Abstract
Results from several microarray-based studies have led to the identification of up-regulated expression levels of the DSG3 gene in pulmonary squamous cell carcinomas (SQCCs). The purpose of this study was to determine the role of DSG3 expression in the diagnosis of SQCCs of the lung and to compare DSG3 with p63, CK5, and CK6, as markers of squamous cell differentiation. Expression of DSG3 mRNA was evaluated in bulk laser capture microdissection-derived microarray data and by quantitative reverse transcription PCR on both SQCCs and adenocarcinomas. Expression levels of p63, CK5, and CK6 were evaluated in microarray data from the same set. An immunohistochemical study using antibodies directed against DSG3, p63, and CK5/6 was also performed. DSG3 was over-expressed in SQCCs but had very limited expression in both adenocarcinomas and non-neoplastic lungs. The microarray data showed that DSG3 had a sensitivity and specificity of 88% and 98%, respectively, in detecting SQCC versus adenocarcinoma. In comparison, sensitivity and specificity was 92% and 82% for p63, and 85% and 96% for CK5, respectively. The correlation coefficient between the microarray and immunohistochemical data for these genes was greater than or equal to 0.9. Using immunohistochemistry, sensitivity and specificity of DSG3 for lung cancers were 98% and 99%, respectively. Therefore, DSG3 can be a useful ancillary marker to separate SQCC from other subtypes of lung cancer.
Lung cancer is the number one cause of cancer-related death for both men and women worldwide. The high lethality of lung cancer is multifactorial, mainly due to diagnosis at advanced stage and lack of effective therapy. Currently, treatment strategies are predominantly guided by separating lung carcinomas into small cell lung cancer and non-small cell lung cancer. However, non-small cell lung cancer represents a heterogeneous group of cancers, with many histological subtypes. This heterogeneity is also reflected at the molecular level by differences in gene expression1,2 and are likely to affect patient’s response to therapy. The advent of novel therapies has started to impact survival.3,4,5 Indeed, Bevacizumab, a monoclonal antibody against vascular endothelial growth factor (Avastin, Genentech), in combination with a standard platin-based chemotherapy regiment has been shown to improve overall survival and delay the time to progression in patients with advanced non-small cell lung cancer, making Bevacizumab one of the few drugs to significantly impact lung cancer survival.6,7 However, a randomized phase II study conducted by Johnson et al showed some patients with squamous cell carcinoma (SQCC) experienced life-threatening pulmonary hemorrhage.8 These results have even led to the exclusion of SQCC cases from ongoing or new phase III studies.6 Therefore, distinguishing SQCC from other subtypes can be very crucial for making therapeutic decisions. In small biopsy specimens or with more poorly differentiated tumors, distinguishing SQCC from other subtypes can be challenging to the pathologist. In a study by Popp et al, the overall agreement in the final histological subtype was 88.9% for bronchial biopsy alone and for squamous cell carcinoma, 80.5%.9 Further subtyping undifferentiated tumor was considered extremely challenging, with histological agreement for “large” cell carcinoma on biopsy being only 23.1%.9
Unfortunately, only rare markers have reportedly distinguished SQCC from adenocarcinoma (AD). Most often, a combination of cytokeratin (CK) 5/6 and p63 are used. Individually, their sensitivity and specificity for SQCC has a wide range.10,11,12,13,14 A combination of these markers was tested in a small number of lung cancers (total 36 cases) and positivity for both markers was present in only 73% (11/15) of SQCCs, but also in 17% (2/12) of ADs.15 Kargi et al reported a specificity of 100% but a sensitivity of 69%.11 Including tumors from different organs, the overall specificity for the combination of the two markers was 96% but with a sensitivity of 77%.15 Therefore, there likely remains a need to identify markers that would be both more specific and sensitive for the diagnosis of SQCC of the lung.
Using a nucleotide microarray data from a previous study on SQCC16 we searched for genes that were highly expressed, specific to SQCC, and had an available commercial antibody that would permit us to perform a thorough validation study. This search identified DSG3 as a promising biomarker for the diagnosis of SQCC. The purpose of this study was first to assess the overall sensitivity and specificity of DSG3 in identifying SQCC in all organs, then more specifically in primary lung cancer. To achieve this, only specimens from surgical resection were used and tumors were well to moderately differentiated since routine H&E-stained sections were our only gold standard. We also compared the sensitivity and specificity of CK5, CK6, and p63 to DSG3 by analysis of our laser capture microdissection (LCM) derived microarray data.
Materials and Methods
Design
DSG3 was identified as a potential marker for differentiating SQCC of the lung from other subtypes by a discovery-evaluation-validation approach. Discovery was performed using a differential transcriptomic analysis of expression profiles derived from SQCC tissues from the lung, non-neoplastic lung tissues, and tissues from approximately 50 organs. We hypothesized that a specific and sensitive marker would be highly expressed in SQCC of the lung, not expressed in non-neoplastic lung, and not expressed or show low expression in tissues from other organs. As this discovery phase used data from microarray experiments and expressed sequence tags (EST), differential expression in lung (SQCC versus non-neoplastic lung tissue) was also validated by an reverse transcription (RT)-PCR assay designed to measure transcript level of DSG3. Evaluation of DSG3 as a differentiation marker for SQCC of the lung and comparison with other markers was performed at the mRNA level by analyzing microarray data. An immunohistochemical study was then performed to evaluate DSG3 at the protein level. The assay was then validated in a large set of various cancers and its performance characteristics were calculated by standard statistical analysis.
Discovery of Genes Specific to SQCC of the Lung
Highly expressed genes for SQCC of the lung were identified by analyzing gene expression data obtained from 18 SQCC samples from a previous study.16 We identified genes that had a “Present” detection call by dChip 1.3 (http://biosun1.harvard.edu/complab/dchip/) in at least 17 (94%) of these cases. Highly and differentially expressed genes were identified by further filtering this list for genes that were not expressed in non-neoplastic lung tissues assessed by “Absent” detection call in at least 8 of 9 (89%) of non-neoplastic lung tissue by dChip.
The selected genes were then assessed for organ specificity using an algorithm previously described.17 Briefly, this algorithm computes a specificity index (SPi) for a gene based on the representation of that gene in the human EST libraries. SPi had a range from 0 to 1, with 1 representing genes that are putatively highly specific to pulmonary SQCC. Because the number of ESTs from SQCC libraries was low (<10,000 sequences total) and could potentially deter the accurate estimation of expression levels of genes, the number of ESTs for each gene was adjusted proportionally to the expression level of a house keeping gene (HSP90AB1) from the microarray data in the EST libraries. We then ranked the selected genes based on the high SPi. DSG3 was one of the top five highly expressed and specific candidate genes. Since there was a high quality commercial antibody available for DSG3, we focused our investigation on the protein expression of DSG3 in SQCC.
Sample Set for mRNA Expression Level Measurement of SQCC Biomarkers
Expression levels of DSG3 transcript were measured by quantitative RT-PCR in 32 SQCC collected by LCM and 20 bulk adjacent non-neoplastic lung (Table 1). Of the 32 SQCC, 26 had sufficient RNA to also perform expression profiling studies by microarray. This set was complimented with LCM samples from 57 pulmonary AD and bulk samples from 10 of these, and 9 LCM and 6 bulk specimens of adjacent non-neoplastic tissue (Table 1).
Table 1.
Details of Case Selection for the Validation of DSG3 Expression by RT-PCR or Microarray
| RT-PCR only | RT-PCR and Microarray | Microarray only | |
|---|---|---|---|
| Squamous cell carcinoma | 6 | 26 | 18 (Bulk)16 |
| MI | 6 | 4 | |
| MIII | 0 | 7 | |
| FI | 0 | 8 | |
| FIII | 0 | 7 | |
| Adenocarcinoma | 57 (10 matching bulk) | ||
| MI | 13 | ||
| MIII | 13 | ||
| FI | 8 | ||
| FIII | 23 | ||
| Adjacent Non-neoplastic | 20 (Bulk) | 9 | |
| SQCC Adjacent | 20 (Bulk) | ||
| AD Adjacent | 9 (6 matching bulk) |
MI = male stage I; MIII = male stage III; FI = female stage I; FIII = female stage III; SQCC = squamous cell carcinoma; AD = adenocarcinoma.
All samples were collected by LCM, except as stated otherwise in the table.
One of ten μl total RNA from LCM collected SQCC or 200 ng of total RNA from non-neoplastic adjacent bulk tissues were used in Superscript III reverse transcription system (Invitrogen, Carlsbad, CA) to make cDNA. Quantitative RT-PCR experiments were done on an ABI 7900 HT system (Applied Biosystems, Foster City, CA). The optimum concentration for DSG3 and β-actin (used as normalizer gene) primer sets were determined by standard curves generated using serial dilutions of a pooled cDNA sample from the SQCC validation cohort. Forward primer (5′-TGATCTGTCCCATTTCCAGTGT-3′) and reverse primer (5′-TCATATTAGACGGGAGCAAGGA-3′) for DSG3 were used at a final concentration of 0.15 pM in the qPCR reactions.
Expression Profiling in SQCC and AD of the Lung
LCM Collection Procedure
Close to 5000 LCM pulses were used for each sample. Total RNA was isolated by PicoPure kit (Arcturus Corp, Mountain View, CA) for SQCC or by the Micropure kit (Qiagen Corp, Valencia, CA) for AD. The quality and quantity of the RNA from the LCM samples were controlled by the Agilent bioanalyzer and the Ribogreen assay (Cat # R11490, Invitrogen Corp., Carlsbad, CA) or by a quantitative PCR assay based on the ratio of concentration of 3′ to middle transcript of β-actin.18 Total RNA (10 ng) from LCM collected samples were labeled in a two round linear amplification/labeling process according to the Small Sample Preparation protocol (Affymetrix Corp, Santa Clara, CA). Affymetrix chips were scanned according to the manufacturer’s protocol.
Bulk Collection Procedure
Total RNA from the bulk tissues were isolated by the RNeasy kit (Qiagen). The quality and quantity of the RNA samples from the bulk tissues were controlled by the Agilent bioanalyzer and a NanoDrop spectrophotometer. Total RNA from bulk tissues were labeled according to the standard Affymetrix protocol.
Microarray Data Analysis
Labeled cRNA were hybridized to U133PLUS2. GC-RMA expression values (Log2 transformed) were calculated by the open source software package R (http://www.r-project.org/). To determine the detection threshold values for gene expression, we generated histograms of signal intensities for probesets corresponding to p63, CK5, CK6, and DSG3. The histogram for each of the probesets had a peak at the signal intensity around 2, which dropped to zero at signal intensity of 4 (see supplemental Figure S1 at http://ajp.amjpathol.org). Therefore, a signal intensity of 4 was selected as the detection threshold.
Immunohistochemical Study
Case selection
A retrospective search was performed using computerized records at Mayo Clinic from 1994 to 2005. Different tumors, including glioma, melanoma, mesothelioma, and carcinoma, from variable organs were identified. Archived H&E stained slides of 420 cases were retrieved and reviewed to confirm the initial diagnosis. After exclusion of cases based on diagnosis or insufficient material (only specimens obtained from surgical resection were included), the final total number of cases included was 414, from 23 different organ systems.
Immunohistochemistry
The study was done with a commercially available antibody to DSG3 (5G11 clone, ABCAM; 1/100) on representative 4-μm sections of formalin-fixed, paraffin embedded tissue. Antigen retrieval was performed by steam EDTA treatment, which consists of graded alcohol deparaffinization, methanol/hydrogen peroxide block, followed by pressure treatment in buffered EDTA at 100°C for 30 minutes. Immunostaining was performed using the Advance platform (Dako) with the Dako Autostainer (Dako, Carpintera, CA). Positive controls included normal skin and squamous cell carcinomas of the lung with known high mRNA expression of DSG-3. The negative control was a mouse IgG1 serum substitution for the primary antibody (DSG3).
Only membranous staining was considered positive and immunoreactivity was assessed according to the intensity of the stain (1+ for mild, 2+ for moderate, and 3+ for marked) and percentage of positive tumor cells, assessed from 0 to 100%, in increments of 5%. The scoring was done by two pathologists and all cases re-reviewed by both to obtain a consensual score.
Correlation between the Results of Microarray and Immunohistochemical Study
The purpose of this experiment was to determine the correlation between the RNA expression and protein expression of DSG3, CK5, CK6, and p63, to further validate our results and show their clinical validity. A subset of SQCC (18 cases) and AD (19 cases) used in our microarray study was assessed by immunohistochemistry. DSG3 immunostaining was performed and scored as describe above. The other immunohistochemical stains were performed on representative 4-μm sections of formalin-fixed, paraffin-embedded tissue from the lung tumors using antibodies to CK 5/6 (Zymed, San Francisco, CA, clone D516B4; 1:200 dilution) and p63 (Biocare Medical, Concord, CA, clone BC4A4; 1:100 dilution). Heat-induced epitope retrieval was performed in a heated 1 mmol/L EDTA pH 8.0 solution for 30 minutes. Antigen-antibody reactions were visualized using a polymer based detection system (Dako) using diaminobenzidine as the chromogen. The positive control used for both CK5/6 and p63 was normal prostatic tissue and the negative control was a mouse IgG1 serum substitution for the primary antibodies. Immunoreactivity was assessed according to the intensity of the stain (1+ for mild, 2+ for moderate, and 3+ for marked) and percentage of positive tumor cells, assessed from 0 to 100%, in increments of 5%. The log10 of the product of the intensity multiplied by the percentage of positive cells was then calculated for each marker and was used to calculate correlation coefficients with the microarray data.
Statistical Analysis
Sensitivity and specificity of DSG3, CK5, CK6, and p63 were calculated by conventional method, using pathological classification as the gold standard. Confidence interval (95%CI) of sensitivity and specificity was estimated using the single proportion mean and SE under a binomial distribution.19
This study was approved by the Institutional IRB and Biospecimen Subcommittee.
Results
DSG3 mRNA Expression in SQCC of the Lung
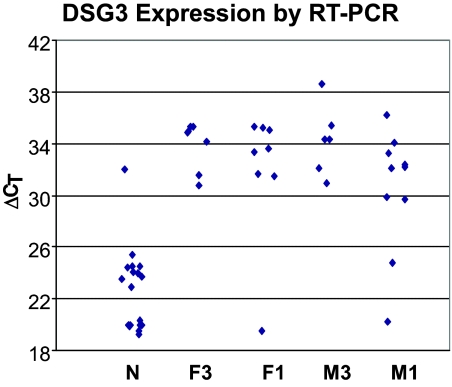
The analysis of organ/tissue distribution as determined by a analysis of public EST libraries,17 in combination with the bulk squamous microarray data,16 identified DSG3 as having moderate to high expression in 17 of 18 (94%) SQCC cases, undetected expression in 8 of 9 non-neoplastic lung tissues, and infrequent expression in other organs assessed by a SPi of greater than 0.65. High expression of DSG3 in SQCC was confirmed in the validation study by quantitative RT-PCR on independent samples that included both bulk and LCM collected tissue from both tumor and non-neoplastic cells (Figure 1 and Table 1). We observed a significant (P < 0.0001) over-expression of DSG3 in SQCC, as compared with the non-neoplastic lung.
Figure 1.
DSG3 is highly overexpressed in SQCC compared with the adjacent non-neoplastic tissue by qRT-PCR. Data points represent DSG3 expression in bulk samples from SQCC and adjacent non-neoplastic samples (N), and in LCM samples from female stage III (F3), female stage I (F1), male stage III (M3), and male stage I (M1) patients. Vertical axis represents β-actin normalized expression by qRT-PCR calculated as ΔCT = 40 − (CT−DSG3 − CT−β-actin). Each cycle difference in the y axis represents ∼twofold change. On the average, DSG3 is overexpressed in SQCC by >30-fold.
Comparison of CK5, CK6, p63, and DSG3 mRNA Expression in AD and SQCC of the Lung
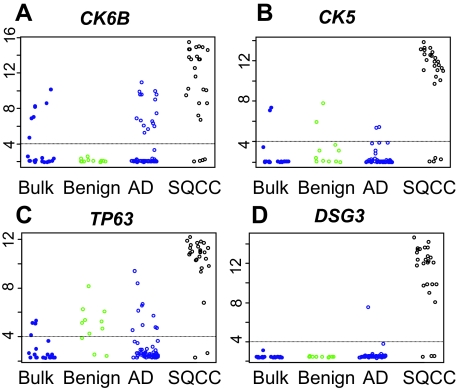
In the lung cancer LCM microarray data, we identified a high expression of DSG3 in SQCC (log2 signal intensity mean = 10.9, SD = 3.5) and a low expression of DSG3 in AD (log2 signal intensity mean = 2.6, SD = 0.7). We determined the specificity and sensitivity of p63, CK5, CK6, and DSG3 in SQCC versus AD based on the microarray data. For each gene, we identified the most sensitive probeset (the one with the largest SD across all samples) (Figure 2). We examined CK5 and CK6 because the current immunoassay presumably recognizes epitopes from both keratins. Among the different keratin 6 isoforms, CK6B produced the highest sensitivity and specificity for SQCC and was selected for analysis. Expression of CK6B was high in most SQCCs (Figure 2A), but we also detected a moderate to high expression of CK6B in 16 of 57 ADs (28%). CK5 had a more specific expression for SQCC (Figure 2B). Similarly, the expression of p63 was high in most SQCCs, but we also identified moderate to high levels of p63 in10 of 57 (18%) ADs (Figure 2C). DSG3 had a high expression (>8.0 log2 intensity) in more than 88% of the SQCCs (Figure 2D). Only one of 57 ADs (<2%) had DSG3 expression above the threshold. Therefore DSG3 alone had a better specificity for SQCCs than p63 or CK5 and a similar specificity to p63 and CK5 combined (Table 2). Combining DSG3 and p63 achieved a specificity of 100% with a sensitivity of 88%. Adding CK5 to these two markers slightly compromised sensitivity.
Figure 2.
Expression of CK6B (A), CK5 (B), p63 (C), and DSG3 (D) in the micro array data. Blue, black, and green colors represent expression in AD, SQCC, and in non-neoplastic lung adjacent to AD. The on/off threshold values for genes (horizontal lines) were found by plotting histograms of expression values and identifying the noise floor (see supplemental Figure S1 at http://ajp.amjpathol.org).
Table 2.
Estimated Performance Characteristics of p63, CK5, CK6,and DSG3 Based on Microarray Data
| Gene | Reporter | AD+N | SQCC−N | Sensitivity* % | Specificity* % |
|---|---|---|---|---|---|
| CK6B | 209126_x_at | 16 | 4 | 85 | 72 |
| CK5 | 201820_at | 2 | 4 | 85 | 96 |
| p63 | 209863_s_at | 10 | 2 | 92 | 82 |
| DSG3 | 235075_at | 1 | 3 | 88 | 98 |
| p63 and CK5 | MIN(209863_s_at, 201820_at) | 4 | 1 | 85 | 98 |
| DSG3 and p63 | MIN(235075_at, 209863_s_at) | 3 | 0 | 88 | 100 |
| DSG3 and CK5 | MIN(235075_at, 201820_at) | 2 | 3 | 92 | 95 |
| DSG3, p63 and CK5 | MIN(235075_at, 209863_s_at, 201820_at) | 5 | 0 | 81 | 100 |
AD+ = all AD cases with gene expression levels above the noise threshold (signal intensity ≥4) out of a total of 57 (supplemental Figure S1 at http://ajp.amjpathol.org).
SQCC− = all SQCC cases with gene expression levels below the noise threshold (signal intensity <4) out of a total of 26.
Sensitivity and specificity were calculated based on the expression in AD and SQCC samples.
To confirm the clinical relevance of these findings, the microarray results were compared with the immunohistochemical results in a subset of these cases. The correlation coefficient for DSG3 and p63 was 0.94 and 0.90, respectively (see supplemental Figure S2 at http://ajp.amjpathol.org). Interestingly, the results for CK5/6 antibody showed a correlation coefficient of 0.9, with microarray signal intensities for CK5 and only 0.75 with CK6B. Since the correlation between CK5 expression in the microarray data and the CK5/6 antibody immunostains was much higher than with CK6B, only the CK5 microarray data were used to estimate the specificity and sensitivity for SQCCs (Table 2). Any cross reactivity with CK6B was ignored in these estimates.
DSG3 Expression in SQCC by Immunostaining
Results of the immunohistochemical study are detailed in Tables 3 and 4. Of 166 SQCCs, 164 (98.8%) expressed DSG3 (Table 5). Immunostaining was membranous and generally diffuse with intensities of 2 to 3 + (Figure 3A–C). Only 33 (13.3%) of non-SQCC cases were positive. These cases included 17 AD from various organs [pancreas (8), colon (5), lung (1), breast (1), stomach (1), and ovary (1)], seven adenoid cystic carcinomas, four mucoepidermoid carcinomas, three urothelial carcinomas, and two thymomas. However, the positive breast carcinoma was a basaloid type cancer and showed distinct squamous differentiation by light microscopy. Immunoreactivity in all mucoepidermoid carcinomas was limited to the areas of squamous differentiation. The three urothelial carcinomas had extensive squamous differentiation and one was re-classified as squamous cell carcinoma of the bladder. In adenocarcinomas, the expression of DSG3 when present was focal (Table 4). Interestingly, in the pancreas, DSG3 expression was present in normal ductal epithelium and expression of DSG3 was focal and weak to moderate (1 to 2+) in ductal types of adenocarcinoma.
Table 3.
Detailed Results of DSG3 Expression in Different Tumor Types and Organs
| Organ | Tumor type | Number of cases | Immunopositive | % |
|---|---|---|---|---|
| Lung | SQCC | 65 | 64 | 98.5 |
| Lung | AD | 47 | 1 | 2.1 |
| Lung | Large cell carcinoma | 36 | 0 | 0 |
| Lung | SCLC | 9 | 0 | 0 |
| Lung | Carcinoid | 10 | 0 | 0 |
| Pleura | Mesothelioma | 8 | 0 | 0 |
| Lymph node | Metastatic SQCC | 20 | 19 | 95 |
| Brain | Astrocytoma | 9 | 0 | 0 |
| Brain | Oligodendroglioma | 9 | 0 | 0 |
| Skin | SQCC | 12 | 12 | 100 |
| Skin | Malignant melanoma | 10 | 0 | 0 |
| Head-Neck | SQCC | 5 | 5 | 100 |
| Head-Neck | Mucoepidermoid ca | 4 | 4 | 100 |
| Head-Neck | Adenoid cystic ca | 10 | 7 | 70 |
| Breast | AD | 7 | 1 | 14.3 |
| Tonsil | SQCC | 10 | 10 | 100 |
| Tongue | SQCC | 8 | 8 | 100 |
| Larynx | SQCC | 9 | 9 | 100 |
| Thymus | Thymoma | 10 | 2 | 20 |
| Esophagus | SQCC | 15 | 15 | 100 |
| Stomach | AD | 10 | 1 | 10 |
| Pancreas | AD | 10 | 8 | 80 |
| Colon | AD | 9 | 5 | 55.6 |
| Anus | SQCC | 4 | 4 | 100 |
| Liver | Hepatocellular ca | 9 | 0 | 0 |
| Bladder | Urothelial ca | 10 | 3 | 30 |
| Prostate | Adenocarcinoma | 9 | 0 | 0 |
| Vulva | SQCC | 9 | 9 | 100 |
| Cervix | SQCC | 9 | 9 | 100 |
| Ovary | Adenocarcinoma | 10 | 1 | 10 |
| Testis | Seminoma | 10 | 0 | 0 |
| Testis | Embryonal carcinoma | 2 | 0 | 0 |
| TOTAL | 414 | 197 | ||
AD = adenocarcinoma; SQCC = squamous cell carcinoma; SCLC = small cell carcinoma; ca = carcinoma.
Table 4.
Detailed Expression of DSG3 in All Positive Cases
| Organ | Tumor type | DSG3 (+) (N) | Positive cells (%)
|
|
|---|---|---|---|---|
| Mean | Range | |||
| Lung | SQCC | 64 | 59.8 | 5–100 |
| Lung | AD | 1 | <5 | 5 |
| Lymph node | SQCC | 19 | 80.4 | 5–100 |
| Skin | SQCC | 12 | 91.8 | 70–100 |
| Head-Neck | SQCC | 5 | 93.6 | 90–98 |
| Head-Neck | Mucoepidermoid ca | 4 | 87.5 | 30–85 |
| Head-Neck | Adenoid cystic ca | 7 | 17.8 | 5–60 |
| Breast | AD | 1 | 70 | 70 |
| Tonsil | SQCC | 10 | 74.4 | 5–100 |
| Tongue | SQCC | 8 | 85.6 | 30–100 |
| Larynx | SQCC | 9 | 86.9 | 30–100 |
| Thymus | Thymoma | 2 | 25 | 10–40 |
| Esophagus | SQCC | 15 | 59.7 | 5–100 |
| Stomach | AD | 1 | 10 | 10 |
| Pancreas | AD | 8 | 16.8 | 5–75 |
| Colon | AD | 5 | 24 | 5–40 |
| Anus | SQCC | 4 | 82 | 20–98 |
| Bladder | Transitional cell ca | 3 | 58.3 | 5–90 |
| Vulva | SQCC | 9 | 81.4 | 10–100 |
| Cervix | SQCC | 9 | 75.6 | 60–100 |
| Ovary | AD | 1 | <5 | 5 |
SQCC = squamous cell carcinoma; AD = adenocarcinoma; ca = carcinoma.
Table 5.
Summary of Results of DSG3 Expression for All Tumors
| SQCC | Non-SQCC | Total | |
|---|---|---|---|
| DSG3-positive | 164 | 33 | 197 |
| DSG3-negative | 2 | 215 | 217 |
| Total | 166 | 248 | 414 |
Sensitivity = 99% (95% CI: 97.5% to 100%); Specificity = 87% (95% CI: 82.8% to 91.2%), SQCC = squamous cell carcinoma.
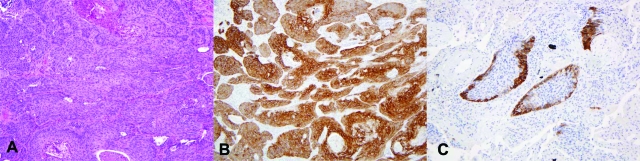
Figure 3.
Expression of DSG3 in squamous cell carcinoma. Example of squamous cell carcinoma of the lung (A) showing strong and diffuse membranous staining with DSG3. Similar staining is seen in the majority of squamous cell carcinomas (B). Some squamous cell carcinomas show focal staining (C). A: H&E, ×100; B: DSG3, ×100; C: DSG3, ×200.
In the lung, 64 out of 65 (98.5%) cases of SQCC were positive for DSG3, whereas only 1 out of 48 (2%) ADs was focally positive (Figure 4A–C). Large cell carcinomas, small cell lung cancer, carcinoid tumors, and malignant mesotheliomas were all negative. Therefore specificity and sensitivity were calculated as 99% (95% CI: 95.6% to 100%) and 98% (95% CI: 97.0% to 100%) (Table 6).
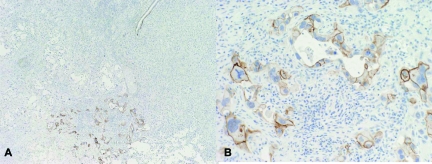
Figure 4.
Expression of DSG3 in pulmonary adenocarcinoma. A single pulmonary adenocarcinoma showed focal moderate staining with DSG3. A: DSG3, ×40; B: DSG3, ×200.
Table 6.
Summary of Results of DSG3 for Lung Tumors
| SQCC of lung | Non-SQCC lung tumors | Total | |
|---|---|---|---|
| DSG3-positive | 64 | 1 | 65 |
| DSG3-negative | 1 | 91 | 92 |
| Total | 65 | 92 | 157 |
Sensitivity = 98% (95% CI: 95.6% to 100%); Specificity = 99% (95% CI: 97.0% to 100%), SQCC = squamous cell carcinoma.
Discussion
In our study, DSG3 appears to be a sensitive and specific marker for SQCC, particularly for the lung. In primary tumors from different sites, DSG3 staining had a sensitivity of 99% and a specificity of 87% for SQCC. Only rare non-SQCCs expressed DSG-3, most of which showed areas of squamous differentiation, with the exception of ADs of the colon and pancreas. Although the expression in these tumors was usually focal, more diffuse staining was seen in rare cases. Interestingly, in the pancreas, DSG3 expression was present in the normal basal layer of the ducts and perhaps explains its expression within ductal AD.
In primary lung tumors, DSG3 expression remained very sensitive and specific. Staining was positive in 98% of SQCCs and negative in 99% of non-SQCC cases. This sensitivity and specificity is overall better than what has been previously reported for CK 5/6 and p63.12,15,20,21 Indeed, reported sensitivity and specificity values for p63 and CK 5/6 have a considerably large range (Table 7). For p63, reported sensitivity ranges anywhere between 78 to 100% and specificity from 35% to 100%. In the two studies with the largest number of lung cancers, the reported sensitivity and specificity are lower than for DSG3 in this study.20,21
Table 7.
Calculated Sensitivity and Specificity of CK5/6 and p63 Based on the Expression in Lung Adenocarcinoma and Squamous Cell Carcinomas
| Study | AD/SQCC (n) | Sensitivity (%)
|
Specificity (%)
|
||
|---|---|---|---|---|---|
| CK5/6 | p63 | CK5/6 | p63 | ||
| Pelosi et al21 | 95/118 | na | 92 | na | 84 |
| Wang et al14 | 23/30 | na | 100 | na | 35 |
| Kaufmann et al15 | 12/15 | 93 | 80 | 67 | 58 |
| Camilo et al10 | 17/18 | 47 | 78 | 44 | 100 |
| Pu et al13 | 10/15 | 100 | 80 | 90 | 70 |
| Kargi et al11 | 10/39 | 79 | 82 | 80 | 100 |
| Wu et al29 | 0/13 | na | 100 | na | na |
| Li et al12 | 9/12 | 89 | na | 92 | na |
| Au et al20 | 93/123 | na | 96 | na | 70 |
na = not assessable.
Minimum and maximum specificity values for p63 and CK5/6 are shown in bold.
When combining CK5/6 and p63, the study by Kaufman et al showed an increased specificity of 96%, but the sensitivity was only 77%.15 Increasing the specificity to 99% resulted in a further drop of the sensitivity to 66%. However, these values were calculated on cancers from all organs, not only for lung. Kargi et al focused on lung cancers and reported a specificity of 100% and sensitivity of 82% but their sample size, compared with our study, was small including only 39 SQCC and 10 ADs.11 Furthermore, they did not include subtypes such as large cell carcinoma.
By quantitative assays that measured mRNA, we also observed a very high expression of DSG3 in SQCCs and mostly absent DSG3 expression in ADs and non-neoplastic lung. These findings are supported by expression profiling studies reported by Inamura et al.22 Our microarray data further supported a higher specificity of DSG3 compared with either CK5 or p63. Only when combining both, did CK5 and p63 achieve a comparable specificity. However, combining DSG3 and p63 achieved the best result, increasing the specificity to 100% without losing sensitivity, which remained at 88%. These results are better than those reported for the combination of CK5/6 and p63 in the diagnosis of SQCC.
In a recent study, Fukuoka et al23 suggested that DSG3 was a prognostic marker. Their survival analysis indicated that positive DSG3 staining was significantly correlated with a favorable prognosis in both non-small cell lung cancer and carcinoid tumors. However, DSG3 did not appear as sensitive and specific. Indeed, DSG3 was found to be expressed in 79.5% of SQCCs and 54.8% of ADs. The difference in sensitivity of SQCCs compared with our study could be explained by their tissue microarray methodology, which used only one core of tumor. Our results showed that DSG3 staining can be heterogeneous, seen in as little as 5% of cells. However, the high DSG3 expression in AD is more difficult to reconcile and differs from several published oligonucleotide microarray data22,24,25 and our LCM-derived microarray data on lung. Furthermore expression of DSG3 in ADs would not be predicted based by its biological role. Indeed, desmogleins are one of the major glycoproteins of the desmosomal structure. They are calcium-dependent adhesion molecules and belong to cadherin superfamily, which link to cytokeratins via desmoplakins and plakoglobin. Therefore, their expression is membranous and not cytoplasmic. Furthermore, DSG3 in particular, has an important role in cellular adhesion of stratified epithelia, such as squamous epithelium.26,27,28
The limitations to our study are that DSG3 expression by immunohistochemistry was assessed only on large surgical specimens, and future studies looking at specificity and sensitivity of DSG3, with further comparison with CK5/6 and p63, on small biopsy specimens need to be done. Also, tumors included in our series were at least moderately differentiated, to guarantee diagnostic accuracy and provide a gold standard to assess specificity and sensitivity of DSG3. Although none of our large cell carcinomas, which epitomize undifferentiated cancers, expressed DSG3 by immunohistochemistry (as well as by mRNA level, data not shown), expression of DSG3 in poorly differentiated cancers will also need to be further assessed and also contrasted to CK 5/6 and p63 expression. Also, our microarray data suggests the possibility that other markers may play an important role in tumor differentiation and will require further studying.
In summary, DSG3 is a promising diagnostic marker to distinguish SQCC from other subtypes of lung cancers.
Footnotes
Address reprint requests to Marie Christine Aubry, M.D., Department of Laboratory Medicine and Pathology, Mayo Clinic, 200 First St. SW, Rochester, MN 55905. E-mail: aubry.mariechristine@mayo.edu.
Supported in part by NIH-R01 grants to P.Y. (CA80127 and CA84354).
C.D.S.-H. and F.K. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol. 2004;31:4–19. doi: 10.1053/j.seminoncol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, West M, Harpole DH, Jr, Nevins JR. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- Gatzemeier U. Targeting the HER1/EGFR receptor to improve outcomes in non-small-cell lung cancer. Oncology (Williston Park) 2003;17:7–10. [PubMed] [Google Scholar]
- Schallier D, Neyns B, Fontaine C, Steene JV, De Mey J, Meysman M, De Greve J. A novel triplet regimen with paclitaxel, carboplatin and gemcitabine (PACCAGE) as induction chemotherapy for locally advanced unresectable non small cell lung cancer (NSCLC). Lung Cancer. 2007;56:247–254. doi: 10.1016/j.lungcan.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Tiseo M, Loprevite M, Ardizzoni A. Epidermal growth factor receptor inhibitors: a new prospective in the treatment of lung cancer. Curr Med Chem Anticancer Agents. 2004;4:139–148. doi: 10.2174/1568011043482106. [DOI] [PubMed] [Google Scholar]
- Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- Sandler A, Herbst R. Combining targeted agents: blocking the epidermal growth factor and vascular endothelial growth factor pathways. Clin Cancer Res. 2006;12:4421s–4425s. doi: 10.1158/1078-0432.CCR-06-0796. [DOI] [PubMed] [Google Scholar]
- Johnson DH, Fehrenbacher L, Novotny WF, Herbst RS, Nemunaitis JJ, Jablons DM, Langer CJ, DeVore RF, 3rd, Gaudreault J, Damico LA, Holmgren E, Kabbinavar F. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 2004;22:2184–2191. doi: 10.1200/JCO.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Popp W, Rauscher H, Ritschka L, Redtenbacher S, Zwick H, Dutz W. Diagnostic sensitivity of different techniques in the diagnosis of lung tumors with the flexible fiberoptic bronchoscope. Comparison of brush biopsy, imprint cytology of forceps biopsy, and histology of forceps biopsy. Cancer. 1991;67:72–75. doi: 10.1002/1097-0142(19910101)67:1<72::aid-cncr2820670114>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Camilo R, Capelozzi VL, Siqueira SA, Del Carlo Bernardi F. Expression of p63, keratin 5/6, keratin 7, and surfactant-A in non-small cell lung carcinomas. Hum Pathol. 2006;37:542–546. doi: 10.1016/j.humpath.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Kargi A, Gurel D, Tuna B. The diagnostic value of TTF-1. CK 5/6, and p63 immunostaining in classification of lung carcinomas. Appl Immunohistochem Mol Morphol. 2007;15:415–420. doi: 10.1097/PAI.0b013e31802fab75. [DOI] [PubMed] [Google Scholar]
- Li Q, Bavikatty N, Michael CW. The role of immunohistochemistry in distinguishing squamous cell carcinoma from mesothelioma and adenocarcinoma in pleural effusion. Semin Diagn Pathol. 2006;23:15–19. doi: 10.1053/j.semdp.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Pu RT, Pang Y, Michael CW. Utility of WT-1, p63. MOC31, mesothelin, and cytokeratin (K903 and CK5/6) immunostains in differentiating adenocarcinoma, squamous cell carcinoma, and malignant mesothelioma in effusions. Diagn Cytopathol. 2008;36:20–25. doi: 10.1002/dc.20747. [DOI] [PubMed] [Google Scholar]
- Wang BY, Gil J, Kaufman D, Gan L, Kohtz DS, Burstein DE. P63 in pulmonary epithelium, pulmonary squamous neoplasms, and other pulmonary tumors. Hum Pathol. 2002;33:921–926. doi: 10.1053/hupa.2002.126878. [DOI] [PubMed] [Google Scholar]
- Kaufmann O, Fietze E, Mengs J, Dietel M. Value of p63 and cytokeratin 5/6 as immunohistochemical markers for the differential diagnosis of poorly differentiated and undifferentiated carcinomas. Am J Clin Pathol. 2001;116:823–830. doi: 10.1309/21TW-2NDG-JRK4-PFJX. [DOI] [PubMed] [Google Scholar]
- Sun Z, Yang P, Aubry MC, Kosari F, Endo C, Molina J, Vasmatzis G. Can gene expression profiling predict survival for patients with squamous cell carcinoma of the lung? Mol Cancer. 2004;3:35. doi: 10.1186/1476-4598-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasmatzis G, Klee EW, Kube DM, Therneau TM, Kosari F. Quantitating tissue specificity of human genes to facilitate biomarker discovery. Bioinformatics. 2007;23:1348–1355. doi: 10.1093/bioinformatics/btm102. [DOI] [PubMed] [Google Scholar]
- Kube DM, Savci-Heijink CD, Lamblin AF, Kosari F, Vasmatzis G, Cheville JC, Connelly DP, Klee GG. Optimization of laser capture microdissection and RNA amplification for gene expression profiling of prostate cancer. BMC Mol Biol. 2007;8:25. doi: 10.1186/1471-2199-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood BR, Sterne JAC, Kirkwood BR. Malden, Mass: Blackwell Science; Essential medical statistics. 1987:pp 83–84, 163–164. [Google Scholar]
- Au NH, Gown AM, Cheang M, Huntsman D, Yorida E, Elliott WM, Flint J, English J, Gilks CB, Grimes HL. P63 expression in lung carcinoma: a tissue microarray study of 408 cases. Appl Immunohistochem Mol Morphol. 2004;12:240–247. doi: 10.1097/00129039-200409000-00010. [DOI] [PubMed] [Google Scholar]
- Pelosi G, Pasini F, Olsen Stenholm C, Pastorino U, Maisonneuve P, Sonzogni A, Maffini F, Pruneri G, Fraggetta F, Cavallon A, Roz E, Iannucci A, Bresaola E, Viale G. p63 immunoreactivity in lung cancer: yet another player in the development of squamous cell carcinomas? J Pathol. 2002;198:100–109. doi: 10.1002/path.1166. [DOI] [PubMed] [Google Scholar]
- Inamura K, Fujiwara T, Hoshida Y, Isagawa T, Jones MH, Virtanen C, Shimane M, Satoh Y, Okumura S, Nakagawa K, Tsuchiya E, Ishikawa S, Aburatani H, Nomura H, Ishikawa Y. Two subclasses of lung squamous cell carcinoma with different gene expression profiles and prognosis identified by hierarchical clustering and non-negative matrix factorization. Oncogene. 2005;24:7105–7113. doi: 10.1038/sj.onc.1208858. [DOI] [PubMed] [Google Scholar]
- Fukuoka J, Dracheva T, Shih JH, Hewitt SM, Fujii T, Kishor A, Mann F, Shilo K, Franks TJ, Travis WD, Jen J. Desmoglein 3 as a prognostic factor in lung cancer. Hum Pathol. 2007;38:276–283. doi: 10.1016/j.humpath.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Chen Y, Pietas A, Schluns K, Pacyna-Gengelbach M, Deutschmann N, Padilla-Nash HM, Ried T, Petersen I. Gene expression profiles in human non-small and small-cell lung cancers. Eur J Cancer. 2003;39:1936–1947. doi: 10.1016/s0959-8049(03)00419-2. [DOI] [PubMed] [Google Scholar]
- Kettunen E, Anttila S, Seppanen JK, Karjalainen A, Edgren H, Lindstrom I, Salovaara R, Nissen AM, Salo J, Mattson K, Hollmen J, Knuutila S, Wikman H. Differentially expressed genes in non small cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149:98–106. doi: 10.1016/S0165-4608(03)00300-5. [DOI] [PubMed] [Google Scholar]
- Buxton RS, Magee AI. Structure and interactions of desmosomal and other cadherins. Semin Cell Biol. 1992;3:157–167. doi: 10.1016/s1043-4682(10)80012-1. [DOI] [PubMed] [Google Scholar]
- Merritt AJ, Berika MY, Zhai W, Kirk SE, Ji B, Hardman MJ, Garrod DR. Suprabasal desmoglein 3 expression in the epidermis of transgenic mice results in hyperproliferation and abnormal differentiation. Mol Cell Biol. 2002;22:5846–5858. doi: 10.1128/MCB.22.16.5846-5858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North AJ, Bardsley WG, Hyam J, Bornslaeger EA, Cordingley HC, Trinnaman B, Hatzfeld M, Green KJ, Magee AI, Garrod DR. Molecular map of the desmosomal plaque. J Cell Sci. 1999;112:4325–4336. doi: 10.1242/jcs.112.23.4325. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang B, Gil J, Sabo E, Miller L, Gan L, Burstein DE. p63 and TTF-1 immunostaining. A useful marker panel for distinguishing small cell carcinoma of lung from poorly differentiated squamous cell carcinoma of lung. Am J Clin Pathol. 2003;119:696–702. doi: 10.1309/P5AB-R5KQ-89RN-JTFH. [DOI] [PubMed] [Google Scholar]