effects of the Nipponbare allele at this locus were −1.9 (2005) and −2.2 (2007) days, respectively.
Figure 3.
Histograms of days to heading in the F5 RIL populations. Horizontal bars indicate the values of parental Nipponbare and EG4.
Figure 4.
QTL likelihood curve of the LOD score of the heading date on chromosome 6.
Although some genes concerning heading date such as Hd3a and RFT1 have been identified on short arm of chromosome 6, they are located in the interval between markers RM3414 and MK6_3 according to their sequences15 which is apparently excluded from the QTL peak detected in this study (Fig. 4).
Rather, this QTL locus fell into the same region of the recently identified locus Hd17.50 Hd17 was mapped using backcrossed inbred lines derived from Nipponbare (japonica) x Koshihikari (japonica), and the Koshihikari allele at this locus was indicated to increase the days to heading compared with Nipponbare allele by a few days. The Gimbozu allele identified in our study has almost the same effect on heading date versus Nipponbare, indicating that this locus is also Hd17. This fact also indicates that mPing-SCAR markers established here are comparable to large scale analysis using SNP/SSR markers.
In addition, it should be noted that classical genetic research had elucidated that the difference in flowering time between Gimbozu (including EG4) and Nipponbare is partly due to a flowering locus E2, with Gimbozu carrying an allele dominant (E2) to that of Nipponbare (e2).51 Koshihikari is also known to carry the dominant E2 allele.51 Thus, Gimbozu and Koshihikari probably have the same allele at this locus. Matsubara et al.50 suggested that the E2 locus corresponds to either Hd16 (between 32 855–33 966 kb on chromosome 3) or Hd17 (between 1964 and 2292 kb on chromosome 6). However, in the absence of information on chromosomal location, they were unable to determine which is the E2 locus. In this study, however, we detected one QTL overlapping Hd17 but none corresponding to Hd16. Taken together, we propose that Hd17 is the previously described E2 locus.52,53
However, in 2005, we could not detect any other QTLs although the F5 RILs showed transgressive segregation in heading date. This might be because F5 populations should be segregating for traits, leading to disruption of data analysis. In fact, the analysis using F7 RIL populations in 2007 detected some minor QTL peaks (LOD > 3.71) on chromosomes 1, 2, 3 and 12 (Supplementary Figure S2). Alternatively, the minor QTLs are in involved in response to environmental cues other than day length, such as temperature because the average temperature during the growing season was higher in 2005 than in 2007. In 2005, there were 68 days with average temperature of 25°C or higher while there were only 55 days in 2007. To further identify minor QTL loci, plants may need to be grown in growth chambers that can strictly control the growth conditions.
3.6. QTL analysis on culm length
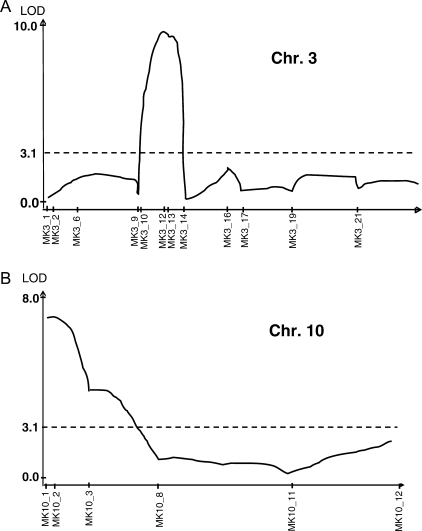
We also conducted QTL analysis on culm length using 96 F7 RILs derived from Nipponbare and EG4. The average plant heights of EG4 and Nipponbare were 84 and 70 cm, respectively. The continuous and transgressive segregation was also detected, indicating involvement of more than two loci (Fig. 5). Two putative QTLs were detected on chromosomes 3 and 10 (Fig. 6A and B). The largest QTL (LOD score >9.5) was near mPing-SCAR marker MK3_13 and the other (LOD score >7.5) was near MK10_2. Significant threshold of LOD values (P < 0.05) was 3.14. The additive effects of these QTLs on plant height are −2.8 cm (chromosome 3, Nipponbare allele) and +2.4 cm (chromosome 10, Nipponbare allele). The transgressive segregation on the culm length can mainly be explained by these two loci, although minor QTL (LOD score >3.14) was also detected (near MK9_4 on chromosome 9) (data not shown). Interestingly, no genes or QTL associated with plant height have been previously reported in these QTL regions. Thus, the use of mPing-SCAR markers may have led to the detection of novel loci or alleles within japonica cultivars.
Figure 5.
Histograms of culm length in the F7 RIL populations. Horizontal bars indicate the values of parental Nipponbare and EG4.
Figure 6.
QTL likelihood curves of the LOD score of culm length. (A) On chromosome 3. (B) On chromosome 10.
3.7. In conclusions
In this study we report the construction of a linkage map, on which QTL analysis could successfully be performed, using RIL populations derived from a cross between Nipponbare (japonica) × Gimbozu EG4 (japonica) (Fig. 1; see Supplementary Table S1) and insertion site polymorphisms of the mPing transposon.37 Unlike other DNA markers used for temperate japonica crossings, mPing-SCAR markers were highly polymorphic and covered almost the whole genomic regions. Although Kwon et al.27 obtained 221 markers in total, not a few markers formed clusters and left many gaps of >30 cM as well. In contrast, we obtained 183 markers with no clusters and attained better coverage of genomic regions than ever. Especially, the Nipponbare and EG4 are closer than any other japonica/japonica cross combinations reported before. The ratio of polymorphisms in SSR markers between the two was only 7.0%, whereas 10% or higher frequency was obtained in most other combinations (See Table 1). Furthermore, mPing-SCAR markers allowed us to easily map the candidate loci in a very short time. For genotyping each marker, only 15 min electrophoresis in 1% agarose gel is required after PCR, which is apparently simpler and easier than any other molecular markers, such as SSR, SNP and RFLP.
It should be emphasized that all characterized japonica varieties (other than EG4) have, like Nipponbare, ∼50 or fewer mPing copies.34,37 Thus, by crossing EG4 to other japonica varieties, linkage map construction and QTL analyses can easily be achieved. In this regard it is important to note the recent growing demand for rice with eating quality and the use of the japonica variety Koshihikari as a genetic resource in several molecular breeding projects.23,54 These studies should benefit from the addition of EG4 and mPing-SCAR markers to facilitate the identification of loci or genes controlling eating quality. The mPing-SCAR markers may also be valuable in uncovering genetic ‘treasures’ that remain hidden in the results of classical genetic studies pursued in Japan until 1990s. During that time japonica × japonica cross combinations were widely and vigorously undertaken to identify loci that co-segregate with unique phenotypes or traits. At present, 926 mPing insertion sites in the EG4 genome are identified (Naito et al., unpublished data). When mPing-SCAR markers of this number become available, the average distance among these markers will be estimated to be 402 kb [=372 090 kb/926markers (according to RAP-DB: http://rapdb.dna.affrc.gp.jp/)]. Considering the gene density of 1 gene/11.8 kb [=31 439 genes/372 090 kb (by RAP-DB)] in the rice genome, the estimated gene numbers between markers will be 31.4.
Although the progress of next-generation sequencing techniques is remarkable today,55,56 only the sequence information of the whole genome is not enough to clone a gene responsible for a phenotype. However, delimiting candidate genes by ∼30 using mPing-SCAR markers would make it much more efficient to detect the target gene in combination with these sequencing techniques and well-developed database information. Once the candidate region is determined, the few sequence polymorphisms among japonica cultivars also make it far easier to clone the target gene. We are confident that the novel marker system developed in this study will dramatically save time, money and manpower in molecular breeding of rice.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
The study was funded by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan as Research Project Grants-in-Aid for Scientific Research 1830005 and 17380003, and a plant genome grant from the National Science Foundation (to S.R.W.).
Acknowledgements
We thank Qingbo Yuan for his technical assistance and encouragement.
Footnotes
Edited by Masahiro Yano
References
- 1.Yano M. Genetic and molecular dissection of naturally occurring variation. Current Opinion in Plant Biology. 2001;4:130–135. doi: 10.1016/s1369-5266(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 2.Ashikari M., Matsuoka M. Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Science. 2006;11:344–350. doi: 10.1016/j.tplants.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Paterson A. H., Lander E. S., Hewitt J. D., Peterson S., Lincoln S. E., Tanksley S. D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphism. Nature. 1988;335:721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- 4.Edwards M. D., Helentjaris T., Wright S., Stuber C. W. Molecular-marker-facilitated investigations of quantitative trait loci in maize. IV. Analysis based on genome saturation with isozyme and restriction fragment length polymorphism markers. Theor. Appl. Genet. 1992;83:765–774. doi: 10.1007/BF00226696. [DOI] [PubMed] [Google Scholar]
- 5.Stuber C. W., Lincoln S. E., Wolff D. W., Helentjaris T., Lander E. S. Identification of genetics factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin G. B., Brommonschenkel S. H., Chunwonge J., et al. Map-based cloning of a protein kinase gene conferring disease restriction in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 7.DeVicente M. C., Tanksley S. D. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics. 1993;134:585–596. doi: 10.1093/genetics/134.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yano M., Sasaki T. Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 1997;35:145–153. [PubMed] [Google Scholar]
- 9.Nishimura A., Ashikari M., Lin S., et al. Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc. Natl Acad. Sci. USA. 2005;102:11940–1. doi: 10.1073/pnas.0504220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harushima Y., Yano M., Shomura A., et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1988;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurata N., Nagamura Y., Yamamoto K., et al. A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat. Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- 12.McCouch S. R., Teytelman L., Xu Y., et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- 13.Yano M., Katayose Y., Ashikari M., et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi Y., Shomura A., Sasaki T., Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl Acad. Sci. USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima S., Takahashi Y., Kobayashi Y., et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 16.Doi K., Izawa T., Fuse T., et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashikari M., Jianzhong W., Masahiro Y., Sasaki T., Yoshimura A. Rice gebberelline-insentive dwarf mutant gene Dwarf1 encodes the α-subunit of GTP-binding protein. Proc. Natl Acad. Sci. USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi A., Genliang B., Shenghai Y., Tomita K. Detection of quantitative trait loci for white-back and basal-white kernels under high temperature stress in japonica rice varieties. Breeding Science. 2007;57:107–116. [Google Scholar]
- 19.Nasu S., Suzuki J., Ohta R., et al. Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Res. 2002;9:163–171. doi: 10.1093/dnares/9.5.163. [DOI] [PubMed] [Google Scholar]
- 20.Monna L., Ohta R., Masuda H., Koike A., Minobe Y. Genome-wide searching of single-nucleotide polymorphisms among eight distantly and closely related rice cultivars (Oryza sativa L.) and a wild accession (Oryza rufipogon Griff.) DNA Res. 2006;13:43–51. doi: 10.1093/dnares/dsi030. [DOI] [PubMed] [Google Scholar]
- 21.Shirasawa K., Maeda H., Monna L., Kishitake S., Nishio T. The number of genes having different alleles between rice cultivars estimated by SNP analysis. Theor. Appl. Genet. 2007;115:1067–1074. doi: 10.1007/s00122-007-0632-z. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi Y., Hayasaka H., Chiba B., Tanaka I., Shimano T. Mapping quantitative trait loci controlling cool-temperature tolerance at booting stage in temperate japonica rice. Breeding Science. 2001;51:191–197. [Google Scholar]
- 23.Wada T., Uchimura Y., Ogata T., Tsubone M., Matsue Y. Mapping of QTLs for physicochemical properties in japonica rice. Breeding Science. 2006;56:253–260. [Google Scholar]
- 24.Tabata M., Hirabayashi H., Takeuchi Y., Ando I., Iida Y., Ohsawa R. Mapping of quantitative trait loci for the occurrence of white-back kernels associated with high temperatures during the ripening period of rice (Oryza sativa L.) Breeding Science. 2007;57:47–52. [Google Scholar]
- 25.Takeuchi Y., Hori K., Suzuki K., Nonoue Y., Takemoto-Kuno Y. Major QTLs for eating quality of an elite Japanese rice cultivar, Koshihikari, on the short arm of chromosome 3. Breeding Science. 2008;58:437–445. [Google Scholar]
- 26.Kobayashi A., Tomita K. QTL detection for stickiness of cooked rice using recombinant inbred lines derived from crosses between japonica rice cultivars. Breeding Science. 2008;58:419–426. [Google Scholar]
- 27.Kwon S. J., Cho Y. C., Kwon S. W., et al. QTL mapping of agronomic traits using an RIL population derived from a cross between temperate japonica cultivars in rice (Oryza sativa L.) Breeding Science. 2008;58:271–279. [Google Scholar]
- 28.Casa A. M., Brouwer M., Nagel A., et al. The MITE family Heartbreaker (Hbr): molecular markers in maize. Proc. Natl Acad. Sci. USA. 2000;97:10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang N., Bao Z., Temnykh S., et al. Dasheng: a recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics. 2002;161:1293–1305. doi: 10.1093/genetics/161.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park K. C., Lee J. K., Kwon S. J., et al. Distribution of MITE transposons in a rice genetic map. Korean J. Breed. 2004;36:95–102. [Google Scholar]
- 31.Kwon S. J., Hong S. W., Son J. H., et al. CACTA and MITE transposon distributions on a genetic map of rice using F15 RILs derived from Milyang 23 and Gihobyeo hybrids. Mol. Cells. 2006;21:360–366. [PubMed] [Google Scholar]
- 32.Lee J. K., Park J. M., Kim J. H., et al. Genetic mapping of the Isaac-CACTA transposon in maize. Theor. Appl. Genet. 2006;113:16–22. doi: 10.1007/s00122-006-0263-9. [DOI] [PubMed] [Google Scholar]
- 33.Huang X., Lu G., Zhao Q., Liu X., Han B. Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol. 2008;148:25–40. doi: 10.1104/pp.108.121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang N., Bao Z., Zhang X., et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 35.Kikuchi K., Terauchi K., Wada M., Hirano H. The plant MITE mPing is mobilized in anther culture. Nature. 2003;421:167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- 36.Nakazaki T., Okumoto Y., Horibata A., et al. Mobilization of a transposon in the rice genome. Nature. 2003;421:170–172. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- 37.Naito K., Cho E., Yang G., et al. Dramatic amplification of a rice transposable element during recent domestication. Proc. Natl Acad. Sci. USA. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lincoln S. E., Daly M., Lander E. S. Cambridge, MA: Whitehead Institute for Biomedical Research Technical Report; 1992. Construction genetic maps with MAPMAKER/EXP 3.0. [Google Scholar]
- 40.Zeng Z. B. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basten C. J., Weir B. S., Zeng Z. B. Raleigh, NC: Department of Statistics, North Carolina State University; 1998. QTL CARTOGRAPHER: a reference manual and tutorial for QTL mapping. [Google Scholar]
- 42.Churchill G. A., Doerge R. W. Genetics. 1994. Empirical threshold values for quantitative trait mapping; pp. 963–971. 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 44.Lisch D., Chomet P., Freeling M. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics. 1995;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harushima Y., Kurata N., Yano M., et al. Detection of segregation distortions in an indica–japonica rice cross using a high-resolution molecular map. Theor. Appl. Genet. 1992;92:145–150. doi: 10.1007/BF00223368. [DOI] [PubMed] [Google Scholar]
- 46.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Z., Buell C. R., Wing R. A., Gu M., Jiang J. Toward a cytological characterization of the rice genome. Genome Res. 2001;11:2133–2141. doi: 10.1101/gr.194601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Q., Zhang Y. J., Hao P., et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Q., Zhang Y., Cheng Z., et al. A fine map of the rice chromosome 4. Genome Res. 2002;12:817–823. doi: 10.1101/gr.48902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsubara K., Kono I., Hori K., et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theor. Appl. Genet. 2008;117:935–945. doi: 10.1007/s00122-008-0833-0. [DOI] [PubMed] [Google Scholar]
- 51.Okumoto Y., Tanisaka T., Yamagata H. Heading-time genes of the rice varieties grown in the Tohoku-Hokuriku region in Japan. Jpn. J. Breed. 1992;42:121–135. [Google Scholar]
- 52.Yamagata H. Heading-time genes of rice, E1, E2 and E3. Rice Genet. Newsl. 1984;1:100–101. [Google Scholar]
- 53.Yamagata H., Okumoto Y., Tanisaka T. Rice Genetics. The Philippines: International Rice Research Institute; 1986. Analysis of genes controlling heading time in Japanese rice; pp. 351–359. [Google Scholar]
- 54.Blakeney A. B., Lewin L. G., Reinke R. F. Australia: Rural Industries Research and Development Corporation; 2001. Quality rice for north Asia. [Google Scholar]
- 55.Blow N. The personal side of genomics. Nature. 2007;449:627–630. doi: 10.1038/449627a. [DOI] [PubMed] [Google Scholar]
- 56.Hutchison C. A., III DNA sequencing: bench to beside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.