Abstract
The use of epigenetic differences between maternal whole blood and fetal (placental) DNA is one of the main areas of interest for the development of noninvasive prenatal diagnosis of aneuploidies. However, the lack of detailed chromosome-wide identification of differentially methylated sites has limited the application of this approach. In this study, we describe an analysis of chromosome-wide methylation status using methylation DNA immunoprecipitation coupled with high-resolution tiling oligonucleotide array analysis specific for chromosomes 21, 18, 13, X, and Y using female whole blood and placental DNA. We identified more than 2000 regions of differential methylation between female whole blood and placental DNA on each of the chromosomes tested. A subset of the differentially methylated regions identified was validated by real-time quantitative polymerase chain reaction. Additionally, correlation of these regions with CpG islands, genes, and promoter regions was investigated. Between 56 to 83% of the regions were located within nongenic regions whereas only 1 to 11% of the regions overlapped with CpG islands; of these, up to 65% were found in promoter regions. In summary, we identified a large number of previously unreported fetal epigenetic molecular markers that have the potential to be developed into targets for noninvasive prenatal diagnosis of trisomy 21 and other common aneuploidies. In addition, we demonstrated the effectiveness of the methylation DNA immunoprecipitation approach in the enrichment of hypermethylated fetal DNA.
Prenatal diagnosis is currently performed using conventional cytogenetic or DNA analysis, which require fetal genetic material to be obtained by amniocentesis, chorionic villus sampling, or chordocentesis. However, these are invasive procedures and are associated with a significant risk of fetal loss (0.5 to 1% for chorionic villus sampling and amniocentesis).1 For this reason, there is an urgent need for the development of diagnostic procedures that do not put the fetus at risk (commonly termed noninvasive prenatal diagnosis).
The discovery of free fetal DNA (ffDNA) in the maternal circulation during pregnancy2 has become a focus for alternative approaches toward the development of noninvasive prenatal tests. ffDNA has been successfully used for the determination of fetal sex and fetal RhD status in maternal plasma.3,4 Nevertheless, direct analysis of the limited amount of ffDNA (3 to 6%)5 in the presence of excess of maternal DNA is a great challenge for the development of noninvasive testing for fetal aneuploidies.
Recent advances in this field have shown that physical and molecular characteristics of the ffDNA can be used for its discrimination from circulating maternal DNA or as a means of fetal DNA enrichment.6,7 One of the most interesting developments has been the size-fractionation of plasma DNA to enrich for fetal DNA because fetal DNA is generally shorter in length than maternal DNA6 in the circulation. Furthermore, additional studies were conducted based on evidence that the ffDNA in maternal plasma is of placental origin.8,9 Thus epigenetic differences between maternal whole blood and placental DNA7 were used to detect hypomethylated maspin (SERPINB5) gene sequences in maternal plasma derived from the fetus (the SERPINB5 gene is known to be hypomethylated in placenta and hypermethylated in whole blood).10 Subsequently, a small number of additional differential fetal epigenetic molecular markers have been described including the RASSF1A gene on chromosome 311 as well as markers on chromosome 21.12,13
Although these studies have clearly demonstrated that epigenetic differences between fetal DNA (placental DNA obtained from chorionic villus sampling) and maternal whole blood DNA may serve as potential fetal molecular markers for noninvasive prenatal diagnosis, only a limited number of genomic regions have been identified or tested so far. A number of studies have focused on single gene promoter regions10,11 whereas others have investigated CpG islands on chromosome 21,12,13 which however cover only a small fraction of the chromosome.14
Current methods developed using ffDNA for noninvasive prenatal diagnosis are subject to a number of limitations. The two main methods being investigated are the use of methylation-sensitive restriction enzymes to remove hypomethylated maternal DNA thus allowing direct polymerase chain reaction (PCR) analysis of ffDNA and the use of sodium bisulfite conversion to allow the discrimination of differential methylation between maternal and fetal DNA. The requirement for regions of differentially methylated DNA containing a restriction site for recognition by methylation-sensitive restriction enzymes12 limits the number of regions suitable for testing. On the other hand, the use of sodium bisulfite conversion followed by methylation-specific PCR or methylation sensitive single nucleotide primer extension and/or bisulfite sequencing,10,11,12,13 has two main problems. Firstly, the accurate analysis of the methylation status after bisulfite conversion depends on the complete conversion of unmethylated cytosines to uracils, a condition rarely achieved. Secondly, the degradation of DNA obtained after bisulfite treatment15 complicates even further the testing and quantification of extremely low amounts of fetal DNA.
To overcome the above limitations, we have coupled a newly developed technique, methylated DNA immunoprecipitation (MeDiP)16 with high-resolution tiling oligonucleotide array analysis to enable chromosome-wide identification of DNA methylation patterns in a high-throughput approach. We have analyzed extensively the methylation patterns of chromosomes 13, 18, 21, X, and Y in female whole blood and placental DNA (the placental tissue was chosen for testing based on the fact that the source of ffDNA in maternal plasma is placenta8,9) Here we describe the identification of differentially methylated regions as potential molecular targets for noninvasive prenatal diagnosis as well as the development of specific fetal epigenetic molecular markers for the diagnosis of the most common aneuploidies1 such as the trisomy 13, 18, and 21 (associated with the Patau, Edwards, and Down (DS[MIM190685]) syndromes, respectively) (http://www.ncbi.nlm.nih.gov/Omim/) as well as sex chromosome abnormalities (such as the XXY-Klinefelter, XYY, XXX, and X-Turner syndromes).
Materials and Methods
Human Samples
The samples used in this study were obtained from AMS Biotechnology (Europe) Ltd. (Oxon, UK). These samples, originally sourced from the Biochain Institute (Hayward, CA), were subject to consent using ethical internal review board-approved protocols. In total, five female normal whole blood samples and five normal placental DNA samples were used in this study. Three of the placental DNA samples were obtained from first trimester pregnancies of which one derived from a female fetus pregnancy and two from male fetus pregnancies. The remaining two placental DNA samples were derived from third trimester pregnancies of which one of them was obtained from a female fetus pregnancy and the other one from a male fetus pregnancy.
MeDiP Assay Combined with LM-PCR Amplification
The MeDiP assay16 was followed by ligation-mediated PCR (LM-PCR)17,18 as described but with additional modifications. Briefly, 2.5 μg of DNA in a total volume of 100 μl was initially sheared by sonication into fragments of ∼300 bp to 1000 bp in size using the Virsonic300 sonicator (Virtis, Gardiner, NY). The fragmented DNA was blunt-ended after combination with 1× NEB buffer 2 (New England BioLabs, Ipswich, UK), 10× bovine serum albumin (New England BioLabs), 100 mmol/L dNTP mix (GE Healthcare, Little Chalfont, UK), T4 DNA polymerase (3 U/μl; New England BioLabs) and distilled water up to a final volume of 120 μl. After 20 minutes of incubation at 12°C, the reaction was cleaned up using the DNA Clean and Concentrator-5 (Zymo Research Corp, Glasgow, UK) according to the manufacturer’s protocol. However, the final elution step was performed in 30 μl of EB buffer provided by the QiAquick PCR purification kit (Qiagen, West Sussex, UK). The cleaned sample was then mixed with 40 μl of annealed linkers (50 μmol/L) (Sigma Genosys, Gillingham, UK) prepared as previously described,17,18 T4 DNA ligase 10× buffer (Roche, Mannheim, Germany), 6 μl of T4 DNA ligase (5 U/μl, Roche), and distilled water up to a final volume of 101 μl. The sample was then incubated overnight at 16°C to allow ligation of the adaptors. Purification of the sample was performed using the DNA Clean and Concentrator-5 as described above. The eluted sample was then combined with 100 mmol/L dNTP mix (GE Healthcare), 1× PCR gold buffer (Roche),1.5 mmol/L MgCl2 (Roche), and 5 U AmpliTaq polymerase (Applied Biosystems, Foster City, CS). The sample was then incubated at 70°C for 10 minutes to fill in DNA overhangs and purified using the DNA Clean and Concentrator-5 as described above. Fifty ng of DNA was removed and retained for use as input genomic control DNA. The remaining ligated DNA sample (700 ng to 1.2 μg) was subjected to MeDiP as described previously, after scaling down the reaction accordingly.16 The immunoprecipitated DNA sample was then cleaned up using the DNA Clean and Concentrator-5 (using 700 μl of binding buffer) according to the manufacturer’s instructions.
Ligation-mediated PCR (LM-PCR) was performed using 10 ng of each input and immunoprecipitated DNA fraction using the Advantage-GC Genomic PCR kit (Clontech, Saint-Germain-en-Laye, France) The thermal cycling conditions applied were 95°C for 2 minutes, 20 cycles at (94°C for 30 seconds and 68°C for 3 minutes) and 68°C for 10 minutes. After PCR amplification, the reaction was purified using the QIAquick PCR purification kit (Qiagen) and eluted in 50 μl of distilled water preheated to 50°C.
Array Hybridizations
The input and the immunoprecipitated DNA fractions from a female whole blood DNA sample, a third trimester placental DNA sample and a first trimester placental DNA sample (both placental samples were obtained from male fetus pregnancies) were sent to Roche NimbleGen Inc. (Reykjsvik, Iceland) for hybridization. The array platforms used were high-resolution tiling oligonucleotide arrays specific for chromosomes 13, 18, 21, X, and Y with a median probe spacing of 225 bp for chromosome 13, 170 bp for chromosome 18, 70 bp for chromosome 21, 340 bp for chromosome X, and 20 bp for chromosome Y. Briefly, the input genomic control DNA and the immunoprecipitated DNA of each sample were differentially labeled with fluorescent dyes (Cy3, Cy5) and were then co-hybridized on the oligonucleotide arrays. The raw data can be accessed in ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) with accession number E-TABM-507.
Data Analysis of the Tiling Oligonucleotide Arrays
To calculate the relative methylation differences between placenta and whole blood, the oligonucleotide array normalized log2 ratio values obtained from whole blood DNA (immunoprecipitated versus input fraction), were subtracted from the log2 ratio values (immunoprecipitated versus input fraction) obtained from placental DNA using the R statistical computing environment (http://www.rproject.org). Positive differences represent relative hypermethylation in placental DNA, negative differences relative hypomethylation. General Feature Format files were then created for the subtracted log2 ratio data and the results were viewed using the SignalMap viewer (NimbleGen System) together with genes, CpG islands, and CG content across chromosomes 13, 18, 21, X, and Y. Gene data were downloaded from the Ensembl genome browser (NCBI build36) (http://www.ensembl.org.) whereas the CpG islands and CG content were downloaded from the UCSC genome browser, (NCBI build36) (http://genome.ucsc.edu.). For the automated selection of regions with significant differential methylation enrichment between whole blood and placenta, we applied the Smith-Waterman dynamic programming algorithm adapted for Array-CGH (SW-ARRAY), which had previously been used for copy number variation calling.19 This algorithm was used to identify segments or islands of consecutive oligonucleotides showing methylation enrichment or methylation depletion in placental DNA compared with whole blood DNA. Each of the oligonucleotide array data were analyzed by chromosome arm. Threshold values (t0) for SW-ARRAY of between 0.2 and 1 were tested to identify the optimal value for calling relative high-score segments or islands based on the identification of at least two consecutive oligonucleotides having the same methylation status and showing the highest score among the scores obtained with different threshold values (t0). A direct comparison between the differentially methylated regions identified by SW-ARRAY and the position of genes, promoter regions (which were defined as the region up to 2 kb upstream of the 5′ end of each gene) and CpG islands was performed.
Although the above analysis identifies differentially methylated regions of the genome, for the use of MeDiP as an enrichment method before quantitative assays of copy number (ie, for the identification of trisomy 21 for example) for noninvasive prenatal diagnosis, it is important that selected targets were hypermethylated in the fetus. Thus, MeDiP would be used to deplete maternal hypomethylated DNA allowing assay of fetal hypermethylated DNA. Therefore, we identified all regions where the immunoprecipitated versus input ratios were positive in the placenta and negative in the whole blood samples (hypermethylation in the placenta). For other assay systems being investigated by others, hypomethylation of DNA in the placenta is required and so we also recorded all regions where the immunoprecipitated versus input ratios were negative in the placenta and positive in the whole blood samples. To achieve this, the oligonucleotide array normalized log2 ratio values obtained from whole blood DNA (immunoprecipitated versus input fraction), were subtracted from the log2 ratio values (immunoprecipitated versus input fraction) obtained from placental DNA only where the sign of the ratios were different. A small number of the most differentially hypermethylated regions in the placenta (eight regions on chromosome 21 and one region on chromosome 18 plus the SERPINB5 promoter region) were selected from this data set for further validation and assay development by using an arbitrary differential ratio cut off of greater than 1.0 (at least one of the consecutive probes showing a log2 ratio value >1).
Confirmation of Tiling Oligonucleotide Array Data by Real-Time Quantitative PCR
The methylation enrichment of the selected regions on chromosome 21 and chromosome 18 were confirmed by real-time quantitative PCR. For this purpose we used five female whole blood DNA samples and five placental DNA samples of both first and third trimester gestational age. Primers specific for the regions of interest were designed using the primer3 web site (http://frodo.wi.mit.edu/.). The primer design was optimized to give a product of 80 to 150 bp in length as recommended for optimal SYBR Green fluorescence melting curve analysis. Additionally, the Tm of all of the primers was chosen to be close to 60°C. The uniqueness of both primer sequences and PCR product sequences was confirmed by mapping these onto the human reference sequence using Blat. Primers were purchased from Sigma Genosys.
Each quantitative PCR reaction was performed in a final volume of 25 μl with 20 ng of either immunoprecipitated methylated DNA or input genomic DNA. We used the SYBR Green PCR master mix (Eurogentec, Seraing, Belgium) and an ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CS). Initially the optimal concentration of each primer was selected after testing a series of dilutions (150 to 900 nmol/L) using normal genomic DNA. For each selected primer concentration, standard curves were calculated on 1:2 serial dilutions (200 ng down to 3.125 ng) and finally, the sample reactions were performed in triplicate. The amplification program consisted of 2 minutes at 50°C and 10 minutes at 95°C, followed by 40 cycles of denaturation for 15 seconds at 95°C and annealing/extension for 1 minute at 60°C. After amplification, melting curve analysis was performed by heating the reaction mixture from 60 to 95°C at a rate of 0.2°C/second. Amplification reactions were routinely checked for nonspecific products by agarose gel electrophoresis. To evaluate the enrichment of target sequences after MeDiP, we calculated the ratios of the calculated cycle difference (ΔCt) between the test DNA (immunoprecipitated or input DNA) (Cttest) and an untreated normal genomic DNA (Ctcontrol) (ΔCt = Ctcontrol − Cttest) in the immunoprecipitated DNA versus input genomic DNA. In addition, to evaluate the methylation enrichment of placental samples compared with whole blood we calculated the ratio of placental immunoprecipitated DNA versus whole blood immunoprecipitated DNA. Moreover, control regions with similar DNA methylation status in whole blood and placental DNA samples as indicated by our oligonucleotide array results were used in every run to test for possible MeDiP or PCR amplification bias. The median variability and median reproducibility of methylation enrichment between individuals or between technical replicate experiments was obtained by calculating the average value of all of the ratio values of the different samples tested or of the technical replicates of a specific region (average IP/INPUT) and then each ratio value was divided by the average ratio value to obtain the reproducibility of each test and expressed as a percentage (IPi/INPUTi)/mean(IPi/INPUTi, IPii/INPUTii, …) = Ri IPi: immunoprecipitated DNA of the “ith” sample, INPUTi: input DNA of the “ith”sample, Ri = reproducibility of the “ith” sample). The median reproducibility was obtained by calculating the median of all of the different reproducibility values: median (Ri, Rii, …). Finally, the variability was calculated using the following formula: (1 − Ri) × 100 = Vi and median variability: median (Vi, Vii, …) (Vi = variability of the “ith” sample).
Results
Selection of Differentially Methylated Marker Candidates
The normalized oligonucleotide array log2 ratio values obtained for chromosomes 13, 18, 21, X, and Y in both placenta and whole blood DNA samples were initially used to calculate the ratio difference (relative methylation) between placental and whole blood DNA. We used SW-ARRAY to identify differentially methylated regions between whole blood and placenta. The optimal SW-ARRAY threshold for the identification of regions was found to be 0.9 for all chromosomes. The differentially methylated regions identified by SW-ARRAY analysis in first and third trimester compared with whole blood DNA are shown in Supplemental Tables 1 to 5 respectively for chromosomes 13, 18, 21, X, and Y (see Supplemental Tables 1 to 5 at http://ajp.amjpathol.org). We found that the number of regions identified to be hypermethylated was approximately equal to the number of regions being hypomethylated in all five chromosomes tested (Table 1). Additionally the total number of differentially methylated regions is comparable in first and third trimester placentas (Table 1). A direct correlation of the identified loci with the position of genes, promoter regions as well as the position of CpG islands is shown in Supplemental Tables 1 to 5 (http://ajp.amjpathol.org).
Table 1.
Differentially Methylated Regions in Placental Compared with Whole Blood DNA Samples Across Chromosomes 13, 18, 21, X, and Y
| CHR* | Trimester | No. of hyper† | No. of hypo‡ | Total no. | % Of hyper§ | % Of hypo¶ |
|---|---|---|---|---|---|---|
| 13 | First | 1310 | 1336 | 2646 | 49.5 | 50.5 |
| 13 | Third | 1311 | 1318 | 2629 | 49.9 | 50.1 |
| 18 | First | 1967 | 1888 | 3855 | 51.0 | 48.9 |
| 18 | Third | 1957 | 1944 | 3901 | 50.2 | 49.8 |
| 21 | First | 1063 | 1015 | 2078 | 51.1 | 48.8 |
| 21 | Third | 1042 | 1040 | 2082 | 50.0 | 49.9 |
| X | First | 1992 | 1951 | 3943 | 50.5 | 49.5 |
| X | Third | 1995 | 1989 | 3984 | 50.1 | 49.9 |
| Y | First | 1192 | 1120 | 2312 | 51.6 | 48.4 |
| Y | Third | 1986 | 1990 | 3976 | 49.9 | 50.0 |
Chromosome.
Number of hypermethylated regions.
Number of hypomethylated regions.
Percentage of hypermethylated regions.
Percentage of hypomethylated regions.
The 17.5 to 43.6% of the identified regions were located within genes and their methylation status varied between chromosomes as well as between trimesters (Table 2). For chromosomes 13 and Y, the majority of the differentially methylated regions (located within genes) are hypomethylated in first trimester placenta but the majority became hypermethylated in third trimester placentas. In chromosomes 21 and X, most of the regions were hypomethylated in both first and third trimester placentas whereas in chromosome 18 equal numbers of regions were found to be hypermethylated and hypomethylated (Table 2). Additionally, a small percentage (1.6 to 11.0%) of the identified regions (both genic and nongenic regions) were overlapping with CpG islands (Table 3). The majority of the CpG islands identified to be differentially methylated were located within genes (including the appropriate promoter sites defined as the regions up to 2 kb upstream the 5′ end of each gene) with a significant number of these (up to 65.5%) located specifically within promoter regions. The methylation status of these regions in each chromosome and in first and third trimester placentas is shown in Table 4.
Table 2.
Differentially Methylated Regions Located Within Genes in Chromosomes 13, 18, 21, X, and Y
| CHR* | Trimester | No. of hyper† | No. of hypo‡ | Total no. | % Of total§ | % Of hyper¶ | % Of hypo∥ |
|---|---|---|---|---|---|---|---|
| 13 | First | 374 | 671 | 1045 | 39.5 | 35.8 | 64.2 |
| 13 | Third | 554 | 480 | 1034 | 39.3 | 53.6 | 46.4 |
| 18 | First | 734 | 728 | 1463 | 37.9 | 50.2 | 49.8 |
| 18 | Third | 746 | 673 | 1419 | 36.4 | 52.6 | 47.4 |
| 21 | First | 362 | 545 | 907 | 43.6 | 39.9 | 60.1 |
| 21 | Third | 338 | 486 | 824 | 39.6 | 41.0 | 59.0 |
| X | First | 700 | 884 | 1584 | 40.2 | 44.2 | 55.8 |
| X | Third | 604 | 897 | 1501 | 37.7 | 40.2 | 59.8 |
| Y | First | 196 | 228 | 424 | 18.3 | 46.2 | 53.8 |
| Y | Third | 387 | 310 | 697 | 17.5 | 55.5 | 44.5 |
Chromosome.
Number of hypermethylated regions within genes.
Number of hypomethylated regions within genes.
Percentage of differentially methylated regions within genes.
Percentage of hypermethylated regions within genes.
Percentage of hypomethylated regions within genes.
Table 3.
Methylation Status of CpG Islands Located Within the Differentially Methylated Regions in Chromosomes 13, 18, 21, X, and Y
| CHR* | Trimester | Regions within CpGs† | % Total‡ |
|---|---|---|---|
| 13 | First | 250 | 9.4 |
| 13 | Third | 142 | 5.4 |
| 18 | First | 182 | 5.4 |
| 18 | Third | 143 | 3.7 |
| 21 | First | 162 | 7.8 |
| 21 | Third | 91 | 4.4 |
| X | First | 435 | 11.0 |
| X | Third | 415 | 10.4 |
| Y | First | 46 | 2.0 |
| Y | Third | 63 | 1.6 |
Number of methylated regions overlapping with CpG islands in chromosomes 13, 18, 21, X, and Y.
Chromosome.
Number of regions overlapping with CpG islands.
Percentage differentially methylated regions overlapping with CpG islands.
Table 4.
Methylation Status of CpG Islands Located Within the Differentially Methylated Regions in Chromosomes 13, 18, 21, X, and Y
| CHR* | Trimester | Hyper/hypo (+/−)† | Genic CpGs | Nongenic CpGs | Total no. | Promoter CpGs | % Of promoter CpGs‡ |
|---|---|---|---|---|---|---|---|
| 13 | First | + | 17 | 7 | 24 | 11 | 45.8 |
| 13 | First | − | 226 | 84 | 310 | 102 | 32.9 |
| 13 | Third | + | 74 | 26 | 100 | 31 | 31.0 |
| 13 | Third | − | 57 | 25 | 82 | 21 | 25.6 |
| 18 | First | + | 13 | 5 | 18 | 6 | 33.3 |
| 18 | First | − | 166 | 90 | 256 | 65 | 25.4 |
| 18 | Third | + | 30 | 12 | 42 | 15 | 35.7 |
| 18 | Third | − | 102 | 60 | 162 | 28 | 17.3 |
| 21 | First | + | 0 | 1 | 1 | 0 | 0.0 |
| 21 | First | − | 220 | 41 | 261 | 121 | 46.4 |
| 21 | Third | + | 4 | 4 | 8 | 1 | 12.5 |
| 21 | Third | − | 90 | 37 | 127 | 49 | 38.6 |
| X | First | + | 2 | 1 | 3 | 0 | 0.0 |
| X | First | − | 592 | 103 | 695 | 414 | 59.6 |
| X | Third | + | 5 | 1 | 6 | 0 | 0.0 |
| X | Third | − | 609 | 93 | 702 | 460 | 65.5 |
| Y | First | + | 3 | 2 | 5 | 0 | 0.0 |
| Y | First | − | 49 | 21 | 70 | 35 | 50.0 |
| Y | Third | + | 28 | 33 | 61 | 3 | 4.9 |
| Y | Third | − | 12 | 11 | 23 | 8 | 34.8 |
Methylation status of genic, nongenic, and promoter CpG islands in chromosomes 13, 18, 21, X, and Y.
Chromosome.
Hypermethylated/hypomethylated.
Percentage of promoter CpGs compared with the total number of differentially methylated CpG islands.
Validation of the Tiling Oligonucleotide Array Results
To validate the array analysis, we first analyzed the methylation status of a previously investigated region by real-time quantitative PCR. We found that the SERPINB5 promoter region was hypomethylated in placenta and hypermethylated in whole blood, which is in agreement with the study of Chim and colleagues.10 To further validate the methylation differences identified by high-resolution array analysis, an additional nine selected regions, including the SERPINB5 promoter region10 (Table 5), were analyzed by real-time quantitative PCR. Various primers were designed to cover each region. Each primer was tested before use and the optimal concentration for each one was determined (Table 6).
Table 5.
The Primers Designed for the Regions Tested on Chromosome 18 and Chromosome 21
| Chr region* | Position (bp) | Location type | Gene involved |
|---|---|---|---|
| CHR18(A) | 55090279 to 55090959 | Intragenic CpG island | RAX |
| CHR21(A) | 39279691 to 39279971 | Intergenic region | |
| CHR21(B) | 44160993 to 44161543 | Intragenic region | AGPAT3 |
| CHR21(C) | 33320530 to 33320815 | CpG island within promoter region | OLIG2 |
| CHR21(D) | 42189235 to 42189849 | Promoter region | C21orf25 |
| CHR21(EI) | 42355366 to 42355908 | Intergenic region | |
| CHR21(EII) | 42357141 to 42357401 | Intergenic region | |
| CHR21(H) | 32268787 to 32269137 | Intragenic region | HUNK |
| CHR21(I) | 44079218 to 44079733 | Intergenic region |
The tested regions on chromosome 18 and chromosome 21.
Table 6.
The Primers Designed for the Regions Tested on Chromosome 18 and Chromosome 21
| Chr region* | Primer name | Forward primer | Reverse primer | Position (bp) | Pr size† | Selected conc‡ |
|---|---|---|---|---|---|---|
| SERBINB5 | SERBINB5(1) | 5′-TATTTCACCTTCCGGTCCTG-3′ | 5′-GTTGTCTGCACAGCTTCCAA-3′ | 59294886 to 59295011 | 126 bp | 300 nmol/L |
| SERBINB5(2) | 5′-TACTTTTTGTGCCACCAACG-3′ | 5′-CAGGACCGGAAGGTGAAATA-3′ | 59294811 to 59294905 | 95 bp | 900 nmol/L | |
| SERBINB5(3) | 5′- TCCAGGTCTTTGTGCTCCTC-3′ | 5′-TACCCCACCTTGCTTACCTG-3′ | 59295190 to 59295275 | 86 bp | 300 nmol/L | |
| CHR18(A) | CHR18(A-1) | 5′-TAATGAGCTGTCGGCCTCTT-3′ | 5′-ATCTTCGATGTTCCCTGTGC-3′ | 55090284 to 55090425 | 142 bp | 300 nmol/L |
| CHR18(A-2) | 5′-TGTGCCTCTCCCTTGAGACT-3′ | 5′-AAATTGCAGCCAATGCTTCT-3′ | 55090427 to 55090524 | 98 bp | 300 nmol/L | |
| CHR18(A-3) | 5′-CGTGAATTCCGAGAAGCATT-3′ | 5′-GGCAAGGTCAACCTACCAGA-3′ | 55090494 to 55090605 | 112 bp | 300 nmol/L | |
| CHR21(A) | CHR21(A1) | 5′-GCTGGACCAGAAAGTGTTGAG-3′ | 5′-GTGTGCTGCTTTGCAATGTG-3′ | 39279856 to 39280004 | 149 bp | 300 nmol/L |
| CHR21(A2) | 5′-AAACACCGGGTGATAAGCAG-3′ | 5′-GGCCACTCACGCTCTTTTTA-3′ | 39279723 to 39279841 | 119 bp | 300 nmol/L | |
| CHR21(B) | CHR21(B1) | 5′-GATTGGTGTGTTTGGACCAG-3′ | 5′-AGAGGCAAGAGCAGAAACCA-3′ | 44161027 to 44161157 | 131 bp | 900 nmol/L |
| CHR21(B2) | 5′-GGTTTCTGCTCTTGCCTCTC-3′ | 5′-GGAGCAGCCTGAGGATCTTA-3′ | 44161139 to 44161279 | 141 bp | 300 nmol/L | |
| CHR21(B3) | 5′-GGTCGAGTTTTTGGTGGTGT-3′ | 5′-CCACCGTCACTGTTCCTAGA-3′ | 44161178 to 44161323 | 146 bp | 300 nmol/L | |
| CHR21(B4) | 5′-CCTCGTGCTCGTGTCTGTAT-3′ | 5′-GAGGAAACAGCTTGGCTCTG-3′ | 44161239 to 44161371 | 133 bp | 300 nmol/L | |
| CHR21(C) | CHR21(C1) | 5′-TAAGGTGGATCCGTTTGAGG-3′ | 5′-TTCGGGCTTTCAGTTAGGTG-3′ | 33320544 to 33320681 | 138 bp | 450 nmol/L |
| CHR21(C2) | 5′-CGTCCCCCTCGCTACTATCT-3′ | 5′-CAACGCTCCCTGAAATAACC-3′ | 33320716 to 33320829 | 114 bp | 300 nmol/L | |
| CHR21(C3) | 5′-CTGTTGCATGAGAGCAGAGG-3′ | 5′-CGTCCCCCTCGCTACTATCT-3′ | 33320735 to 33320829 | 95 bp | 900 nmol/L | |
| CHR21(D) | CHR21(D1) | 5′-TGTGCAGGATATTTGGCAAG-3′ | 5′-CTGTGCCGGTAGAAATGGTT-3′§ | 42189555 to 42189683 | 129 bp | 300 nmol/L |
| CHR21(D2) | 5′-TGCAGGATATTTGGCAAGGT-3′ | 5′-CTGTGCCGGTAGAAATGGTT-3′§ | 42189557 to 42189683 | 127 bp | 450 nmol/L | |
| CHR21(D3) | 5′-CACAGCACTGTCAGGAGGAA-3′ | 5′-GTCCTAGGAGTGCAGCCTGT-3′¶ | 42189223 to 42189343 | 121 bp | 300 nmol/L | |
| CHR21(D4) | 5′-CAGCACTGTCAGGAGGAACA-3′ | 5′-GTCCTAGGAGTGCAGCCTGT-3′¶ | 42189225 to 42189343 | 119 bp | 300 nmol/L | |
| CHR21(EI) | CHR21(EI-1) | 5′-TGAATCAGTTCACCGACAGC-3′ | 5′-GAAACAACCTGGCCATTCTC-3′ | 42355712 to 42355815 | 104 bp | 900 nmol/L |
| CHR21(EI-2) | 5′-GGCCAGGTTGTTTCAGATTG-3′ | 5′-TTCCGGCAGAGTTTATTTGG-3′ | 42355802 to 42355908 | 107 bp | 300 nmol/L | |
| CHR21(EII) | CHR21(EII-1) | 5′-CCGTTATATGGATGCCTTGG-3′ | 5′-AAACTGTTGGGCTGAACTGC-3′ | 42357215 to 42357341 | 127 bp | 750 nmol/L |
| CHR21(H) | CHR21(H1) | 5′-ACAGCGACGTGATCAACACT-3′ | 5′-CGCACTTACCCCTGACAAAT-3′ | 32268803 to 32268909 | 107 bp | 150 nmol/L |
| CHR21(H2) | 5′-CCACATCCTGGCCATCTACT-3′ | 5′-TTCCACAGACAGCAGAGACG-3′ | 32268843 to 32268943 | 101 bp | 300 nmol/L | |
| CHR21(I) | CHR21(I1) | 5′-TGAGCTCACAGGTCTGGAAA-3′ | 5′-CCCCACAGGGTTCTGGTAAT-3′ | 44079235 to 44079322 | 88 bp | 300 nmol/L |
| CHR21(I2) | 5′-CCCTGTGGGGTTTTTATTGAT-3′ | 5′-AGGTTTGTGAGGGTCCATGA-3′ | 44079313 to 44079415 | 103 bp | 300 nmol/L | |
| CHR21(I3) | 5′-CATGGACCCTCACAAACCTC-3′ | 5′-GTGGTCTGGGTGTGGAGAAT-3′ | 44079397 to 44079535 | 139 bp | 300 nmol/L |
The primer characteristics for the regions tested on chromosome 18 and chromosome 21.
Chromosomal region.
Product size.
Selected concentration.
Common primers.
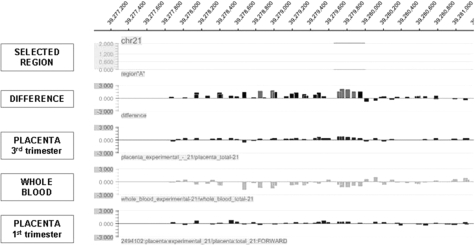
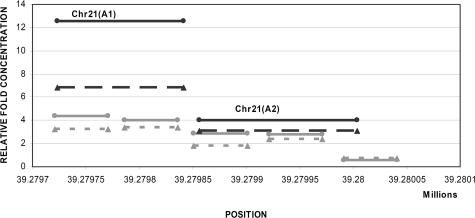
As an example, the oligonucleotide array results for a region located on chromosome 21 (Figure 1) can be compared with the real-time quantitative PCR results as shown in Figure 2. The same whole blood and placental DNA samples were used for PCR validation as had been used for oligonucleotide array hybridizations. Both methods reported enrichment for methylation in placenta compared with whole blood. Additionally, both oligonucleotide array and real-time quantitative PCR demonstrated a lower degree of methylation enrichment in first trimester placental DNA sample compared with third trimester placental DNA sample. Nevertheless, the methylation enrichment observed in the first trimester placenta was higher compared with the methylation enrichment obtained from the whole blood DNA sample. We found concordance of differential methylation for all nine selected regions as well as the SERPINB5 promoter region between array analysis and quantitative PCR. However, the relative fold enrichment obtained using real-time quantitative PCR was generally found to be greater than the enrichment obtained by oligonucleotide array analysis.
Figure 1.
DNA methylation enrichment of CHR21(A) region assayed by oligonucleotide array. Methylation difference between third trimester placenta and whole blood and individual methylation status of whole blood, first trimester and third trimester placental DNA samples.
Figure 2.
Comparison of the DNA methylation enrichment of CHR21(A) from oligonucleotide arrays and real-time quantitative PCR using whole blood, first trimester and third trimester placental DNA samples. The y axis indicates the relative fold enrichment of placenta when compared with whole blood DNA sample and the x axis indicates the chromosomal position in bp. The gray lines represent the oligonucleotides covering the specific region on chromosome 21 whereas the black lines represent the PCR products [CHR21(A1) and CHR21(A2)] Biomarkers, Genomics, Proteomics, and Gene Regulation when real-time quantitative PCR was applied. The dotted lines represent the results obtained from a first trimester placenta whereas the solid lines represent the results obtained from a third trimester placenta.
Evaluation of MeDiP-LMPCR Efficiency
The experimental reproducibility of the MeDiP procedure was assessed by performing technical replicates of a single placental DNA sample with the methylation status of two regions on chromosome 21 being assessed by real-time quantitative PCR. The median reproducibility was found to be 98.62% and 95.84% for the regions shown in Supplemental Figure 1, A and B, respectively (http://ajp.amjpathol.org).
Evaluating the Methylation Status of Selected Regions in Different Individuals
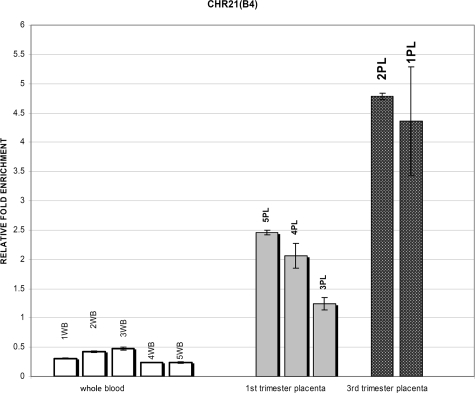
To assess the normal variability in methylation status between individuals, we applied the real-time quantitative PCR assay for the nine selected regions to five different whole blood and three different first trimester placentas. The methylation status of two third trimester placentas was tested in only four of the selected regions. Further investigation of interindividual variability of third trimester placentas was not necessary in this study in which we preferably tested first trimester placentas as their gestational age is the most appropriate for the development of noninvasive prenatal diagnosis. The results of the CHR21(B4) region are shown in Figure 3. All five whole blood samples were found to be hypomethylated compared with the five placental DNA with a variability of DNA methylation enrichment between individuals as shown in Table 7. Less methylation enrichment was observed in first trimester placentas compared with third trimester placentas confirming the oligonucleotide array results (Figure 3). A lower level of variability was also observed between different placentas of the same gestational age (Table 7).
Figure 3.
DNA methylation enrichment of CHR21(B4) by real-time quantitative PCR on MeDiP and input replicated DNA samples. Open bars, whole blood samples; gray bars, first trimester placental DNA samples; solid black bars, third trimester placental DNA samples. The error bars indicate the SD between technical replicates. WB, whole blood; PL, placenta.
Table 7.
Percentage Variability of Methylation between Different Samples of the Same Tissue Origin (Whole Blood or Placenta) and Gestational Age (Placenta) Calculated for Multiple Regions Located in Chromosomes 18 and 21
| Chr | Region | Primer tested | WB_aver* | 1st TR PL_aver† | 3rd TR PL_aver‡ | WB var (%)§ | 1st TR PL var (%)¶ | 3rd TR PL var (%)∥ |
|---|---|---|---|---|---|---|---|---|
| 18 | CHR18(A) | CHR18(A-2) | 0.12 | 1.72 | 1.90 | 6.2 | 1.3 | 0 |
| 18 | CHR18(A-3) | 0.08 | 1.89 | 2.30 | 5.9 | 10.2 | 0 | |
| 21 | CHR21(A) | CHR21(A1) | 0.43 | 1.49 | 2.08 | 0.1 | 10.7 | 0 |
| 21 | CHR21(A2) | 0.21 | 1.23 | 2.67 | 4.9 | 4.6 | 0 | |
| 21 | CHR21(B) | CHR21(B3) | 0.38 | 2.09 | 4.85 | 12.7 | 3.3 | 0 |
| 21 | CHR21(B4) | 0.34 | 1.92 | 4.57 | 7.6 | 7.6 | 0 | |
| 21 | CHR21(C) | CHR21(C1) | 0.25 | 1.51 | 1.3 | 1.1 | ||
| 21 | CHR21(C2) | 0.30 | 1.53 | 3.0 | 5.3 | |||
| 21 | CHR21(C3) | 0.28 | 1.88 | 3.9 | 1.5 | |||
| 21 | CHR21(D) | CHR21(D2) | 0.66 | 6.13 | 3.3 | 4.5 | ||
| 21 | CHR21(D3) | 0.27 | 2.71 | 1.9 | 6.2 | |||
| 21 | CHR21(D4) | 0.35 | 2.58 | 4.8 | 4.8 | |||
| 21 | CHR21(EI) | CHR21(EI-1) | 0.05 | 1.85 | 58.3 | 18.7 | ||
| 21 | CHR21(EI-2) | 0.07 | 1.86 | 45.2 | 15.1 | |||
| 21 | CHR21(EII) | CHR21(EII-1) | 0.36 | 1.92 | 3.78 | 10.2 | 22.4 | 0 |
| 21 | CHR21(H) | CHR21(H1) | 0.57 | 2.03 | 2.4 | 0.8 | ||
| 21 | CHR21(H2) | 0.50 | 1.90 | 1.0 | 5.3 | |||
| 21 | CHR21(I) | CHR21(I1) | 0.41 | 1.67 | 0.3 | 0.1 | ||
| 21 | CHR21(I2) | 0.30 | 1.48 | 3.8 | 4.8 |
The mean of five different whole blood samples.
The mean of three 1st trimester placentas.
Mean of two 3rd trimester placentas.
Median variability between the whole blood samples.
Median variability between the 1st trimester placental samples.
Median variability between the 3rd trimester placental samples.
The majority of the regions tested so far by real-time quantitative PCR showed methylation variability between placental DNA samples of different gestational ages (Table 7). However, not all of the regions demonstrated this effect. One example is a region located on chromosome 18 [CHR18(A)] shown in Table 7 and Supplemental Table 6 (view Supplemental Table 6 at http://ajp.amjpathol.org). Only minor differences of the methylation level between first and third trimester placentas were found for this region. Additionally, we identified regions with opposite DNA methylation status between first and third trimester placentas as well as other regions showing differential methylation (examples shown in Table 8).
Table 8.
Example of Regions Found to Have Opposite Methylation Status between 1st and 3rd Trimester Placental DNA or Being Differentially Methylated at a Specific Gestational Age Compared with Whole Blood DNA
| Chr* | Gene name | Meth. status 1st TR_PL† | Meth. status 3rd TR_PL‡ |
|---|---|---|---|
| 13 | ALG5 | Hypomethylated | Nondifferentially methylated |
| 13 | C13orf3 | Nondifferentially methylated | Hypomethylated |
| 13 | EBPL | Hypomethylated | Nondifferentially methylated |
| 13 | BRCA2 | Hypomethylated | Hypermethylated |
| 13 | C13orf21 | Hypomethylated | Hypermethylated |
| 18 | CPLX4 | Nondifferentially methylated | Hypomethylated |
| 18 | CTDP1 | Hypomethylated | Nondifferentially methylated |
| 18 | MCART2 | Hypomethylated | Nondifferentially methylated |
| 18 | ME2 | Hypomethylated | Nondifferentially methylated |
| 21 | SUMO3 | Hypomethylated | Nondifferentially methylated |
| 21 | WRB | Hypomethylated | Nondifferentially methylated |
Chromosome.
Methylation status of 1st trimester placenta.
Methylation status of 3rd trimester placenta.
Differentially Methylated Control Regions
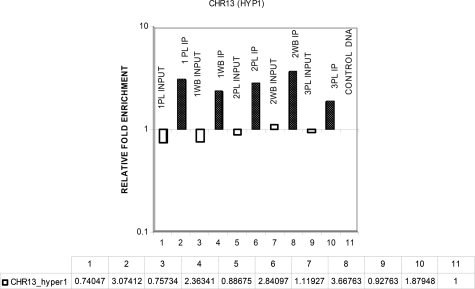
To assess MeDiP efficiency in all experiments, we selected two hypermethylated and two hypomethylated regions that showed a similar degree of differential methylation from the array experiments for use as controls (Table 9). The methylation status of these regions was assessed by real-time quantitative PCR in multiple samples of whole blood and placenta. The results for a region on chromosome 13, which was expected to be hypermethylated in both whole blood and placental DNA samples, are shown in Figure 4. The same DNA samples were used for testing all of the control primers. The median reproducibility of methylation enrichment between individuals was found to be 98.48% for the region CHR13 (HYP1), 96.02% for the region CHR13 (HYP2), 98.71% for the region CHR22(U1), and 99.75% for the region CHR22(U2).
Table 9.
Control Primers Used to Assess the MeDiP Efficiency on Different Samples Using Real-Time Quantitative PCR
| Primer name | Forward primer | Reverse primer | Position (bp) | Pr. size* | Selected conc† | Meth. status‡ |
|---|---|---|---|---|---|---|
| CHR13(HYP1)§ | 5′-CAGGAAAGTGAAGGGAGCTG-3′ | 5′-CAAAACCCAATGGTCAATCC-3′ | 19991387 to 19991465 | 79 bp | 300 nmol/L | Hypermethylated |
| CHR13(HYP2)¶ | 5′-AATGATTGTGCAGGTGGTGA-3′ | 5′-GAGCGCCTTGAGTAGAGGAA-3′ | 20191970 to 20192091 | 122 bp | 300 nmol/L | Hypermethylated |
| CHR22(U1)∥23 | 5′-AAGGTGCCCAATTCAAGGTA-3′ | 5′-CTTCCCCACCAGTCTTGAAA-3′ | 30214952 to 30215055 | 104 bp | 300 nmol/L | Hypomethylated |
| CHR22(U2)**23 | 5′-TGAGAGCGGATGACAGATTG-3′ | 5′-GGTCCCTCCCTTTTCTGTCT-3′ | 35582634 to 35582737 | 104 bp | 300 nmol/L | Hypomethylated |
Product size.
Selected concentration.
Methylation status.
Hypermethylated region 1 on chromosome13.
Hypermethylated region 2 on chromosome13.
Hypomethylated region 1 on chromosome 22.
Hypomethylated region 2 on chromosome 22.
Figure 4.
DNA methylation enrichment of CHR13(HYP1) using real-time quantitative PCR. Open bars, input DNA compared with whole blood; solid black bars, immunoprecipitated DNA compared with whole blood; 1PL and 2PL, third trimester placentas; 3PL, first trimester placenta; WB, whole blood; PL, placenta.
Discussion
We have demonstrated in this study that the application of MeDiP followed by high-resolution tiling oligonucleotide array analysis enables the identification of differentially methylated DNA sequences between whole blood and placenta. The high sensitivity and specificity of the methodology allowed us to select previously unreported candidate fetal epigenetic molecular markers located on chromosomes 13, 18, 21, X, and Y (see Supplementary Tables 1 to 5, respectively, at http://ajp.amjpathol.org) Furthermore, MeDiP allowed us to enrich directly regions, which were shown to be hypermethylated in placental DNA compared with whole blood. Thus by using the unbound fraction of DNA eluted after immunoprecipitation it would be possible to enrich for differentially hypomethylated fetal regions. Looking ahead, the epigenetic differences between fetal and maternal DNA identified in this study will facilitate their discrimination within the plasma sample. More importantly, the high sensitivity and specificity of this methodology will facilitate the enrichment of fetal DNA within maternal plasma and therefore the technical challenges that are involved in the development of noninvasive prenatal diagnosis of aneuploidies will be minimized. Such epigenetic markers will be used for noninvasive prenatal diagnosis using either SNP allele-specific ratio or by direct comparison with a fetal-specific methylation marker on a reference chromosome. Furthermore, the application of the MeDiP methodology has overcome the limitations of current methylation-based methods developed for noninvasive prenatal diagnosis such as the use of methylation-sensitive restriction enzymes or bisulfite conversion methodologies.8,9,10,11 However, these methods will remain important for further validation of the targets identified in this study.
In addition to the current approaches toward noninvasive prenatal diagnosis of aneuploidies, a new methodology has been described recently by two groups.20,21 Their approach is based on the application of next generation sequencing performed using the Solexa/Illumina platform. By direct sequencing of maternal free DNA they were able to show a small increase in read depth across chromosome 21 in aneuploid pregnancies. Although this methodology looks promising, it is a technologically demanding approach, which is not easily adapted for high-throughput, low-cost prenatal screening. Furthermore, the vast majority of diagnostic laboratories do not have access to or can afford next generation sequencing technologies so it is uncertain whether this methodology could be actively implemented in worldwide clinical practice in the near future.
An important feature of our study is the use of high-resolution, chromosome-wide analysis of differential DNA methylation. To perform the initial wide chromosome screen of the chromosomes of interest (chromosomes 13, 18, 21, X, and Y), we used a female whole blood and two placental samples of different gestational age. The chromosome-wide screening has revealed regions showing differential methylation between female whole blood and placental samples. Real-time quantitative PCR was then applied to confirm the existence of differential methylation by using multiple MeDiP products of the same sample. By this way we were able to exclude any experimental variability. We then tested the methylation status of selected regions in multiple samples and we were able to confirm that the differential methylation of those regions between whole blood and placenta was a true biological event and not an artifact caused by any experimental or handling variability. The effectiveness of the MeDiP methodology in identifying true biological events has also been demonstrated by a recent study in which a high correlation of the results was found when using MeDiP and bisulfite sequencing methodologies.22 In total, we selected eight of the candidate fetal epigenetic molecular markers identified on chromosome 21 and one on chromosome 18 for validation and further investigation. All regions were hypermethylated in placental DNA samples and were selected based on the presence of an opposite methylation status when compared with whole blood.
Application of the MeDiP approach has enabled us to identify differentially methylated regions in addition to those found at CpG islands, gene promoter regions, or restriction enzyme recognition sites as previously reported.10,11,12,13 Additionally, although a recent study has described DNA methylation profiles in a genome-wide and tissue-specific manner, the analysis has concentrated only on genic regions and CpG islands.22 Unfortunately, because of the different resolution of the array platforms used in this recent study and described here and the alternative analysis approaches used, direct comparison of the methylation status between the studies has not been successful.
Moreover, it is of great interest to note that most of the differentially methylated regions identified are located within nongenic regions (Table 2). Furthermore, the majority of the identified differentially methylated regions between placenta and whole blood are located within low CpG density regions and nonpromoter regions.
In this study, we have detailed a large number of previously unreported fetal epigenetic regions, which have the potential to be developed into molecular markers for noninvasive prenatal diagnosis. Additionally, we have demonstrated how the use of the 5-methylcytidine antibody combined with the high sensitivity and specificity of immunoprecipitation allow the enrichment of hypermethylated fetal DNA and, potentially for the enrichment of hypomethylated fetal DNA for use in noninvasive prenatal testing. The molecular markers we have described will be of great use to groups developing various methods for noninvasive prenatal diagnosis of fetal aneuploidy based on methylation differences.
Acknowledgments
We thank Richard Redon, Armand Valsesia, Tomas Fitzgerald, Daniel Andrews, and Stephen Clayton for assistance with bioinformatics analysis; and Loudmila Kousoulidou, Carolina Sismani, and Paola Evangelidou for helpful discussion.
Footnotes
Address reprint requests to Dr. Philippos C. Patsalis. Chief Executive Medical Director; The Cyprus Institute of Neurology and Genetics; Nicosia, P.O. Box 23462, 1683, Cyprus. E-mail: patsalis@cing.ac.cy.
Supported by the SAFE Network of Excellence European Commission Funded 6th Framework Package Project No. LSHB-CT-2004-503243) and the Wellcome Trust (to N.P.C., H.F., V.R., and S.B.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Current addresses of H.F.: Roche Diagnostics UK Limited, West Sussex, UK; V.R.: Barts and The London School of Medicine and Dentistry; Institute of Cell and Molecular Science, London, UK; and S.B.: UCL Cancer Institute; University College London; London, UK.
References
- Hultén MA, Dhanjal S, Pertl B. Rapid and simple prenatal diagnosis of common chromosome disorders advantages and disadvantages of the molecular methods FISH and QF-PCR. Reproduction. 2003;126:279–297. doi: 10.1530/rep.0.1260279. [DOI] [PubMed] [Google Scholar]
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, Poon PM, Redman CW, Wainscoat JS. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med. 1998;339:1734–1738. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Avent ND, Costa JM, Van der Schoot CE. Noninvasive prenatal diagnosis of fetal Rhesus D: ready for Prime(r) Time. Obstet Gynecol. 2005;106:841–844. doi: 10.1097/01.AOG.0000179477.59385.93. [DOI] [PubMed] [Google Scholar]
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- Poon LL, Leung TN, Lau TK, Chow KC, Lo YM. Differential DNA methylation between fetus and mother as a strategy for detecting fetal DNA in maternal plasma. Clin Chem. 2002;48:35–41. [PubMed] [Google Scholar]
- Masuzaki H, Miura K, Yoshiura K-I, Yoshimura S, Niikawa N, Ishimaru T. Detection of cell free placental DNA in maternal plasma: direct evidence from three cases of confined placental mosaicism. J Med Genet. 2004;41:289–292. doi: 10.1136/jmg.2003.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flori E, Doray B, Gautier E, Kohler M, Ernault P, Flori J, Costa JM. Circulating cell-free fetal DNA in maternal serum appears to originate from cyto- and syncytio-trophoblastic cells. Case report. Hum Reprod. 2004;19:723–724. doi: 10.1093/humrep/deh117. [DOI] [PubMed] [Google Scholar]
- Chim SS, Tong YK, Chiu RW, Lau TK, Leung TN, Chan LY, Oudejans CB, Ding C, Lo YM. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci. 2005;102:14753–14758. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RW, Chim SS, Wong IH, Wong CS, Lee WS, To KF, Tong JH, Yuen RK, Shum AS, Chan JK, Chan LY, Yuen JW, Tong YK, Weier JF, Ferlatte C, Leung TN, Lau TK, Lo KW, Lo YM. Hypermethylation of RASSF1A in human and rhesus placentas. Am J Pathol. 2007;170:941–950. doi: 10.2353/ajpath.2007.060641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old RW, Crea F, Puszyk W, Hultén MA. Candidate epigenetic biomarkers for non-invasive prenatal diagnosis of Down syndrome. Reprod Biomed Online. 2007;15:227–235. doi: 10.1016/s1472-6483(10)60713-4. [DOI] [PubMed] [Google Scholar]
- Chim SS, Jin S, Lee TY, Lun FM, Lee WS, Chan LY, Jin Y, Yang N, Tong YK, Leung TY, Lau TK, Ding C, Chiu RW, Lo YM. Systematic search for placental DNA-methylation markers on chromosome 21: toward a maternal plasma-based epigenetic test for fetal trisomy 21. Clin Chem. 2008;54:500–511. doi: 10.1373/clinchem.2007.098731. [DOI] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Oberley MJ, Tsao J, Yau P, Farnham PJ. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- Price TS, Regan R, Mott R, Hedman A, Honey B, Daniels RJ, Smith L, Greenfield A, Tiganescu A, Buckle V, Ventress N, Ayyub H, Salhan A, Pedraza-Diaz S, Broxholme J, Ragoussis J, Higgs DR, Flint J, Knight SJ. SW-ARRAY: a dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res. 2005;16:3455–3464. doi: 10.1093/nar/gki643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RWK, Chan KCA, Gao Y, Lau VYM, Zheng W, Leung TY, Foo CHF, Xie B, Tsui NBY, Lun FMF, Zee BCY, Lau TK, Cantor CR, Lo YMD. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan KV, Down AT, Thorne PN, Flicek P, Kulesha E, Gräf S, Tomazou ME, Bäckdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJ, Carter NP, Tavaré S, Beck S. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]