Summary
Lysine 56 acetylation in the helical core of histone H3 opens yeast chromatin and enables histone gene transcription, DNA replication, DNA repair, and prevents epigenetic silencing. While K56Ac is globally abundant in yeast and flies its presence has been uncertain in mammals. We show here using mass spectrometry and genome wide analyses that K56Ac is present in human embryonic stem cells (hESCs) overlapping strongly at active and inactive promoters with the binding of the key regulators of pluripotency NANOG, SOX2 and OCT4. This includes also the canonical histone gene promoters and those for the hESC-specific microRNAs. K56Ac then relocates to developmental genes upon cellular differentiation. Thus K56Ac state more accurately reflects the epigenetic differences between hESCs and somatic cells than other active histone marks such as H3 K4 tri-methylation and K9 acetylation. These results suggest that K56Ac is involved in the human core transcriptional network of pluripotency.
Introduction
Histone modifications play important roles in eukaryotic cellular physiology affecting DNA based functions (Kouzarides, 2007). Most known modifications occur at the N-termini of histone tails that extend from the nucleosome core. These modifications may function by regulating higher order chromatin structures or by regulating binding surfaces for regulatory proteins (Kouzarides, 2007). However, histone tail modifications do not appear to directly affect the structure of the nucleosome itself (Luger et al., 1997).
Unlike acetylation sites at the N termini that extend from the nucleosome core, K56Ac occurs in the α-N helical region of histone H3 near the entry-exit sites of the DNA superhelix. There, the acetylation of K56 is believed to lead to the unfolding of nucleosomes and chromatin (Xu et al., 2005; Masumoto et al., 2005; Xu et al., 2007). Yeast H3 K56Ac is very abundant (~28% of H3 in yeast contains K56Ac, Xu et al., 2005) and occurs in a promoter specific manner and also globally. At histone gene promoters a putative HAT Spt10 is required for S phase specific K56 acetylation that in turn is necessary for SWI/SNF mediated nucleosome remodelling and efficient gene transcription (Xu et al., 2005). In contrast K56Ac also occurs globally in a promoter independent manner through the HAT Rtt109, on newly synthesized histones in S phase (Masumoto et al., 2005; Downs, 2008). Through promoter-specific and global mechanisms K56 acetylation in yeast regulates not only gene activity (Xu et al., 2005; Schneider et al., 2006; Williams et al., 2008) but also histone replacement (Rufiange et al., 2007), DNA replication, DNA repair, chromatin assembly (Downs, 2008) and silencing at heterochromatin (Xu et al., 2007; Miller et al., 2008).
Given the importance of K56Ac in yeast, it is of great interest to know whether K56Ac exists in mammals. While yeast and Drosophila histone H3 K56Ac were detected by Western blotting or mass spectrometry (Ozdemir et al., 2005; Xu et al., 2005; Masumoto et al., 2005), this was not the case in mammals (Zhang et al., 2003; Xu et al., 2005). Even with more sensitive mass spectrometry of histones from human embryonic kidney (HEK293) cells its identification has been uncertain (Garcia et al., 2007a), although a small fraction of mammalian K56 has been reported to be methylated (Zhang et al., 2003; Garcia et al., 2007a). Therefore, we re-examined the presence of K56 acetylation in human cells using two new approaches. In the first, we used targeted mass spectrometry in which we specifically interrogated the sample for the presence of the acetylated K56 peptide. In the second we performed chromatin immunoprecipitation on microarrays (ChIP-on-Chip) using a highly specific antibody for acetylated K56. These studies were then adapted to an analysis of K56 acetylation during human cell differentiation.
Human embryonic stem cells (hESCs) are human pluripotent cells capable of differentiating into all adult cell lineages (Thomson et al., 1998). The pluripotent state is determined by key regulators of pluripotency such as transcription factors NANOG, SOX2 and OCT4 (Chambers and Smith, 2004; Jaenisch and Young, 2008). These factors (NSO for brevity) recognize the promoters of a number of genes to form a core transcriptional network in ESCs that consists of active regulators that contribute to the ESC pluripotency or self-renewal, as well as those that are repressed but ready for activation upon differentiation (Jaenisch and Young, 2008). We asked here whether there is an association of K56Ac with the core transcriptional network in hESCs and how K56 acetylated gene targets change during differentiation.
Results
K56Ac is present in human cells
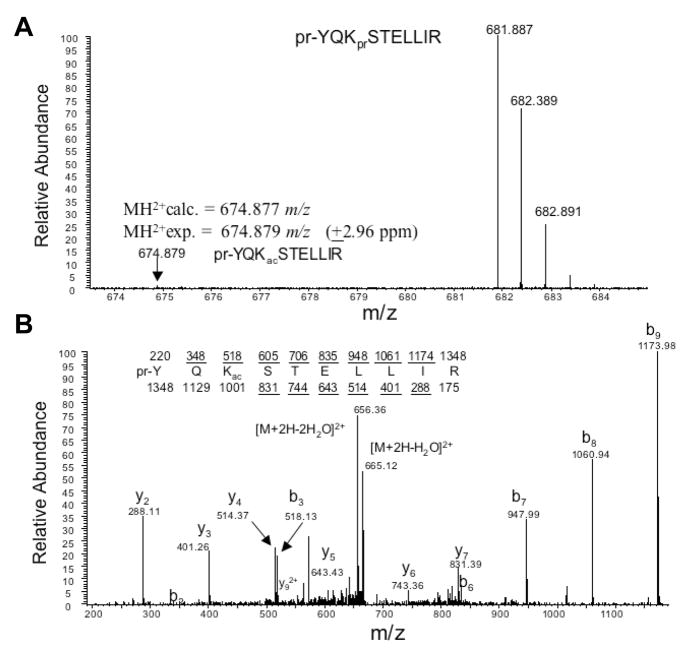
To first ask whether K56Ac is present in human cells we isolated nuclear histones from Hela cells and subjected the proteins to a sensitive spectrometry analysis using a recently developed protocol in which propionylated histone H3 tryptic digest was analyzed (Garcia et al., 2007b). We then specifically interrogated the sample for the presence of peptide containing acetylated K56 (Supplementary Methods). The targeted approach improves the chances of detecting a quality spectrum of the peptide of interest. A low abundance of K56Ac (~1% of total H3) was observed in Hela cells (Figure 1A), compared to the abundance of several other known H3 N terminal acetylation sites (ranging from 3.8%–22.9%) in Hela cells (Horwitz et al., 2008). Its identity was further verified by the tandem mass spectrum analysis (Figure 1B). We conclude that a small but significant fraction of Hela cell histone H3 is acetylated at K56.
Figure 1. K56Ac is present in human cells.
A) Histones were isolated from Hela cells and analyzed by mass spectrometry. Full mass spectrum is shown for unmodified (pr-YQKprSTELLIR, 681 m/z) and acetylated (pr-YQKacSTELLIR, 674 m/z) peptides from HPLC separated elutions (pr = propionyl amide, 56 Da and ac = acetyl, 42 Da). Due to the propionic anhydride treatment which only reacts with unmodified and monomethylated lysine residues, the unmodified peptide has a slightly heavier mass than the acetylated peptide. Accurate mass on the peptide at 674.887 m/z clearly indicates that the modification is an acetylation and not a tri-methylation mark.
(B) Tandem mass spectrum of the acetylated peptide (pr-YQKacSTELLIR) in (A) shows that the acetylation modification is localized to the K56 residue.
Genome wide localization of K56Ac in hESCs and somatic cells
To examine the acetylation of K56 during human cell differentiation we wished to test for the presence of K56Ac in a hESC line HSF1. Using the mass spectrometry approach described above we found that approximately 1% of HSF1 histone H3 is acetylated at K56 (Figure S1) as described for Hela cells. We then employed high-resolution ChIP-on-Chip techniques to determine where K56Ac occurs in the hESC genome. Chromatin immunoprecipitation was performed on HSF1 cell extracts using a highly specific anti-K56Ac antibody that has been tested not only by ELISA but validated also by using ChIP and Western blots with yeast K56 mutant strains (Xu et al., 2005). Input and precipitated DNA were amplified, labelled and hybridized to microarrays (Agilent) covering promoter regions of approximately 17,000 human genes (Supplementary Methods). To confirm our data we also profiled K56Ac in another independently derived hESC line HSF6. The Pearson correlation (r) of promoter average signal between the two different hESC lines is 0.86, indicating that these data are highly reproducible. In addition we examined K56Ac using an alternate microarray design (Affymetrix Human Tiling Array 2.0R) for HSF6 on chromosomes I and VI and the result is highly consistent with the HSF6 data obtained using the Agilent microarray (r= 0.80, data not shown). Finally, to show that the ChIP signal we obtained is specific for K56Ac, we performed ChIP on candidate genes in the presence of competitive peptides. We found (Figure S2) that the high enrichment for K56Ac at the candidate promoters (e.g. HIST1H1E, HIST1H3D, NANOG, SOX2, OCT4) is unaffected in the presence of unacetylated peptide. In contrast, the high enrichment is reduced to background levels in the presence of K56 acetylated peptide. These experiments confirm that our α-K56Ac antibody is highly specific for ChIP and for ChIP-on-Chip and that K56Ac is present at distinct gene promoters in human embryonic stem cells.
To identify specific human gene classes that are acetylated at K56 in HSF1 and HSF6, we examined 15,581 promoters covered by sufficient valid probes (Supplementary Methods). Using a peak finding method (The Whitehead Neighborhood Model, see Supplementary Methods), we found that 9.5% (8824 kb) and 10.3% (9621kb) of these promoter regions are identified as K56Ac positive loci corresponding to 0.38% and 0.42% of the genome in HSF1 and HSF6 respectively (using a 500bp window). This is comparable to the abundance of K56Ac in HSF1 as measured by mass spectrometry (Figure S1A). We first wished to identify the most acetylated targets in the genome based on the average levels of K56Ac across the promoter regions in both hESCs. An examination of the top 1% genes (156 genes, see Table S1 for complete gene list) of the entire genome that are acetylated at K56 includes almost the entire family of canonical histone genes (53 out of total 61 genes, p-value = 1.4E-101). It also contains a high concentration of regulators of pluripotency including genes coding for the key transcription factors NANOG, SOX2 and OCT4 (Chambers and Smith, 2004; Jaenisch and Young, 2008), as well as many pluripotency-related genes that either have ESC specific expression or play important roles in ESC pluripotency and self-renewal regulation (e.g. TDGF1, GDF3, LEFTY1, ZIC3, DPPA4, TERF1, GJA1) (Sato et al., 2003; Boyer et al., 2005; Adewumi et al., 2007). Finally we also found 13 microRNA genes in the top 1% genes, among which 11 are known to be preferentially expressed in hESCs (Suh et al., 2004; Laurent et al., 2008)(Table S1). These data demonstrate a conserved role of K56Ac at histone genes and its presence at genes of the core transcriptional network that regulates pluripotency.
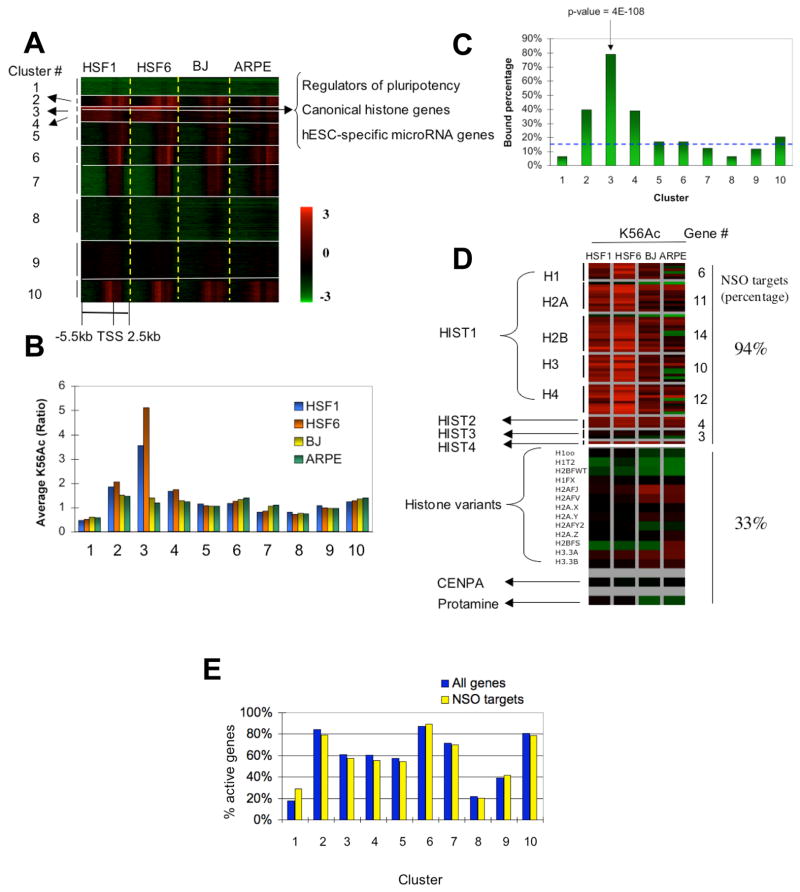
To obtain a more comprehensive description of K56Ac localization in the hESC genome, all 15,581 genes were grouped by acetylation pattern into 10 clusters using K-means clustering (Supplementary Methods) for hESCs (HSF1 and HSF6) as shown in the heat map in Figure 2A. The highest average level of acetylation is found in cluster 3 and to a lesser extent in clusters 2 and 4 (Figure 2B). Detailed gene lists (Table S2) and gene ontology (GO) output (Table S3) for each cluster are included in Supplemental Data. Strikingly of the top 1% (156) of genes most acetylated at K56 in the entire genome, 95% (148) including all representative genes described above are found in cluster 3 (n = 246, see Table S4 for summarized gene lists). This small cluster has a uniquely broad spread of K56Ac across the entire promoter region and the region downstream of the transcription start site, a pattern that is distinct from that in most other hESC gene clusters. Clusters 2 and 4 are also somewhat hyperacetylated. GO analysis shows that cluster 2 is enriched in genes that function in the regulation of transcription (p-value = 2.5E-5) and translation (p-value = 3.3E-5) while cluster 4 is heavily enriched in genes involved in transcription (p-value = 5.1E-12) and development (p-value = 1.8E-9) (see Table S3 for GO). As shown in Figure 2B other clusters have much lower average levels of acetylation.
Figure 2. Genome-wide localization of histone H3 K56Ac by ChIP-on-Chip in hESCs and differentiated cells.
(A) Histone H3 K56Ac is present in human cells and occurs at distinct gene promoters. 15581 genes with at least 25% of the regions (2kb) covered by valid probes were classified into 10 groups by K-means clustering based on K56Ac patterns in hESCs (HSF1 and HSF6). K56Ac pattern in somatic cells (BJ and ARPE) were then mapped to the same gene groups for comparison. Each row represents the same promoter region in HSF1, HSF6, BJ and ARPE. Each single promoter region ranges from –5.5 kb to +2.5 kb with respect to the TSS and is represented as 16 step-wise 500 bp windows. Red, green and black represent relative enrichment (log ratio) compared to input DNA as shown in the color bar. Grey represents missing values. The major gene classes enriched in cluster 3 are shown at the arrow on the right.
(B) The average K56Ac enrichment for clusters 1–10 is shown. The mean of K56Ac enrichment (promoter average) of all genes in each cluster is computed and plotted as bar graphs for HSF1 (blue), HSF6 (orange), BJ (yellow) and ARPE (green).
(C) Percentages of genes bound by the NSO regulators in K56Ac clusters 1–10 are shown. Genes that are bound by at least one of the proteins NANOG, SOX2 and OCT4 were counted and used to compute the percentage of pluripotency key regulator targets in each cluster. Blue dashed line represents the average enrichment of the NSO regulator targets across all clusters (background level).
(D) Canonical histone genes are heavily acetylated at K56 and targeted by the key regulators for pluripotency. Promoter average K56Ac signals for canonical histone genes HIST1-4, variant histone genes, centromere protein A (CENPA) and protamine are shown for HSF1, HSF6, BJ and ARPE. For canonical histone genes, only regions 1 kb upstream and 0.5 kb downstream of the TSS were used due to their short gene lengths (median 460 bp) and closely adjacent chromosomal positions (the median distance between neighbouring genes is 4.3 kb). The percentages of genes that are bound by NANOG, SOX2 or OCT4 (NSO) are also shown for both canonical histone genes and variant histone genes on the right.
(E) K56Ac preferentially marks active genes but also occurs at inactive genes. The percentages of active genes among all genes (blue) or among genes that are bound by the NSO regulators (yellow) in each cluster are shown as bar graphs.
We next wished to know how hESC K56Ac patterns are altered in differentiated cells. We compared K56 acetylation in HSF1 and HSF6 to that of two diverse somatic cell lines: foreskin fibroblast (BJ) and adult retinal pigment epithelial (ARPE) cell lineages. As demonstrated in Figure 2A and Figure 2B, K56Ac in cluster 3 is strongly depleted in the somatic cell lines (two samples t-test p-value = 3.4E-101). Clusters 2 and 4 show a significant but less dramatic decrease in K56Ac in the differentiated cells (p-value = 1.6E-50 and 2.1E-67 respectively). The depletion of K56Ac from these clusters in differentiated cells is not due to histone loss (Figure S3). In contrast to clusters 2, 3 and 4, several other gene clusters (such as clusters 6 and 10) are weakly acetylated in a similar manner in both pluripotent cells and lineage-committed cells (Figure 2B). GO analysis shows that these genes participate in processes such as RNA metabolism, biosynthesis and the cell cycle (Table S3). In summary, these data illustrate that the genes of cluster 3, and to a lesser extent clusters 2 and 4, are preferentially deacetylated at H3 K56 in differentiated lineages.
High levels of K56Ac mark the pluripotency transcriptional network in hESCs
Why genes in clusters 2, 3 and 4 are preferentially deacetylated at K56 in somatic cells is explained in part by examining their identity. Given that many of the pluripotency-related genes in cluster 3 (e.g. TDGF1, NODAL, GDF3, LEFTY1, ZIC3, DPPA4, GJA1, DNMT3B) are also bound by NANOG, SOX2 or OCT4 (including themselves) (Boyer et al., 2005; Marson et al., 2008), we asked if the NSO proteins generally occupy promoters acetylated at K56. By matching a combined binding profile of the NSO proteins (Boyer et al., 2005; Marson et al., 2008) to each of the 10 clusters, we found that 79% of genes in cluster 3 are bound by one or more of the NSO regulators. This enrichment is extremely significant (p-value = 4E-108) as compared to only 16% of a random gene set (Figure 2C). An even higher percentage (136/156, 87%) of promoters in the top 1% genes of the genome are found to be NSO targets. There is also a significant although lower fraction of genes in cluster 2 (39.4% of n = 705, p-value = 7.5E-54) and in cluster 4 (38.8% of n = 903, p-value = 4.9E-66) that interact with one or more of the NSO proteins. In contrast, no more than 20% of genes in other clusters are bound by at least one NSO protein (Figure 2C). Conversely, of a total of 2479 genes bound by NANOG, SOX2 or OCT4, 71% contain one or more bound probes for K56Ac (42% are acetylated at K56 greater than 1.5 fold) in at least one hESC line (p-value = 3E-185). In summary, we observe a highly significant co-localization between high levels of K56Ac and NSO regulator binding at hESC gene promoters.
Histone genes in mammalian cells consist of two major classes: canonical histone genes coding for H1, H2A, H2B, H3 and H4, and variant histone genes such as H3.3, H2A.X and H2A.Z (Marzluff et al., 2002). The canonical human histone genes are located in 4 separate chromosomal domains: HIST1 (53 genes), HIST2 (4 genes), HIST3 (3 genes), and HIST4 containing a single H4 gene. We found that most of these genes are preferentially acetylated at K56 in hESCs (Figure 2D). Of the 61 canonical histone genes, the majority (53) are found in cluster 3 (p-value = 2.2E-89). Interestingly most of these genes (50/53 or 94%, p-value=1E-36) also bind NANOG, SOX2 or OCT4. It is worth noting that the preferential presence of NSO targets in cluster 3 is not biased by histone genes, as the percentage of NSO targets in cluster 3 changes slightly from 79% to 75% after excluding histone genes. Unlike the canonical histone genes, the variant histone genes including centromere protein A (CENPA) and protamine genes are relatively depleted of K56Ac and few are bound by NANOG, SOX2 or OCT4 (5/15 or 33%, p-value = 0.07). Interestingly, K56Ac consistently marks fewer canonical histone genes in somatic cells (49 in BJ, 33 in ARPE) (Figure 2D) but is enriched at several variant histone genes (H2AFJ, H2AFV, H3.3) somewhat more in these lines. This may reflect the selective usage of histone genes during cell differentiation (Holmes et al., 2005). Therefore, K56 is highly acetylated at canonical histone genes as part of the core transcriptional network of hESCs.
We found that cluster 3 also contains a number of genes for microRNA (miRNA), a type of non-coding regulatory 22-nt long RNA that has been demonstrated to play critical roles in many biological processes including development and also regulation of ESCs (Bartel, 2004; Marson et al., 2008). Strikingly, 12 out of 18 miRNA genes in cluster 3 including three polycistrons mir-302 (hsa-mir-302a/302b/302c/302d/367), mir-371 (hsa-mir-371/372/373) and mir-498 (hsa-mir-498/512-1/512-2/520e), are known to be expressed only in hESCs (Suh et al., 2004; Laurent et al., 2008). Moreover, while only 8% of a random set of miRNA genes are bound by OCT4 (Marson et al., 2008), OCT4 occupies mir-302 group and mir-371 group corresponding to 44% of miRNA genes in cluster 3 (p-value = 1E-5). Thus, a significant fraction of the hESC-specific miRNAs in cluster 3 are highly acetylated at K56 and are selectively recognized by the key regulators of pluripotency.
K56Ac is largely an active mark but also occurs at repressed genes
To ask whether the preferential presence of K56Ac on the promoters of the core transcriptional network of hESCs is simply due to an association with gene activity, we compared K56Ac to the gene transcription levels in hESCs in each cluster. Using a combined expression dataset from microarrays (Sato et al., 2003; Abeyta et al., 2004), we found that although clusters 2, 3 and 4 contain many active genes, they also include significant fractions of inactive genes (16%, 39% and 40% respectively). This is also true when we restrict the analysis to those genes that are bound by one or more NSO proteins (Figure 2E). For example, high levels of K56Ac are observed at the promoters of GATA4, GATA6, OTX1, ONECUT1, ISL1 and WNT3, which are known to be bound by NSO proteins in hESCs and are preferentially expressed only upon hESC differentiation (Boyer et al., 2005; Hyslop et al., 2005; Marson et al., 2008; Zhang et al., 2008). Globally, we find that K56Ac marks ~23% of inactive genes in hESCs. An even larger fraction (~41% inactive genes) is obtained using different expression profiling datasets by Massively Parallel Signature Sequencing (MPSS) (Brandenberger et al., 2004; Wei et al., 2005). In fact, K56Ac shows only a weak correlation with gene expression genome wide as shown in the scatter plots (Figure S4, r = 0.18 for both HSF1 and HSF6). A relatively higher correlation was observed when we compared K56Ac to a published Pol II binding dataset (Lee et al., 2006) (r = 0.50 for HSF1 and r = 0.45 for HSF6), possibly due to the occupancy of initiating Pol II at many inactive genes in mammals (Guenther et al., 2007). Therefore, K56Ac in hESCs is largely an active mark but also occurs at a large fraction of repressed genes, suggesting that K56Ac is not simply a marker for gene activity.
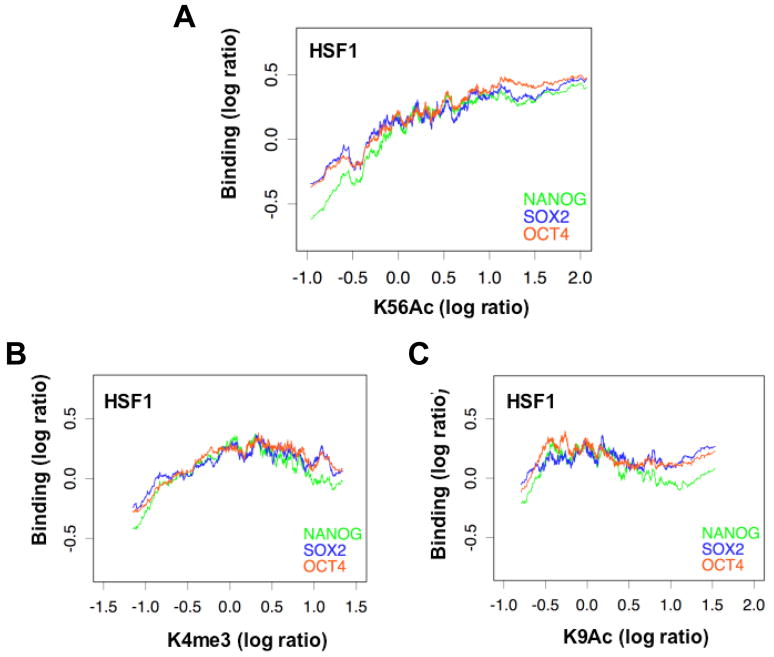
K56Ac correlates positively with NANOG, SOX2 and OCT4 binding along promoters in hESCs
As the occurrence of K56Ac is unlikely to be simply caused by transcription, it is alternatively possible that it is mediated by the NSO regulatory factors which are known to bind both active and inactive genes (Boyer et al., 2005). To probe this question we examined the relationship between K56Ac and NSO binding at their gene targets in hESCs. Using published data (Boyer et al., 2005), we have found that average K56Ac level positively correlates with the binding levels of NSO proteins on the promoters of their targets as shown in the moving average plot for HSF1 (Figure 3A). To ask whether this is a general phenomenon that is observed for other histone marks that are related to gene activity, we performed ChIP-on-chip for K4me3 and K9Ac similar to K56Ac in both hESCs and somatic cell lines (see Supplementary Methods “Validation of microarray results”). Unlike K56Ac, K4me3 and K9Ac do not show positive correlations with the binding of the NSO regulators on these same genes (Figure 3B–C). Similar observations were obtained for HSF6 (Figure S5).
Figure 3. K56Ac correlates with NANOG, SOX2 and OCT4 binding on their targets in hESCs.
A total of 2479 genes were used that are bound by at least one of the three regulators NANOG, SOX2 and OCT4. Moving averages of NANOG (green), SOX2 (blue) and OCT4 (orange) binding levels (log ratio) are plotted against the promoter average levels of K56Ac (A), K4me3 (B) and K9Ac (C) for HSF1, with window size = 100 genes and moving step = 1 gene.
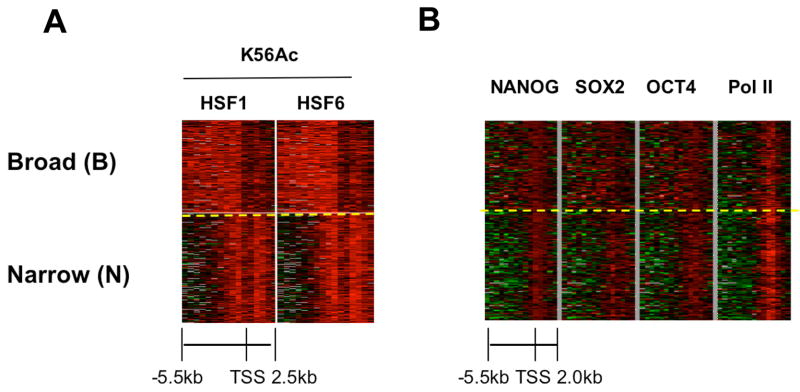
We then asked whether the physical location of K56Ac along the gene promoters overlaps with that of the key regulators of pluripotency. One unique feature of clusters 3 (and also cluster 4) is the presence of K56Ac along the entire promoter and not only at the TSS. This is not due to the broad distribution of nucleosomes, as histone H3 is relatively depleted from these promoters (Figure S3). Therefore, we further classified the K56Ac enriched genes in hESCs based on their spatial acetylation patterns. As genes with low levels of K56Ac generally do not associate with the NSO regulators (Figure 3A), we focused on genes with high levels of K56Ac whose average signal across the promoter is 1.5 fold or more in both hESC lines (n = 1461). To reduce the bias of clustering introduced by missing probe signals, only genes with valid probe coverage no less than 75% (6 kb) were used (n = 778). These K56Ac enriched genes in hESCs were classified into two groups by K-means clustering. As shown in both the heat map (Figure 4A) and the average signal plot (Figure S6) one of the groups (n = 364) has a broad K56Ac domain covering the entire 5.5 kb region upstream of the TSS and was termed ‘B’ for ‘Broad’. The other pattern (n = 414) has a strong K56Ac peak only around the TSS. We termed this pattern ‘N’ for ‘Narrow’ (for complete gene lists see Table S5). No significant difference in overall expression levels was observed between genes in the B and N groups (Figure S7). Interestingly, we find that NSO targets are preferentially present in the B group where 60% of the genes are bound by NANOG, SOX2 or OCT4, while 46% of genes in the N group are bound by one or more of these regulators. The difference between the B and the N groups in marking the core transcriptional network is significant (p-value = 2E-4) while both groups are bound in a similar manner (9.3% and 10.6% respectively) by a cell cycle regulator E2F4 (Boyer et al., 2005; data not shown). We then wished to compare the physical locations of K56Ac along the gene promoters in the B and N group with those of the NSO regulators. By mapping the binding of NSO proteins in the B and N group, we find that NSO occupancy along target promoters (Boyer et al., 2005) to a large extent parallels K56Ac spreading in the B or N patterns (Figure 4B and Figure S8A-C). This is not similarly observed for RNA polymerase II (Pol II) that is enriched primarily at the TSS (Figure 4B “Pol II” and Figure S8D). These data uncover a spatial correspondence between K56Ac and the binding of the NSO regulators along the affected promoters.
Figure 4. NANOG, SOX2 and OCT4 binding parallels the presence of K56Ac along the gene promoters in hESCs.
(A) Identification of two distinct K56 acetylation spatial patterns on gene promoter targets in hESCs. K56Ac hESC targets (histone genes and miRNA genes excluded due to their short gene lengths) were clustered into the B (Broad)-group (364 genes) and N (Narrow)-group (414 genes), using K-means clustering based on K56Ac spatial distributions in hESCs. (B) NANOG, SOX2 and OCT4 binding parallels the presence of K56Ac (A) at the B and N group genes. The NANOG, SOX2, OCT4 and Pol II binding levels are shown using step-wise 500 bp windows spanning the promoters where each row represents one gene.
K56Ac defines epigenetic differences between hESCs and somatic cells better than either H3K4me3 or H3K9Ac
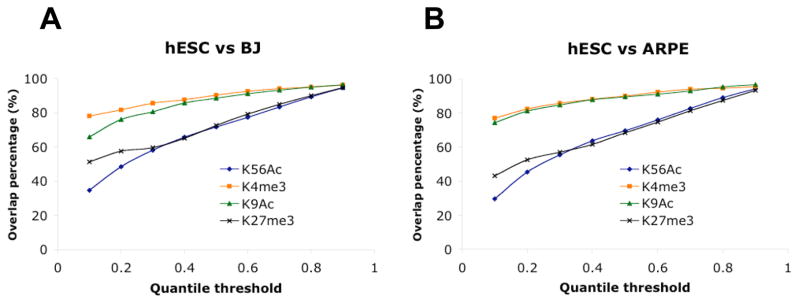
As K56Ac is closely connected with the core transcriptional network in pluripotent cells, we further compared K56Ac to other ‘active’ marks K4me3 and K9Ac by testing which histone mark best distinguishes the epigenetic states of pluripotent and somatic cells. We compared genes most highly enriched in various histone modifications within hESCs and somatic cells. To do so we applied a series of quantile cutoffs (0.1–0.9) to select genes showing highest enrichments (10%–90%) within each cell type and measured the overlap between hESCs and the somatic cell lines. As shown in Figure 5A–B the percentage of hESC K56Ac targets that are also modified by K56 acetylation in somatic cell lines (overlap percentage) is lower than that for either K4me3 or K9Ac. This difference is observed not only at the lowest quantile threshold (0.1) but for all except the highest threshold examined (0.9). In contrast to active marks K4me3 and K9Ac, H3 K27me3 is a repressive mark that is known to control cell fate and differentiation and displays a dramatic difference in presence between ESCs and somatic cells (Buszczak and Spradling, 2006). To compare K56Ac to K27me3 using similar analyses we performed ChIP-chip for H3 K27me3 in HSF1, HSF6, BJ and ARPE (see Supplemental Methods “Validation of microarray results”). As shown in Figure 5A–B, K56Ac distinguishes hESCs versus somatic cells in a manner that is comparable to K27me3. It shows even better discrimination at the top thresholds (0.1 and 0.2) than K27me3. Therefore K56Ac is a largely active mark that defines the epigenetic difference between pluripotent cells and differentiated cells as well if not better than repressive mark K27me3, and much better than known active marks such as K4me3 and K9Ac.
Figure 5. K56Ac defines epigenetic differences between hESCs and somatic cells better than either H3K4me3 or H3K9Ac.
Genes were sorted by their average signal of K56Ac, and a series of quantile cutoffs (0.1–0.9) were applied to select K56Ac targets in hESCs (common to HSF1 and HSF6) and somatic cells. The percentages of hESC K56Ac targets (blue line) that are also shared by BJ (A) or ARPE (B) were plotted against the quantile thresholds. Similar analysis was performed for K4me3 (orange line), K9Ac (green line) and K27me3 (black line).
Developmental genes are hyperacetylated at K56 in differentiated cells
The differential acetylation of K56 in hESCs and somatic cells as shown in Figure 5 suggests the existence of genes that are marked only in somatic cells. To identify genes that might be preferentially acetylated at K56 in somatic cells we selected those (741 genes) that are acetylated in both BJ and ARPE but not in either of the hESC line. GO analysis shows that cohesive classes in these genes include those involved in signal transduction, tissue remodeling and organ development (Table S6). Therefore in differentiated cells K56Ac marks genes that are likely to be important for somatic development.
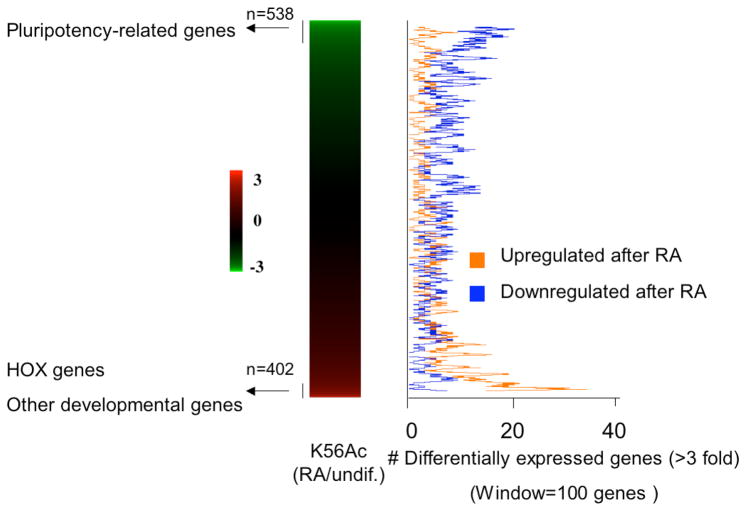
As BJ and ARPE are lineage-committed cells, we asked how K56Ac relocates in cells during the transition from the pluripotent state to the differentiated state. To probe this question we induced the differentiation of HSF6 cells by retinoic acid (RA). After 5 days treatment we were able to observe a morphological change of hESCs (Figure S9A-B) and a significant reduction in the transcript levels of NANOG, SOX2 and OCT4 (Figure S9C), confirming that these cells have undergone differentiation. We profiled K56Ac in these cells similarly using ChIP-on-Chip and compared our data to that of the undifferentiated HSF6 cell line. We found that upon RA treatment, the enrichment of K56Ac fell at least 2 fold at 538 promoters including 60% of genes in cluster 3 (Figure 6, see Table S7 for complete gene list). As expected, pluripotency-related genes NANOG, SOX2, OCT4, TDGF1, PHC1, DPPA4, ZIC3, GDF3, LEFTY1 are among those that display most hypoacetylation (top 1% of the genome). Interestingly, the most deacetylated locus in the genome is the hESC-specific miRNA gene group mir-302 (11 fold reduction in K56Ac), implying a tight regulatory control of this miRNA cluster which is known to be bound by OCT4 (Marson et al., 2008). In contrast, 402 genes are hyperacetylated (2-fold cutoff) at K56 in the differentiated cells (see Table S7 for gene list). The HOX gene family, a homeobox transcription factor family that controls embryonic patterning and is known to be activated by retinoic acid (Langston and Gudas, 1994), is the most hyperacetylated gene class (p-value = 6E-30). We found that 51% of the HOX genes (19 out of 37 on arrays) are among the top 1% most hyperacetylated genes upon RA treatment. Other hyperacetylated genes include those participating in developmental events (p-value = 6E-9) such as organ morphogenesis (p-value = 2E-6) and ectoderm development (p-value = 1E-4). To directly test whether these genes are upregulated when cells lose pluripotency, we compared published gene expression data from RA induced hESCs (Pan et al., 2007) to K56Ac levels. These data clearly demonstrate that, in contrast to pluripotency-related genes that are generally repressed during differentiation, these genes are preferentially upregulated upon RA treatment (Figure 6), reflecting a re-distribution of K56Ac from pluripotency-related genes to developmental genes. We also find that a portion of genes (>60%) that are hyperacetylated upon differentiation do not show upregulation, indicating again that K56Ac does not depend entirely on transcription. Taken together, K56Ac relocates to genes that are important for somatic development in not only differentiated cells but also those that are undergoing differentiation.
Figure 6. Developmental genes are hyperacetylated at K56 upon hESC differentiation induced by retinoic acid.
The K56Ac level changes (RA treated/undifferentiated, log ratio) for the entire genome are shown on the left ranked from the most hypoacetylated genes to the most hyperacetylated genes. Genes that are differentially expressed in hESCs upon RA treatment (3 fold cutoff) were obtained from a published dataset (Pan et al., 2007). The number of upregulated (orange) or downregulated (blue) genes was computed as a moving average (step = 1 gene; window = 100 genes) shown on the right.
Discussion
H3 K56Ac is present in human cells
It has been unclear whether K56Ac, a unique histone modification in the core of histone H3 that exists in yeast and flies, is also present in humans. Using a targeted mass spectrometry approach with improved sensitivity and data quality, together with chromatin immunoprecipitation we now show that, in contrast to the abundant K56Ac in yeast (~28% of H3 is acetylated), K56Ac is present at low abundance in human cells (~1% of H3 is acetylated). In addition, the canonical histone gene promoters are amongst the most acetylated targets in the genome in both yeast and humans, demonstrating the conservation of this cohesive class of K56Ac targets. In summary, we have shown here that K56Ac indeed exists in the genome of human cells.
K56Ac occurrence displays similarities and differences between yeast and humans
K56Ac targets canonical histone genes and other defined gene classes in humans suggesting that as in yeast, K56Ac can occur in a gene-specific manner. Nevertheless, K56Ac also shows striking difference between yeast and humans. In yeast the global acetylation of K56 results from Rtt109 activity on newly synthesized histones that are deposited on replicating chromatin. However our data suggest that this mechanism is not prevalent in humans based on several findings. First, compared to a high abundance in yeast, K56Ac in human cells exists at a very low level, which makes it unlikely to occur on bulk newly synthesized histones. Second, K56Ac does not appear to be tied to cell cycle regulated DNA replication in mammals. NANOG and OCT4 are early replicating genes in both pluripotent and differentiated cells (Azuara et al., 2006), yet we find that NANOG and OCT4 are acetylated at K56 only in hESCs but not in somatic cells (BJ and ARPE). Finally to directly address this question, we probed the cell cycle regulation of K56Ac in Hela cells using a commercial antibody (Epitomics) that is specific for K56Ac in Western blots under the conditions shown as illustrated using K56 mutants (Figure S10A). We show that in contrast to cyclin E, K56Ac shows equal enrichment throughout the cell cycle (Figure S10B). Therefore unlike the situation in yeast, the time of replication does not appear to be the major determinant of K56 acetylation in humans. Nevertheless, we do not exclude the possibility that K56 acetylation may affect DNA repair and chromosomal integrity through other pathways.
Since K56Ac in humans is not global but gene specific, other mechanisms by which K56Ac takes place at their target promoters may be considered. In yeast the levels of K56Ac have been shown to correlate with replication-independent histone replacement (Rufiange et al., 2007), therefore it is possible that in humans K56Ac may result from the active replacement of histones at promoter regions. Metazoan H3 variant H3.3 replaces canonical H3 during transcription (Ahmad and Henikoff, 2002). This raises the interesting possibility that H3.3 may be selectively acetylated at K56 allowing K56Ac to occur at promoters that are subject to H3.3 replacement. However, we do not favor this model as our western blots (using anti-K56Ac antibody from Epitomics) show that all three H3 variants (H3.1, H3.2 and H3.3) appear to be acetylated in human cells (data not shown). Alternatively, a HAT for K56 may be recruited by cell specific transcription factors, such as the transcriptional regulators of pluripotency of hESCs, to the targeted promoters.
K56Ac is linked to the hESC core transcriptional network
We showed that K56Ac in human embryonic stem cells occurs on the targets of the hESC key regulators for pluripotency in a highly significant manner. First, a striking fraction of genes with high levels of K56Ac are bound by NANOG, SOX2 and OCT4. Among the top 1% of genes acetylated in the genome, 87% are bound by at least one NSO regulator. Moreover these genes share a common K56Ac signature as 95% of them are included in a small gene cluster (cluster 3), which has a broad acetylation pattern across the promoter regions. Genes with high levels of K56Ac are also included in two other gene clusters 2 and 4, both of which contain higher than average levels of NSO targets (~40%). Second, compared to canonical histone genes, of which 94% are bound by NANOG, SOX2 or OCT4, K56Ac is relatively depleted from variant histone genes of which only 33% are bound by the key regulators. Similarly, consistent with the absence of the NSO regulators in differentiated cells, genes of clusters 2, 3 and 4 are preferentially deacetylated at K56 in somatic cells. Third, at the promoter regions of the NSO protein targets, K56Ac levels correlate with the binding of NANOG, SOX2 and OCT4, a feature that is not observed for K4me3 and K9Ac. Finally, along the promoters of K56Ac targets, the spatial patterns of K56Ac parallel the spatial binding of NSO proteins. These data indicate that K56Ac in hESCs is closely linked to the core transcriptional network of hESCs.
K56Ac also occurs at other genes involved in certain housekeeping functions. Clusters 6 and 10 are acetylated mainly at the TSSs, contain few targets of NSO proteins and are generally involved in cell proliferation. The low concentration of NSO targets in clusters 6 and 10 are not due to lower gene activity in these clusters as both clusters exhibit comparable or even higher transcription levels compared to clusters 2, 3 and 4 (Figure 2E). It is possible that K56Ac may be involved in other cellular functions that are independent of the NSO regulators. The factors that may be associated with the lower levels of K56Ac at these genes remain to be determined.
K56Ac is largely an active mark but also occurs at inactive genes
An intriguing question is whether the presence of K56Ac on the core transcriptional network of hESCs is simply due to its association with gene activity. We showed that this is unlikely to be the case since even hESC clusters with the highest levels of K56Ac (clusters 2, 3 and 4) contain significant fractions of repressed genes. On a genome wide scale K56Ac shows a weak correlation (r = 0.18) with gene expression in hESCs. This is consistent with the finding that NSO proteins are known to bind both active and inactive genes (Boyer et al., 2005).
Other histone marks such as H3K4me3 and H3K9Ac have also been shown to be present on both active and inactive genes (Guenther et al., 2007). However K56Ac differs from H3K4me3 and H3K9Ac in a significant manner. First, the levels of K56Ac correlate with the binding of NANOG, SOX2 and OCT4, which is not observed for K4me3 or K9Ac (Figure 3). Second, K56Ac best distinguishes pluripotent cells versus somatic cells compared to K4me3 or K9Ac (Figure 5). These data strongly argue that K56Ac is a mark that occurs at both active and inactive genes, but is different from known active marks such as K4me3 and K9Ac.
Since the acetylation of K56 is essential for SWI/SNF binding at yeast histone gene promoters (Xu et al., 2005), K56Ac and the NSO regulators may recruit a SWI/SNF-like nucleosome remodeling complex that is important not only to allow gene activation (Xu et al., 2005) but also in some cases enable gene repression as shown both in yeast and mammals (Martens and Winston, 2003) including ESCs (Kaeser et al., 2008).
K56Ac preferentially marks developmental genes in differentiated cells
Extensive genomic programming occurs when cells are converted from a pluripotent state to a committed state. We find that genes in the hESC core transcriptional network are preferentially acetylated at K56 in the pluripotent cells, however there is a re-distribution of K56Ac in differentiated cells. This is evident when we examined hESCs that were induced to differentiate by retinoic acid treatment, during which developmental genes including the HOX genes are hyperacetylated at least in certain cell populations, underscoring the importance of K56Ac during development. Thus K56Ac is able to distinguish the pluripotent state from a somatic state better than other active histone marks such as K4me3 and K9Ac. Since the NSO regulators are not present in somatic cells this also predicts that other regulators may recruit K56Ac to promoters in somatic cells in a similar yet developmentally distinct manner.
Extending the core transcriptional network of hESCs by examining K56Ac
By marking the core transcriptional network of hESCs K56Ac may predict new targets of the NSO transcriptional regulators. In the top 1% most acetylated genes at K56 in the genome, there are 55 genes that previously were not identified as targets of the NSO regulators (Boyer et al., 2005). 35 of these are found to be bound by OCT4 in a recent study (Marson et al., 2008), including the two hESC-specific miRNA gene groups mir-302 and mir-371 in cluster 3 (Suh et al., 2004). Our data and that of a recent study (Laurent et al., 2008) indicate that the third miRNA gene group mir-498 in cluster 3 may also be uniquely expressed in hESCs. Gene ontology analysis shows that certain K56Ac targets (above 1.5 fold enrichment) which have not yet been shown to be bound by NSO proteins (n = 809) are involved in the regulation of gene expression (p-value = 4E-7). Our data predict that these genes are also potential targets of NANOG, SOX2 or OCT4. Alternatively some of these K56Ac modified promoters may be targeted by other key regulators of pluripotency. For instance, transcription factors KLF4 and c-myc together with SOX2 and OCT4 have been shown to reprogram differentiated cells into ES-like cells (Lewitzky and Yamanaka, 2007; Jaenisch and Young, 2008). By examining a number of factors that are involved in transcriptional regulation of ESCs, two recent studies have dramatically expanded the mouse ESC regulatory network (Kim et al., 2008; Chen et al., 2008). A similar extended pluripotency transcriptional network in humans is yet to be established. In this regard K56Ac which has identified genes in the pluripotency transcriptional network may further help predict new regulators.
Experimental Procedures
Cell culture
Human ESC lines HSF1 and HSF6 were maintained on irradiated mouse embryonic fibroblasts under standard growth conditions (see Supplementary Methods). BJ, ARPE, Hela and HEK293 cells were maintained under standard conditions with details described in Supplementary Methods.
Mass spectrometry
Histone H3 from Hela or HSF1 cells was purified and prepared for mass spectrometry analysis as previously described (Garcia et al., 2007b) with details included in Supplementary Methods.
Chromatin immunoprecipitation (ChIP) and DNA microarray analysis
Detailed descriptions of ChIP are provided in Supplementary Methods. Agilent 244K Human Promoter microarrays (G4489A) were used for ChIP-on-Chip experiments. Protein binding data for Pol II, NANOG, SOX2, OCT4, E2F4, gene expression data and the detailed analysis methods are included in Supplementary Methods.
Acknowledgments
We are grateful to members of the Grunstein laboratory for generous help throughout this work. We thank Arnold Berk and Jennifer Woo for the gifts of Hela and HEK293 cell lines, the UCLA microarray core facility for array services, and Siavash Kurdistani, Roberto Ferrari, Sheng Yin, Wei Sun for discussions and advice. This work was supported by National Institutes of Health grants to M.G.. A.T.C. is the recipient of a STOP CANCER career development award. K.P. is a Kimmel Scholar and is supported by the Margaret E. Early Trust Foundation.
Footnotes
Accession Numbers
Micorarray data are available at GEO under XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R, Khrebtukova I, Thies RS, Miura T, Jingli C, Puri R, Vasicek T, Lebkowski J, Rao M. MPSS profiling of human embryonic stem cells. BMC Dev Biol. 2004;4:10. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125:233–236. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Downs JA. Histone H3 K56 acetylation, chromatin assembly, and the DNA damage checkpoint. DNA Repair (Amst) 2008;7:2020–2024. doi: 10.1016/j.dnarep.2008.08.016. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007a;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007b;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WF, Braastad CD, Mitra P, Hampe C, Doenecke D, Albig W, Stein JL, van Wijnen AJ, Stein GS. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. J Biol Chem. 2005;280:37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyslop L, Stojkovic M, Armstrong L, Walter T, Stojkovic P, Przyborski S, Herbert M, Murdoch A, Strachan T, Lako M. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells. 2005;23:1035–1043. doi: 10.1634/stemcells.2005-0080. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a Novel PBAF-specific SWI/SNF Subunit, Is Required for Target Gene Activation and Repression in Embryonic Stem Cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Langston AW, Gudas LJ. Retinoic acid and homeobox gene regulation. Curr Opin Genet Dev. 1994;4:550–555. doi: 10.1016/0959-437x(94)90071-a. [DOI] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitzky M, Yamanaka S. Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol. 2007;18:467–473. doi: 10.1016/j.copbio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Miller A, Yang B, Foster T, Kirchmaier AL. Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics. 2008;179:793–809. doi: 10.1534/genetics.107.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei HR, Jonsdottir GA, Stewart R, Thomson JA. Whole-Genome Analysis of Histone H3 Lysine 4 and Lysine 27 Methylation in Human Embryonic Stem Cells. Cell Stem Cell. 2007;17:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone h3 exchange occurs predominantly at promoters and implicates h3 k56 acetylation and asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, Yong J, Jiang W, Sun X, Du L, et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]