Abstract
Infantile hemangiomas are characterized by rapid capillary growth during the first year of life followed by involution during early childhood. The natural history of these lesions creates a unique opportunity to study the changes in gene expression that occur in the vessels of these tumors as they proliferate and regress. Here we use laser capture microdissection and genome-wide transcriptional profiling of vessels from proliferating and involuting hemangiomas to identify differentially expressed genes. Relative to normal placental vessels, proliferating hemangiomas were characterized by increased expression of genes involved in endothelial-pericyte interactions, such as angiopoietin-2 (ANGPT2), jagged-1 (JAG1), and notch-4 (NOTCH4), as well as genes involved in neural and vascular patterning, such as neuropilin-2 (NETO2), a plexin domain containing receptor (plexinC1), and an ephrin receptor (EPHB3). Insulin-like growth factor binding protein-3 (IGFBP3) was down-regulated in proliferating hemangiomas. Involuting hemangiomas were characterized by the expression of chronic inflammatory mediators, such as the chemokine, stromal cell-derived factor-1 (SDF-1), and factors that may attenuate the angiogenic response, such as a member of the Down syndrome critical region (DSCR) family. The identification of genes differentially expressed in proliferating and involuting hemangiomas in vivo will contribute to our understanding of this vascular lesion, which remains a leading cause of morbidity in newborn children.
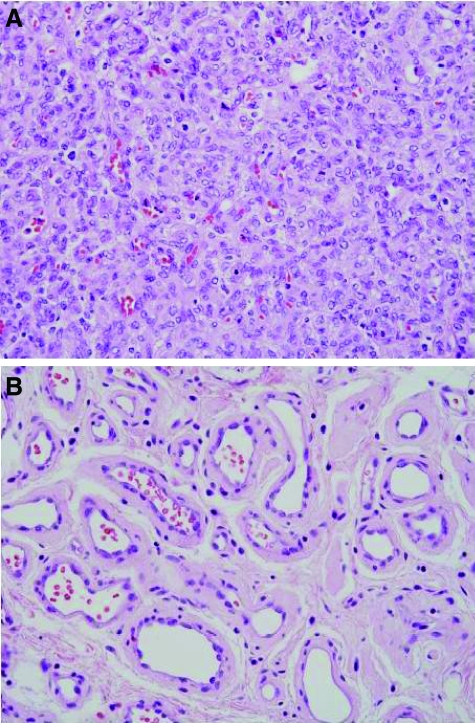
Infantile hemangiomas are extremely common tumors, affecting ∼4 to 10% of all infants.1,2,3 The lesions have unique growth characteristics, typically being absent or barely noticeable at birth, followed by a period of rapid growth and subsequent involution. The proliferating phase is characterized by abundant immature endothelial cells and adjacent pericyte-like cells (Figure 1A), whereas the involuting phase is characterized by fewer and larger capillary-like vessels surrounded by connective tissue (Figure 1B).
Figure 1.
Proliferating and involuting hemangiomas. A: Representative image of vessels in a proliferating hemangioma captured by LCM. B: Representative image of vessels in an involuting hemangioma captured by LCM.
Recent studies provide some insights into the pathogenesis of these vascular tumors.4 First, hemangiomas are clonal lesions. Evidence of nonrandom X inactivation in endothelial cells was obtained from endothelial cells cultured from infantile hemangiomas.5 Similarly, evidence of clonality was seen in cells obtained directly from proliferating phase infantile hemangiomas.6 Although the vast majority of infantile hemangiomas occur sporadically,7 several kindreds have been identified to segregate as an autosomal dominant trait with high penetrance. Fibroblast growth factor receptor-4 (FGFR4), platelet-derived growth factor receptor-β (PDGFRB), and Flt-4 have been suggested as candidate genes for familial infantile hemangioma.8 Also, somatic missense mutations in the kinase insert of the vascular endothelial growth factor receptor-2 (VEGFR2 or FLK1/KDR) gene and the VEGFR3 (FLT4) gene have been found.6 Recently, in some individuals with infantile hemangioma, germline mutations have been identified in the genes encoding VEGFR2 (KDR) and TEM8 (ANTXR1). These mutations in infantile hemangioma result in low VEGFR1 expression, VEGF-dependent activation of VEGFR2 and its downstream signaling pathways, and endothelial proliferation.9
Second, endothelial progenitor cells, in small numbers, are found in proliferating hemangiomas. Early proliferating hemangiomas contain endothelial cells that express CD133 and CD34, markers for endothelial precursor cells, suggesting that the endothelial cells in proliferating hemangiomas may be arrested at an immature stage of vascular development.10,11
Third, some aspects of hemangioma resemble that seen in the placenta. An unexpected set of tissue-specific markers co-expressed by both infantile hemangiomas at all stages of their evolution and placental vessels has suggested that the vasculature of the placenta and hemangioma may share a common cellular origin.12,13
Despite the number of studies on proliferating hemangioma, there is little information on the molecular mechanisms of involution. Once hemangiomas reach their maximum size, they spontaneously regress in a process that is characterized by endothelial apoptosis,14,15 dilation of vascular lumina, flattening of endothelial cells, loss of mitotic activity, thickening of basement membranes, and drop-out of lesional capillaries (Figure 1B). The maturation process results in an involuting hemangioma that consists of large capillary-like vessels dispersed in fibroadipose tissue. Mesenchymal stem cells present in the tumor may give rise to the adipocyte-like cells found in the involuting phase hemangioma.16
The natural evolution of infantile hemangiomas creates a unique opportunity to study the changes in gene expression that take place as the vessels of these tumors proliferate and then regress. Initial expression analysis has been used to try and identify potential regulators of hemangioma progression. These studies identified factors such as insulin-like growth factor-2 (IGF-2) and several interferon inducible genes as potentially important regulators of hemangioma growth and involution.17,18 Additionally, molecular profiling suggested a placental origin for infantile hemangioma.19 However, none of these studies used genome wide arrays and typically involved preparing transcript from random samples of the resected tumor. To more precisely define patterns of gene expression in proliferating and involuting hemangiomas, we used laser capture microdissection (LCM) to selectively isolate the lesional vessels and avoid missing molecular events that are masked by studying random samples from the resected specimen.20 The gene profiling approach with LCM-captured vascular cells provided new insights into some of the signaling pathways that are associated with proliferating and involuting hemangiomas.
Materials and Methods
RNA Quality Assessment
Keeping in mind the age of the patient, cases were selected such that identification of genes involved in the early proliferative and early- and mid-involutive phases would be enriched, in an attempt not to miss molecular events. Infants with proliferating hemangioma were under the age of 12 months and children with involuting hemangioma were 2 to 7 years. To determine the pattern of gene expression in the hemangiomas, blocks containing formalin-fixed, paraffin-embedded lesions were screened for useable RNA. In an initial quality control step, blocks were screened for RNA quantity and quality using a quantitative real-time polymerase chain reaction (PCR) assay with primers designed to two amplicons on the β-actin gene, following the manufacturer’s recommended protocol (Molecular Devices, Mountain View, CA). The 3′ amplicon is designed from a region ∼100 nucleotides and the 5′ amplicon ∼400 nucleotides from the poly A tail, respectively. The assumption in this screen is that the 3′ amplicon represents an indication of RNA quantity and a ratio of 3′:5′ amplicons represents a qualitative indicator of transcript length and transcribability. The samples represent the average status (i.e., length and transcribability) of other RNA molecules in the same block. This assay was performed on a Mx3000p real-time PCR thermal cycler (Stratagene, La Jolla, CA) using the Brilliant SYBR Green QPCR Master Mix (Stratagene). Dilutions of Human Universal RNA (Stratagene) are run from which a standard curve is used to estimate the amount of RNA in the sample. The assay measures the average β-actin cDNA length by quantification of the PCR product yield from the 3′ end and compares this yield to a relatively 5′ sequence. The following primer sequences are used: 3′ primers: HBAC1650: 5′-TCCCCCAACTTGAGATGTATGAAG-3′, HBAC1717: 5′-AACTGGTCTCAAGTCAGTGTACAGG-3′, and HBAC1355: 5′-ATCCCCCAAAGTTCACAATG-3′, and HBAC1472: 5′-GTGGCTTTAGGATGGCAAG-3′. cDNA generated from the uRNA was serially diluted with polyI (Sigma-Aldrich, St. Louis, MO) yielding a standard curve consisting of four standards: 100 ng, 10 ng, 1 ng, and 0.1 ng. cDNA generated during the first strand synthesis reaction served as the 100-ng standard. The standard curve of the 3′ primer set (HBAC1650, HBAC1717) was used to estimate the quantity of RNA in the sample. The ratio of the RNA yield obtained from both sets of PCR primers is the 3′/5′ ratio and was used as an indication of RNA quality. For example, if the cDNA contains both the 3′ and 5′ target sequences, the 3′/5′ ratio would be ∼1. Cross-linked or modified RNAs would have a ratio greater than 1. Based on this ratio, an estimation of the quality of RNA was made. Lesions from which RNAs yielded acceptable 3′ to 5′ ratios were selected for microdissection.
Laser Capture Microdissection
Three blocks each (proliferating hemangioma and normal term placenta) and four blocks of involuting hemangioma were identified with useable RNA in the quality control screen. Multiple, serially sectioned formalin-fixed, paraffin-embedded sections were cut from the blocks at 7 μ thickness, deparaffinized, stained and dehydrated following the manufacturer’s protocol (Paradise reagent system Kit, Molecular Devices). Lesional tissue was microdissected from PEN membrane glass slides (Molecular Devices) using both the UV cutting and IR capture lasers on the Veritas microdissection instrument (Molecular Devices). All attempts were made to capture only lesional vessels so that connective tissue including adipose tissue would be excluded. It was inevitable; however, that intraluminal white cells, some perivascular collagen, and occasional resident cells such as fibroblasts and mast cells would have been included in the samples. Still, these would have made a very minor contribution to the sample and resultant data.
After microdissection, RNA was extracted from the captured cells on the cap by incubating dissected tissue in the RNA extraction buffer (Paradise reagent system kit, Molecular Devices) overnight at 50°C. RNA isolation was performed following the manufacturer’s protocol using the MiraCol purification column as part of the Paradise reagent system kit. Samples were eluted in 12 μl of elution buffer.
RNA Amplification, Hybridization, and Array Analysis
RNA was amplified according to the manufacturer’s suggested protocol (Paradise reagent system kit). Briefly, first-strand synthesis was performed on each RNA yielding cDNAs that incorporate a single-stranded T7 promoter sequence. cDNAs generated in a second-strand synthesis reaction using exogenous primers were purified using the MiraCol purification column provided in the paradise reagent system kit. Amplified and purified cDNAs were submitted to the Partners Health Care Center for Personalized Genetic Medicine (Cambridge, MA) for labeling, quantification, and hybridization.
Samples were labeled with biotinylated probes using the Bioarray high-yield transcription kit following the manufacturer’s protocol (Enzo Biochemical, New York, NY). The concentration of the biotin-labeled cRNA was determined by UV absorbance using a Bio-Tek plate reader (Bio-Tek Instruments, Winooski, VT). In all cases, 20 μg of each biotinylated cRNA preparation was fragmented, assessed by gel electrophoresis, and placed in a hybridization cocktail containing hybridization controls as recommended by the manufacturer. Samples were then hybridized to the Affymetrix Human X3P GeneChip Array (Affymetrix Inc., Santa Clara, CA) at 45°C for 24 hours. Microarrays were washed and stained using the manufacturer’s protocol for the Human X3P GeneChip Array on a Model 450 Fluidics station (Affymetrix Inc.). The Fluidics station process is controlled by the Affymetrix GeneChip Operating System (GCOS).
The Affymetrix Human X3P GeneChip Array is designed for whole-genome expression profiling of RNA from formalin-fixed, paraffin-embedded samples. The target sequences on the X3P array are identical to those used for designing the Human Genome U133 Plus 2.0 GeneChip array, for a total of 47,000 transcripts with 61,000 probe sets, although the probes on the two types of arrays are significantly different. The probe selection region on the X3P array is restricted to the 300 bp at the most 3′ end of the transcripts. In contrast, the standard Affymetrix design selects probe sets within the region 600 bases proximal to the 3′ ends (Affymetrix Inc.). Images from the scanned chips were processed using an Affymetrix Model 7000 scanner with autoloader (Affymetrix Inc.). The Affymetrix GCOS v1.3 operating system (Affymetrix Inc.) controls the Model 7000 scanner and data acquisition functions. Image files were downloaded, imported, and analyzed using the GeneSifter statistical software package (Geospiza, Inc., Seattle, WA).
Statistical Analysis
Statistical t-test analysis was performed in which a pairwise comparison was made between the proliferating hemangioma versus placenta, proliferating hemangioma versus involuting hemangioma, and involuting hemangioma versus placenta (see Supplemental Tables S1, S2, and S3 at http://ajp.amjpathol.org). Data were normalized to the mean. Results were filtered more stringently by imposing a threshold cutoff of 3.0 or greater fold change in expression and a quality call of 1 (P) in all three replicates of at least one group. False-positives were reduced by applying the Benjamini and Hochberg correction coefficient. By plotting the relative intensities of each of the genes from both samples on the same graph, it is possible to directly compare the data (see Supplemental Figure S1 at http://ajp.amjpathol.org). Their similarity (mirror images) suggests that the procedure is reproducible. For example, box plots of the range of intensities of the two samples also show that the data from the two samples is comparable (data not shown). Pearson’s correlation coefficients were calculated from the array data as previously described (see Supplemental Figure S2 at http://ajp.amjpathol.org).
A one-way analysis of variance was performed on the three groups (proliferating hemangioma, involuting hemangioma, and placenta). See Supplemental Table S4 at http://ajp.amjpathol.org. Data were normalized to the mean, then filtered, by imposing a threefold threshold cutoff, a quality call of 1, and application of the Benjamini and Hochberg correction coefficient.
GeneSifter uses Gene Ontology (GO) reports and z-scores to summarize the biological processes, molecular functions, or cellular components, as well as, the KEGG (Kyto Encyclopedia of Genes and Genomes) pathways21 associated with a gene list. The z-score is calculated by subtracting the expected number of genes in a GO term meeting the criterion from the observed number of genes and dividing by the SD of the observed number of genes.22 z-scores can then be used to identify GO terms that are significantly over- or under represented in a gene list.
Immunohistochemistry
Expression of proteins generated by some of the differentially regulated genes was validated by immunohistochemical analysis. Formalin-fixed, paraffin-embedded tissue sections were mounted on microscope slides. Immunohistochemical staining was optimized using a Ventana Discovery XT automated immunohistochemistry slide processing platform according to the manufacturer’s instructions (Ventana Medical Systems, Tucson, AZ). Following the closed loop assay development (CLAD) protocol (Ventana Medical Systems), antibodies were optimized using either the OmniMap or UltraMap DAB Anti-Mouse (HRP) or Anti-Rabbit (HRP) detection kits (Ventana Medical Systems). Antibodies used in this study are listed in Table 1. For each antibody, standard quality control procedures were undertaken to optimize antigen retrieval, primary antibody dilution, secondary antibody detection, and other factors for both signal and noise. Specificity for the antibodies was demonstrated by Western blot analysis.
Table 1.
List of Antibodies
| Antibody | Manufacturer | Source | Concentration |
|---|---|---|---|
| MIB-1 | DAKO | Mouse monoclonal | Predilute |
| PDGFRα | Santa Cruz | Rabbit polyclonal | 1:100 |
| Clusterin | Santa Cruz | Mouse monoclonal | 1:100 |
| SDF-1 | R&D Systems | Mouse monoclonal | 1:100 |
| DSCR1L1 | Abgent | Rabbit polyclonal | 1:100 |
Results
Gene Expression in Proliferating and Involuting Hemangiomas
In an initial quality control step, blocks of formalin-fixed, and paraffin-embedded proliferating and involuting hemangiomas were screened for useable RNA by real-time PCR. Seven primary tumor blocks with useable RNA were identified from recent cases: three proliferating and four involuting hemangiomas, as well as three term placental specimens (see Supplemental Table S5 at http://ajp.amjpathol.org). LCM was used to isolate vessels from the blocks that had useable RNA. Gene expression profiles were measured in amplified RNA from laser-captured microdissected vessels using the recently developed Human Affymetrix GeneChip X3P array, which can be used for whole-genome expression profiling of formalin-fixed, paraffin-embedded samples.23
Comparing proliferating hemangioma with placenta using pairwise analysis, 843 genes showed significant differential regulation (459 up-regulated and 384 down-regulated). The complete list of these genes is found in Supplementary Table S1 (available at http://ajp.amjpathol.org). In the pairwise comparison of proliferating and involuting hemangiomas, only 289 genes showed differential expression (116 up-regulated and 173 down-regulated). The complete list of these genes is found in Supplementary Table S2 (available at http://ajp.amjpathol.org). Differentially expressed genes were placed into biological process gene ontology categories and several highly represented gene ontology categories were found. Rather than presenting the entire data set, we will focus on just some of the interesting signaling systems that may be relevant to the biology of hemangiomas. Expression levels for selected genes (discussed below) were consistent, when reviewed case by case, for the proliferative and involutive phases and generally varied minimally when correlated with patient age within each group (see Supplemental Figure S3 at http://ajp.amjpathol.org).
Cell Proliferation Genes in Proliferating Hemangioma
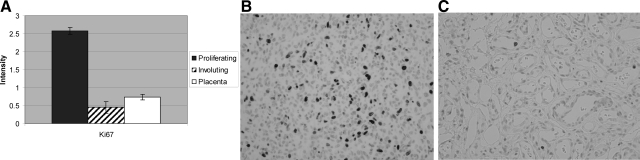
Not unexpectedly, proliferating hemangiomas express genes involved in cellular proliferation. For example, proliferating hemangiomas selectively express the gene for the antigen identified by the monoclonal antibody Ki-67 (MK167) (Figure 2, A–C). Expression of this gene is increased 3.5-fold, relative to levels found in the placenta. These findings confirm the observations of many other groups and help validate the gene expression analysis from formalin-fixed tissue.
Figure 2.
Increased Ki-67 antigen expression in proliferating hemangioma. A: Analysis of variance showing statistically significantly increased MKI67 gene expression in proliferating hemangioma, relative to placenta and involuting hemangioma. B: numerous Ki-67-immunopositive cells in proliferating hemangioma. C: Only rare Ki-67-positive endothelial cells in an involuting hemangioma.
Endothelial-Pericyte Signaling Systems in Proliferating Hemangioma
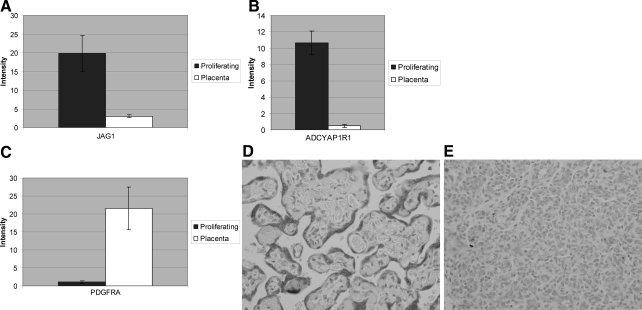
The first biological process category of differentially expressed genes in proliferating hemangioma, relative to placenta, were categories related to endothelial differentiation (z-score, 6.53), angiogenesis (z-score, 4.03), as well as blood vessel development and morphogenesis (z-score, 3.83). Some of the significantly up-regulated genes in these categories included established components of vascular growth factor signaling systems. For example angiopoietin-2 (ANGPT2 or Ang2) is increased 33.36-fold in proliferating hemangioma, relative to placenta. Ang2 is expressed at low levels in many normal adult tissues, but is strongly up-regulated at sites of active vessel remodeling, such as in tumors.24 Jagged 1 (JAG1) (Figure 3A) and Notch 4 (NOTCH4) were increased 6.5- and 3.2-fold, respectively, relative to placenta (Table 2).
Figure 3.
Patterns of gene expression in proliferating hemangiomas, relative to normal placenta. A: Increased expression of Jagged 1 in proliferating hemangioma. B: Increased expression of ADCYAPIR1 in proliferating hemangioma. C: Decreased expression of PDGFRα in proliferating and involuting hemangioma relative to placenta. Pairwise comparison showing statistically significant increased PDGFRα gene expression, relative to placenta. D: PDGFRa is strongly expressed in the trophoblast layer in placental villi and weakly in a few stromal cells. E: Proliferating hemangioma shows weak endothelial cytoplasmic immunoreactivity for PDGFRa.
Table 2.
Genes Identified in Proliferating Hemangioma Versus Placenta in the Endothelial Gene Ontology Group
| Gene description | Gene symbol | Cluster ID | Placenta | SEM | PH | SEM | Fold change |
|---|---|---|---|---|---|---|---|
| Genes up-regulated in proliferating hemangioma | |||||||
| Angiopoietin 2 | ANGPT2 | Hs.553484 | 0.0296 | ±0.0155 | 0.9858 | ±0.2653 | +78.6 |
| Angiopoietin 2 | ANGPT2 | Hs.553484 | 0.6157 | ±0.4018 | 4.8394 | ±0.7700 | +33.36 |
| Matrix-remodeling associated 5 | DKFZp564I1922 | Hs.369422 | 1.6427 | ±0.3292 | 37.8485 | ±5.9250 | +23.04 |
| Insulin-like growth factor binding protein 7 | IGFBP7 | Hs.479808 | 0.3566 | ±0.1678 | 4.916 | ±1.5634 | +13.78 |
| Neuropilin and tolloid-like 2 | NETO2 | Hs.444046 | 0.1026 | ±0.0201 | 1.1525 | ±0.3427 | +11.23 |
| Neuropilin and tolloid-like 2 | NETO2 | Hs.444046 | 0.1802 | ±0.0220 | 0.9866 | ±0.1610 | +5.48 |
| Brain-specific angiogenesis inhibitor 3 | BAI3 | Hs.13261 | 0.187 | ±0.0252 | 1.3583 | ±0.2695 | +7.26 |
| Plexin domain containing 1 | PLXDC1 | Hs.125036 | 0.2664 | ±0.0947 | 1.8975 | ±0.4650 | +7.12 |
| Plexin domain containing 1 | PLXDC1 | Hs.125036 | 0.5977 | ±0.1128 | 2.1092 | ±0.0928 | +3.53 |
| Jagged 1 | JAG1 | Hs.224012 | 3.0387 | ±0.3908 | 19.8124 | ±4.8743 | +6.52 |
| Endothelin receptor type A | EDNRA | Hs.183713 | 3.3445 | ±1.1985 | 16.6182 | ±3.0545 | +4.97 |
| Intercellular adhesion molecule 2 | ICAM2 | Hs.431460 | 1.1697 | ±0.4959 | 4.7548 | ±0.7278 | +4.06 |
| Notch homolog 4 | NOTCH4 | Hs.436100 | 0.958 | ±0.2294 | 3.0168 | ±0.5963 | +3.15 |
| Stabilin 1 | STAB1 | Hs.301989 | 2.1844 | ±0.5872 | 6.8518 | ±0.8059 | +3.14 |
| EPH receptor B3 | EPHB3 | Hs.2913 | 0.8408 | ±0.1470 | 2.6435 | ±0.2397 | +3.14 |
| Latrophilin 1 | LPHN1 | Hs.654658 | 1.9484 | ±4259 | 6.0487 | ±0.4863 | +3.1 |
| Natriuretic peptide receptor A | NPR1 | Hs.490330 | 0.3672 | ±0.0899 | 1.1348 | ±0.1405 | +3.09 |
| Genes down-regulated in proliferating hemangioma | |||||||
| G protein-coupled receptor 37 (endothelin receptor type B-like) | GPR37 | Hs.406094 | 0.9995 | ±0.2430 | 0.0327 | ±0.0299 | −30.57 |
| Insulin-like growth factor binding protein 3 | IGFBP3 | Hs.450230 | 58.0815 | ±8.2921 | 2.2601 | ±0.1342 | −25.7 |
| fms-related tyrosine kinase 1 | FLT1 | Hs.507621 | 0.9922 | ±0.1717 | 0.2611 | ±0.0568 | −22.67 |
| fms-related tyrosine kinase 1 | FLT1 | Hs.507621 | 7.2929 | ±0.3733 | 0.3216 | ±0.0334 | −3.8 |
| Platelet-derived growth factor receptor, α polypeptide | PDGFRA | Hs.74615 | 0.8004 | ±0.2168 | 0.0698 | ±0.0159 | −19.94 |
| Platelet-derived growth factor receptor, α polypeptide | PDGFRA | Hs.74615 | 21.6065 | ±5.9401 | 1.0834 | ±0.3965 | −11.47 |
| Transforming growth factor, β receptor III (betaglycan, 300 kDa) | TGBFGR3 | Hs.482390 | 24.4882 | ±7.4150 | 3.5753 | ±1.1188 | −6.85 |
| Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 | EDG2 | Hs.126667 | 0.1337 | ±0.0204 | 0.0219 | ±0.0065 | −6.12 |
| Insulin-like growth factor binding protein 5 | IGFBP5 | Hs.369982 | 3.0338 | ±0.8238 | 0.5067 | ±0.2263 | −5.99 |
| Endothelin receptor type B | EDNRB | Hs.82002 | 5.4154 | ±1.1597 | 0.9312 | ±0.4842 | −5.82 |
| Platelet-derived growth factor C | PDGFC | Hs.148162 | 2.2281 | ±0.5625 | 0.5379 | ±0.0195 | −5.28 |
| Platelet-derived growth factor C | PDGFC | Hs.148162 | 1.7552 | ±0.1443 | 0.3326 | ±0.0759 | −4.14 |
| Bone morphogenetic protein 4 | BMP4 | Hs.68879 | 3.2881 | ±0.2085 | 0.581 | ±0.1587 | −5.66 |
| Angiopoietin-like 1 | ANGPTL1 | Hs.555903 | 6.0324 | ±0.1531 | 1.1551 | ±0.2386 | −5.22 |
| Vascular cell adhesion molecule 1 | VCAM1 | Hs.109225 | 11.9634 | ±2.6759 | 2.851 | ±0.0521 | −4.2 |
| Bone morphogenetic protein 5 | BMP5 | Hs.296648 | 1.1733 | ±0.0269 | 0.3078 | ±0.1370 | −3.81 |
| Insulin-like growth factor 1 receptor | IGF1R | Hs.20573 | 1.1978 | ±0.1845 | 0.3409 | ±0.1461 | −3.51 |
Data from the arrays is normalized to the mean and a t-test is performed. Differences greater than threefold were considered significant, as outlined in Supplemental Table S1 (available at http://ajp.amjpathol.org). Shown are average intensity values and standard errors of the mean.
Other more novel genes involved in signal transduction are expressed at higher levels in proliferating hemangioma, relative to placenta (Table 2). For example, expression of the gene that encodes type 1 adenylate cyclase-activating polypeptide (pituitary) receptor (ADCYAP1R1, often designated PAC1), is increased more than 10-fold in proliferating hemangioma, relative to placenta (Figure 3B). The ligand for PAC1, pituitary adenylate cyclase-activating polypeptide (PACAP), has been implicated in the regulation of several homeostatic systems in the body, including balancing pro- and anti-inflammatory responses and cardiopulmonary control, as well as being a mediator of adult neural stem cell differentiation. Human tumors overexpress PACAP, suggesting a relationship between PACAP and PAC1 to tumor progression.25,26 Transcript for the endothelin receptor type A (EDNRA) gene, which mediates some of the effects of the vasoconstrictor endothelin-1, was increased 4.97-fold in proliferating hemangiomas, relative to the placental vessels (Table 2). Additionally, the latrophilin 1 gene (LPHN1), which encodes a member of the latrophilin subfamily of G-protein-coupled receptors, was expressed in proliferating hemangioma. This protein functions in both cell adhesion and in neuropeptide signaling.
Somewhat surprisingly, components of some other established signaling systems were significantly down-regulated in proliferating hemangiomas. Notably, transcript for insulin-like growth factor-binding protein-3 (IGFBP3) dropped 25.7-fold, relative to placenta (Table 1 and Supplemental Table S6 available at http://ajp.ajpathol.org). IGFBP-3 has anti-proliferative, as well as, pro-apoptotic effects and suppression of IGFBP-3 has been implicated in the progression of several other tumors.27 Also decreased were levels of transcripts for vascular endothelial growth factor (VEGF) receptor-1 (FLT1 or VEGFR1) (−22.7-fold), which interacts with VEGF-A, -B, and-E, as well as placental growth factor. Of relevance, low levels of VEGFR1 have been found in infantile hemangioma caused by mutations in the genes encoding VEGFR2 and TEM8. This is thought to cause diminished expression of VEGFR1 with VEGF-dependent activation of VEGFR2 resulting in endothelial proliferation.9 Interestingly, VEGFR1 expresses two types of mRNA, one full-length receptor and another for a short protein known as soluble VEGFr1 (sFLT-1). Decreased expression of Flt-1 in proliferating hemangioma, relative to normal placenta, raises the possibility that levels of the angiogenesis inhibitor (sFLT-1) also fall in the proliferating lesion. The platelet-derived growth factor-α (PDGFα) receptor was significantly decreased (−19.9-fold) (Figure 3C), as was transforming growth factor-β receptor III (betaglycan) (TGFGR3) (−6.9-fold), and the endothelial differentiation gene (EDG2) lysophosphatidic acid G-protein-coupled receptor-2 (−6.1-fold) (Table 2). The EDG family of G protein-coupled receptors consists of eight family members that bind lysophospholipid mediators, including lysophosphatidic acid. The family members function to mediate endothelial survival, growth, migration, and differentiation.28 Additionally, several growth factors, including PDGF-C (PDGFC), bone morphogenetic protein-4 and -5 (BMP4 and BMP5) and angiopoietin like-1 (ANGPTL1) were decreased in proliferating hemangioma, relative to placenta (Table 2). Collectively, these findings suggest that vessels in proliferating hemangioma express growth factor signaling systems that are different from those in placenta.
Differential Expression of Neural-Vascular Guidance Proteins in Hemangiomas
A second biological process category of differentially expressed genes in proliferating hemangioma, relative to placenta, were categories related to the biology of the nervous system. These included gene ontology groups for the regulation of axogenesis and neurogenesis (z-score, 5.94), positive regulation of neuron differentiation (z-score, 5.00), and neuron development and differentiation (z-scores, 3.73 and 3.68, respectively). Some of the genes that are found in these Gene Ontology groups are listed in (Table 3).
Table 3.
Genes Identified in Proliferating Hemangioma Versus Placenta in the Neural-Related Gene Ontology Group
| Gene description | Gene symbol | Cluster ID | Placenta | SEM | PH | SEM | Fold change |
|---|---|---|---|---|---|---|---|
| Genes up-regulated in proliferating hemangioma | |||||||
| Neurexin 3 | NRXN3 | Hs.368307 | 0.0475 | ±0.0274 | 0.6723 | ±0.1983 | −14.15 |
| Sema domain | SEMA5B | Hs.210870 | 0.3446 | ±0.0823 | 4.7682 | ±1.4570 | −13.84 |
| Neuropilin and tolloid-like 2 | NETO2 | Hs.444046 | 0.1026 | ±0.0201 | 1.1525 | ±0.3427 | −11.23 |
| Neuropilin and tolloid-like 2 | NETO2 | Hs.444046 | 0.1802 | ±0.0220 | 0.9866 | ±0.1610 | −5.48 |
| Calcium channel, voltage-dependent, β 2 subunit | CACNB2 | Hs.59093 | 0.181 | ±0.0353 | 1.9403 | ±0.1480 | −10.72 |
| Thy-1 cell surface antigen | THY1 | Hs.134643 | 2.4515 | ±0.1842 | 24.686 | ±4.7819 | −10.07 |
| Thy-1 cell surface antigen | THY1 | Hs.134643 | 0.1932 | ±0.1113 | 2.0679 | ±0.3991 | −10.7 |
| Glycoprotein M6B | GPM6B | Hs.495710 | 0.2587 | ±0.0754 | 2.2359 | ±0.4121 | −8.64 |
| Stathmin-like 2 | STMN2 | Hs.521651 | 0.0666 | ±0.0376 | 0.5451 | ±0.1036 | −8.19 |
| Growth-associated protein 43 | GAP43 | Hs.134974 | 0.1623 | ±0.0335 | 1.2836 | ±0.2768 | −7.91 |
| Like-glycosyltransferase | LARGE | Hs.474667 | 0.1282 | ±0.0805 | 0.9798 | ±0.0411 | −7.64 |
| Like-glycosyltransferase | LARGE | Hs.474667 | 0.2785 | ±0.0649 | 0.9296 | ±0.1537 | −3.34 |
| Doublecortin and CaM kinase-like 1 | DCAMKL1 | Hs.507755 | 0.0804 | ±0.0074 | 0.5062 | ±0.1272 | −6.23 |
| Growth differentiation factor 11 | GDF11 | Hs.567411 | 0.1215 | ±0.0606 | 0.6995 | ±0.0654 | −5.76 |
| Calcium channel, voltage-dependent, β 2 subunit | CACNB2 | Hs.59093 | 0.3233 | ±0.1428 | 1.7604 | ±0.3409 | −5.44 |
| Spondin 2, extracellular matrix protein | SPON2 | Hs.302963 | 1.5026 | ±0.6944 | 7.9847 | ±0.9696 | −5.31 |
| Agrin | AGRN | Hs.273330 | 1.1908 | ±0.3151 | 5.8269 | ±1.6378 | −4.89 |
| Neurotrophic tyrosine kinase, receptor, type 2 | NTRK2 | Hs.584783 | 0.201 | ±0.625 | 0.913 | ±0.1898 | −4.54 |
| Syntaphilin | SNPH | Hs.323833 | 0.4756 | ±0.1026 | 1.8957 | ±0.0835 | −3.99 |
| Neuronal PAS domain protein 2 | NPAS2 | Hs.156832 | 0.2201 | ±0.0683 | 0.8405 | ±0.1069 | −3.82 |
| LIM homeobox 6 | LHX6 | Hs.103137 | 0.4948 | ±1.8650 | 1.865 | ±0.3464 | −3.77 |
| Zinc finger homeobox 1b | ZFX1B | Hs.34871 | 0.1723 | ±0.0671 | 0.6075 | ±0.1102 | −3.53 |
| Debrin 1 | DBN1 | Hs.130316 | 0.4122 | ±0.1631 | 1.4353 | ±0.0678 | −3.48 |
| Kinesin family member 1B | KIF1B | Hs.97858 | 0.2501 | ±0.0530 | 0.8569 | ±0.1366 | −3.43 |
| Genes down-regulated in proliferating hemangioma | |||||||
| Neuron cell adhesion molecule | NRCAM | Hs.21422 | 3.1693 | ±0.7344 | 0.11 | ±0.0310 | 28.81 |
| Serpin peptidase inhibitor, clade E | SERPINE2 | Hs.38449 | 3.7492 | ±1.1239 | 0.2653 | ±0.0528 | 14.13 |
| Thrombospondin 1 | THBS1 | Hs.164226 | 4.1411 | ±0.9587 | 0.3096 | ±0.0583 | 13.38 |
| Roundabout, axon guidance receptor, homolog 2 | ROBO2 | Hs.13305 | 0.6957 | ±0.0722 | 0.0754 | ±0.0252 | 9.23 |
| Brain-derived neurotrophic factor opposite strand | BDNF | Hs.502182 | 1.1189 | ±0.2679 | 0.1788 | ±0.0381 | 6.26 |
| Endothelin receptor type B | EDNRB | Hs.82002 | 5.4154 | ±1.1597 | 0.9312 | ±0.4842 | 5.815 |
| Slit homolog 2 | SLIT2 | Hs.29802 | 6.5785 | ±0.3129 | 1.4273 | ±0.3137 | 4.609 |
| Neuron cell adhesion molecule | NRCAM | Hs.21422 | 1.4863 | ±0.3032 | 0.3739 | ±0.2421 | 3.98 |
| Plexin domain containing 1 | PLXDC1 | Hs.125036 | 0.5977 | ±0.1128 | 2.1092 | ±0.0928 | 3.53 |
Data from the arrays is normalized to the mean and a t-test is performed. Differences greater than threefold were considered significant, as outlined in the Supplemental Table S1 (available at http://ajp.amjpathol.org). Shown are average intensity values and standard errors of the mean.
Notable in the list of neuronal gene ontology groups in proliferating hemangioma were components of signaling systems implicated in neural and vascular patterning. These signaling systems include the semaphorins and their neuropilin receptors, ephrins, and their Eph receptors and slits and their Robo (roundabout) receptors.29 Semaphorin 5B (SEMA5B) is up-regulated 13.8-fold in proliferating hemangioma. Expression of neuropilin-2 (NETO2) was increased 11.2-fold in proliferating hemangiomas, relative to placenta. Like the neuropilins, plexins are also receptors for the semaphorins. PlexinC1 (PLXDC1) was increased in proliferating hemangiomas, relative to placenta. Interestingly, other classes of genes involved in guidance of vascular and neural network formation were also differentially regulated. Roundabout axon guidance receptor 2 (ROBO2) and slit-2 (SLIT2) were up-regulated in placenta, 9.2- and 4.6-fold, respectively, relative to proliferating hemangioma (Table 3).
Changes in the expression of netrins and their DCC (deleted in colon carcinoma) or Unc5 receptors were not observed in comparisons of placenta and proliferating hemangioma or in proliferating and involuting hemangioma. Lack of differential expression of these guidance molecules suggests that these genes are dynamically regulated during formation and regression of the vascular lesions.
Chronic Inflammatory Mediators in the Involuting Hemangioma
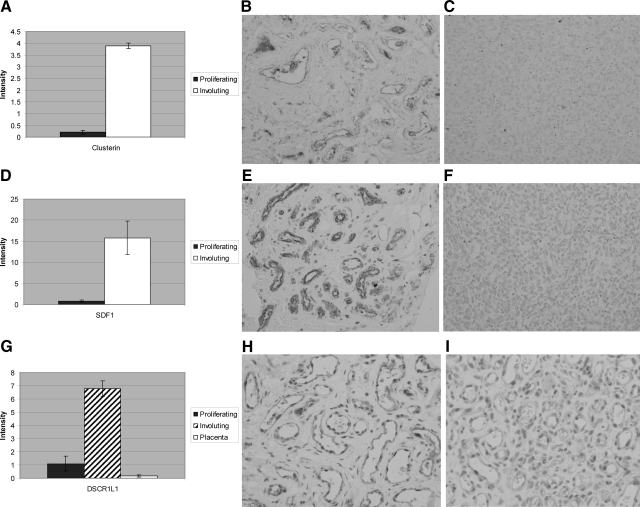
Notable in the biological process categories of differentially expressed genes in involuting hemangioma, relative to proliferating hemangioma, were categories related to chronic inflammation, including immune response (z-score, 2.71) and cytokine- and chemokine-mediated signaling pathways (z-score, 4.23). For example, the clusterin gene is selectively expressed by involuting hemangiomas, consistent with previously reported findings.30 Expression of clusterin transcription increased almost 20-fold in involuting relative to proliferating hemangioma (Figure 4A–C). Clusterin is widely expressed and has been implicated in apoptosis, inhibition of complement-mediated cell lysis, transport of lipoproteins, and the modulation of cell-cell interactions.31 Overexpression of clusterin has been linked to multiple disorders, including a variety of human cancers.31
Figure 4.
Increased protein expression in involuting hemangioma A: Increased expression of clusterin expression in involuting hemangioma relative to proliferating hemangioma. Pairwise comparison showing statistically significantly increased clusterin in involuting hemangioma, relative to proliferating hemangioma. B: A moderate number of clusterin-immunopositive cells in involuting hemangioma. C: Only a few clusterin-immunopositive cells in proliferating hemangioma. D: Many SDF-1 immunopositive endothelial cells are present in involuting hemangioma. E: No SDF-1 immunopositive cells are present in proliferating hemangioma. F: Many SDF-1-immunopositive endothelial cells in involuting hemangioma. G: Increased expression of DSCR1L1 expression in proliferating hemangioma relative to placenta or proliferating hemangioma. Analysis of variance showing statistically significant increased DSCR1L1 in involuting hemangioma, relative to placenta or proliferating hemangioma. H: Endothelial cells show moderate intense immunopositivity for DSCR1L1 in involuting hemangioma. I: Endothelial cells show less intense immunopositivity for DSCR1L1 in proliferating hemangioma.
Some of the significantly up-regulated chemokine genes in involuting hemangioma are shown in Table 4. Most dramatically up-regulated in involuting hemangioma was stromal cell-derived growth factor 1 (SDF-1, or CXCL12) (Figure 4D–F). This chemokine and its unique receptor, CXCR4, are required for normal nervous system and cardiovascular development, as well as for trafficking of normal stem cells and metastasis of cancer stem cells.32,33 Also increased in involuting hemangiomas is chemokine (CC) ligand 15 (CCL15). The cytokine encoded by this gene is chemotactic for monocytes and is thought to act through the CCR1 receptor. Similarly, fractalkine (chemokine (C-X3-C motif) ligand 1 (CX3CL1) is up-regulated 4.5-fold in involuting hemangioma. Fractalkine is a cell surface adhesion molecule that is expressed by vascular smooth muscle cells or by endothelial cells activated by inflammatory cytokines.34 Fractalkine can engage the C-X3-C receptor (CX3CR1) that is expressed by monocytes and macrophages. Additionally, the chemokine (C-C) motif receptor 5 (CCR5) is increased 4.5-fold in involuting hemangioma. This receptor is expressed by macrophages and T cells and it has as ligands, monocyte chemoattractant protein 2 (MCP-2), macrophage inflammatory proteins-1α and -β (MIP-1α and -β), as well as regulated on activation normal T-cell-expressed and -secreted protein (RANTES). Colony-stimulating receptor factor 2 receptor β (CSF2RB), the common signaling subunit of the receptor for GM-CSF, IL-3, and IL-5, is also differentially expressed in involuting, relative to proliferating hemangioma (Table 4). The high-affinity receptor for GM-CSF consists of a cytokine-specific α subunit and the common β subunit.35 If involuting hemangiomas also express the α subunit, a functional cytokine receptor may be generated.
Table 4.
Genes Identified in Involuting Hemangioma Versus Proliferating Hemangioma in the Cytokine and Chemokine Gene Ontology Groups
| Gene description | Gene symbol | Cluster ID | PH | SEM | IH | SEM | Fold change | |
|---|---|---|---|---|---|---|---|---|
| Genes up-regulated in involuting hemangioma | ||||||||
| Chemokine (C-X-C motif) ligand 12 | CXCL12 | Hs.522891 | 0.8233 | ±0.2576 | 15.8125 | ±3.9838 | 19.21 | |
| Chemokine (C-C motif) ligand 15 | CCL15 | Hs.272493 | 0.2674 | ±0.1746 | 2.77 | ±0.5274 | 10.36 | |
| Chemokine (C-X-C motif) ligand 12 | CXCL12 | Hs.522891 | 0.577 | ±0.3545 | 3.9029 | ±0.9987 | 6.76 | |
| Chemokine orphan receptor 1 | CMKOR1 | Hs.471751 | 0.369 | ±0.0250 | 1.9564 | ±0.4700 | 5.3 | |
| Chemokine orphan receptor 1 | CMKOR1 | Hs.471751 | 0.335 | ±0.1330 | 1.3817 | ±0.2787 | 4.12 | |
| Colony stimulating factor 2 receptor, β | CSF2RB | Hs.592192 | 0.8702 | ±0.2074 | 4.4963 | ±0.7623 | 5.17 | |
| Chemokine (C-C motil) receptor 5 | CCR5 | Hs.450802 | 0.123 | ±0.0291 | 0.5581 | ±0.1340 | 4.54 | |
| Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | Hs.531668 | 0.8211 | ±0.4302 | 3.7119 | ±0.3802 | 4.52 | |
| Duffy blood group, chemokine receptor | DARC | Hs.153381 | 0.3705 | ±0.0326 | 1.6034 | ±0.3574 | 4.33 | |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | Hs.478275 | 3.131 | ±0.7497 | 13.0375 | ±1.0947 | 4.16 | |
| Tumor necrosis factor (ligand) superfamily, member 13b | TNSF13B | Hs.525157 | 2.1214 | ±0.3642 | 8.7882 | ±2.0486 | 4.14 | |
| Activated leukocyte cell adhesion molecule | ALCAM | Hs.591293 | 0.6274 | ±0.0471 | 2.5164 | ±0.5349 | 4.01 | |
| CCAAT/enhancer binding protein (C/EBP), γ | CEBPG | Hs.429666 | 0.1457 | ±0.0825 | 0.4861 | ±0.0863 | 3.34 | |
Data from the arrays is normalized to the mean and a t-test is performed. Differences greater than threefold were considered significant, as outlined in the Supplemental Table S2 (available at http://ajp.amjpathol.org). Shown are average intensity values and standard errors of the mean.
Expression of genes that regulate the angiogenic process may play a key role in the resolution of hemangiomas. A member of the Down syndrome critical region (DSCR) family, designated Down syndrome candidate region like 1 (DSCR1L1, also known as MCIP1, Adapt78), was overexpressed 6.2-fold in involuting relative to levels found in proliferating hemangiomas (Figure 4G–I). This protein is an endogenous inhibitor of calcineurin, which in turn, regulates the NFAT pathway.36 Of note, one study notes reduction of DSCR1L1 expression in proliferative hemangioma but no information is present regarding involuting hemangioma.9 Interestingly, another member of the family, DSCR1, which was not differentially expressed in proliferating and involuting hemangiomas (data not shown), is known to attenuate endothelial cell proliferation and angiogenesis.37,38
Discussion
Applying LCM of vessels from formalin-fixed specimens, it is possible to selectively isolate vessels from proliferating and involuting hemangioma. Using techniques and reagents that have been optimized for RNA isolation, unbiased amplification from these fixed samples and an array that is designed to interrogate sequences closer to the 3′ end of the transcript, we demonstrate that genome-wide profiling is possible.23,39
Expression of Endothelial-Pericyte Signaling Systems in Hemangioma
Proliferating hemangiomas were characterized by increased expression of signaling systems involved in endothelial-pericyte interactions. Multiple studies suggest that angiopoietin-Tie2 signaling is involved in the reciprocal communication between endothelial cells and pericytes.40,41 The increased Tie2 transcript expression seen in proliferating hemangioma (Table 1) is consistent with previous findings of increased expression of Tie2 and enhanced response to angiopoietin-1 in hemangioma-derived endothelial cells.42 Ang1 is a pericyte-derived, microvessel-stabilizing signal, whereas Ang2 is expressed by endothelial cells43 and antagonizes Tie2 signaling, leading to vessel destabilization. Up-regulated Ang2 marks the onset of angiogenic sprouting in tumors.44 Although levels of Ang1 and Ang2 are not significantly different, Ang-like-1 transcript levels are decreased in proliferating hemangioma, relative to placenta, raising the possibility that Tie2 signaling may be interrupted and the vessel destabilized.
Both Jagged 1 and Notch 4 are increased in proliferating hemangiomas, raising the possibility that Notch signaling plays a role in the proliferative lesion. Notch signaling is important in establishing arterial/venous identity in the development of vasculature.45 Activation of notch signaling in endothelium induces arterial makers such as neuropilin 1 and ephrin B2, and suppresses venous markers such as EphB4. Conversely, suppression of Notch in endothelial cells is important in establishing venous identity.46
The endothelin receptor type A (EDNRA or ETA-R), which mediates the effect of the vasoconstrictor endothelin-1, is increased in proliferating hemangiomas. Interestingly, there is increasing evidence that this receptor is expressed in a variety of tumor cells and may play a role in angiogenesis, tumor growth, and invasion in vivo.47,48,49
The insulin-like growth factor system may play a key role in regulating proliferating hemangioma. Previous studies have demonstrated that IGF2 is overexpressed and regulates growth in proliferating hemangioma.17 Here we find that transcripts for insulin-like growth factor-binding protein-3 (IGFBP-3) drop 25.7-fold in proliferating hemangioma, relative to placenta (Table 2). IGFBP-3 is the major carrier of insulin-like growth (IGF) factors and antagonizes IGF signaling, resulting in an inhibition of cell growth and induction of apoptosis.50 Supporting its role as a primary growth inhibitor, IGFBP-3 has been shown to be increased by cell growth-inhibitory agents, such as transforming growth factor-β51 and the tumor suppressor gene p53.52 These observations complement previous studies demonstrating increased IGF-2 in proliferating hemangioma17 and suggest that increased expression of IGFBP-3 may prevent apoptosis of the lesions.
Expression of Nerve and Blood Vessel Guidance Cues in Hemangioma
Axonal guidance and vascular patterning share several guidance cues, including the semaphorins and their neuropilin receptors, ephrins and their Eph receptors, slits and their Robo receptors, and netrins and their DCC (deleted in colon carcinoma) or Unc5 receptors.29 Several classes of signaling systems implicated in vascular patterning were up-regulated in proliferating hemangiomas. The first class was the semaphorins and their neuropilin and plexin receptors.
In this study, expression of both plexinC1 (PLXDC1) and neuropilin-2 were increased in proliferating hemangiomas, relative to placenta. Although little is known about plexinC1, neuropilin-2 is expressed in veins and lymph vessels and defects in these vessels are observed in neuropilin-2 mutant mice.53 Because neuropilins are receptors for semaphorins, these findings might be interpreted to reflect altered semaphorin signaling in the lesions. However, neuropilins are also receptors for specific VEGF isoforms, suggesting that changes in the level of the neuropilin receptor may reflect alterations in VEGF signaling.
The second class of signaling systems controlling vascular development increased in proliferating hemangioma was the ephrin-EPH receptor. Increased expression of the Eph receptor B3 was seen in proliferating hemangiomas. Expression of ephrin B ligands and EphB receptors on adjacent endothelial or smooth muscle cells in the same vessels may provide bidirectional signals for establishing contact-dependent communication and promote vessel assembly, sprouting, and maturation.54 These Ephrin-Eph signaling events regulate recruitment of vascular smooth muscle cells toward endothelial channels.
The third class of signaling system regulating vascular and neural patterning are the Slits and their Roundabout (Robo) receptors. Members of the Slit-Robo receptor family are capable of regulating midline axon guidance and more recently have been implicated in angiogenesis and endothelial migration, which are essential for angiogenesis in vivo.55,56,57 Here we find that Slit-2 and Robo2 were down-regulated in proliferating hemangioma, relative to normal placenta. Additionally, Slit-3 was overexpressed in involuting hemangiomas relative to proliferating hemangiomas. Interestingly, Slit-Robo interactions have been implicated in establishing the nonstereotyped vascular network in a tumor model.58 Our findings support this observation and raise the possibility that dysregulated Slit-Robo signaling may be a general feature of the chaotic tumor vascular network. The differential expression of multiple genes involved in several vessel path finding systems suggests that the vascular lesions represent an abnormal vascular network resulting, at least in part, from aberrant guidance cues.
Signaling Systems in the Involuting Hemangioma
The chemokine SDF-1 is expressed at higher levels in involuting, relative to proliferating, hemangiomas. SDF-1 and its unique receptor, CXCR4, are key regulators in stem cell trafficking. It is possible that SDF-1 induces recruitment or retention of endothelial progenitors in involuting hemangiomas. Alternatively, SDF-1 may play a role in the recruitment and retention of mesenchymal stem cells that give rise to the connective tissue components found in the involuting hemangioma. SDF-1 within the tumor environment may orchestrate endothelial cell movement and drive the formation of the more definitive blood vessels characteristic of the involuting hemangioma.
Information about the spontaneous involution of proliferating hemangiomas may provide new insights into attenuation of the angiogenic process. Several studies suggest that the calcium-calcineurin-NFAT pathway may be important in endothelial cells. For example, VEGF acting through the VEGF receptor leads to activation of the NFAT pathway with subsequent expression of endothelial-specific genes (unpublished data).9,59 Additionally, cyclosporine A, a well known inhibitor of calcineurin activity, inhibits endothelial cell migration and tube formation in vitro, as well as corneal neovascularization in mice.60 Endogenous inhibitors of calcineurin, termed Down syndrome critical region 1 (DSCR1) and DSCR1-L1 interact directly with calcineurin and inhibit its activity.61 Interestingly, DSCR1 attenuates endothelial cell proliferation and angiogenesis37,38,62 and it diminishes the expression of inflammatory markers on activated endothelial cells.63 VEGF acts to induce the expression of DSCR1, suggesting that the up-regulation of the members of the DSCR family may represent a mechanism for the negative or feedback regulation of angiogenesis.
In summary, here we combined LCM and genome-wide expression profiling to define mediators involved in proliferating hemangiomas and to identify biological themes associated with the transition from the proliferating to the involuting phase. Our ability to isolate and transcriptionally profile vessels from proliferating and involuting hemangiomas has enabled us to substantiate reports of genes previously implicated in endothelial-pericyte interactions and to uncover biological themes that could be important in orchestrating the molecular brake regulating hemangioma involution.
Acknowledgments
We would like to thank Dr. John Mulliken for contributing to the inspiration for this manuscript.
Footnotes
Address reprint requests to Harry P. Kozakewich, Department of Pathology, Children’s Hospital Boston, 300 Longwood Ave., Boston, MA 02115. E-mail: harry.kozakewich@childrens.harvard.edu.
Supported in part by the National Institutes of Health (grant RO1 HL35716) and the Harvard Medical School (funds from the Wolbach Chair).
We regret the untimely death of Tucker Collins, M.D., Ph.D., on June 8, 2007. He was a great mentor and friend. His enthusiasm and support will be greatly missed.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- North PE, Waner M, Buckmiller L, James CA, Mihm MC. Vascular tumors of infancy and childhood: beyond capillary hemangioma. Cardiovasc Pathol. 2006;15:302–317. doi: 10.1016/j.carpath.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Waner M, North PE, Scherer KA, Frieden IJ, Waner A, Mihm MC. The nonrandom distribution of facial hemangiomas. Arch Dermatol. 2003;139:869–875. doi: 10.1001/archderm.139.7.869. [DOI] [PubMed] [Google Scholar]
- Mulliken JB, Fishman SJ, Burrows PE. Vascular anomalies. Curr Probl Surg. 2000;37:517–584. doi: 10.1016/s0011-3840(00)80013-1. [DOI] [PubMed] [Google Scholar]
- Brouillard P, Vikkula M. Genetic causes of vascular malformations. Hum Mol Genet. 2007;16(Spec No. 2):R140–R149. doi: 10.1093/hmg/ddm211. [DOI] [PubMed] [Google Scholar]
- Boye E, Yu Y, Pafranya G, Mulliken JB, Olsen BR, Bischoff J. Clonality and altered behavior of endothelial cells from hemangiomas. J Clin Invest. 2001;107:745–752. doi: 10.1172/JCI11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JW, North PE, Waner M, Mizeracki A, Blei F, Walker JW, Reinisch JF, Marchuk DA. Somatic mutation of vascular endothelial growth factor receptors in juvenile hemangiomas. Genes Chromosom Cancer. 2002;33:295–303. doi: 10.1002/gcc.10028. [DOI] [PubMed] [Google Scholar]
- Berg JN, Walter JW, Thisanagayam U, Evans M, Blei F, Waner M, Diamond AG, Marchuk DA, Porteous ME. Evidence for loss of heterozygosity of 5q in sporadic haemangiomas: are somatic mutations involved in haemangioma formation? J Clin Pathol. 2001;54:249–252. doi: 10.1136/jcp.54.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JW, Blei F, Anderson JL, Orlow SJ, Speer MC, Marchuk DA. Genetic mapping of a novel familial form of infantile hemangioma. Am J Med Genet. 1999;82:77–83. doi: 10.1002/(sici)1096-8628(19990101)82:1<77::aid-ajmg15>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Jinnin M, Medici D, Park L, Limaye N, Liu Y, Boscolo E, Bischoff J, Vikkula M, Boye E, Olsen BR. Suppressed NFAT-dependent VEGFR1 expression and constitutive VEGFR2 signaling in infantile hemangioma. Nat Med. 2008;14:1236–1246. doi: 10.1038/nm.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Flint AF, Mulliken JB, Wu JK, Bischoff J. Endothelial progenitor cells in infantile hemangioma. Blood. 2004;103:1373–1375. doi: 10.1182/blood-2003-08-2859. [DOI] [PubMed] [Google Scholar]
- Dadras SS, North PE, Bertoncini J, Mihm MC, Detmar M. Infantile hemangiomas are arrested in an early developmental vascular differentiation state. Mod Pathol. 2004;17:1068–1079. doi: 10.1038/modpathol.3800153. [DOI] [PubMed] [Google Scholar]
- North PE, Waner M, Mizeracki A, Mrak RE. A unique microvascular phenotype shared by juvenile hemangiomas and human placenta. Arch Dermatol. 2001;137:559–570. [PubMed] [Google Scholar]
- North PE, Waner M, Brodsky MC. Are infantile hemangiomas of placental origin? Ophthalmology. 2002;109:223–224. doi: 10.1016/s0161-6420(01)00994-0. [DOI] [PubMed] [Google Scholar]
- Iwata J, Sonobe H, Furihata M, Ido E, Ohtsuki Y. High frequency of apoptosis in infantile capillary haemangioma. J Pathol. 1996;179:403–408. doi: 10.1002/(SICI)1096-9896(199608)179:4<403::AID-PATH604>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Razon MJ, Kraling BM, Mulliken JB, Bischoff J. Increased apoptosis coincides with onset of involution in infantile hemangioma. Microcirculation. 1998;5:189–195. [PubMed] [Google Scholar]
- Yu Y, Fuhr J, Boye E, Gyorffy S, Soker S, Atala A, Mulliken JB, Bischoff J. Mesenchymal stem cells in adipogenesis in hemangioma involution. Stem Cells. 2006;24:1605–1612. doi: 10.1634/stemcells.2005-0298. [DOI] [PubMed] [Google Scholar]
- Ritter MR, Dorrell MI, Edmonds J, Friedlander SF, Friedlander M. Insulin-like growth factor 2 and potential regulators of hemangioma growth and involution identified by large-scale expression analysis. Proc Natl Acad Sci USA. 2002;99:7455–7460. doi: 10.1073/pnas.102185799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter MR, Moreno SK, Dorrell MI, Rubens J, Ney J, Friedlander DF, Bergman J, Cunningham BB, Eichenfield L, Reinisch J, Cohen S, Veccione T, Holmes R, Friedlander SF, Friedlander M. Identifying potential regulators of infantile hemangioma progression through large-scale expression analysis: a possible role for the immune system and indoleamine 2,3 dioxygenase (IDO) during involution. Lymphat Res Biol. 2003;1:291–299. doi: 10.1089/153968503322758094. [DOI] [PubMed] [Google Scholar]
- Barnés CM, Huang S, Kaipainen A, Sanoudou D, Chen EJ, Eichler GS, Guo TY, Yu Y, Ingber DE, Mulliken JB, Beggs AM, Folkman J, Fishman SJ. Evidence by molecular profiling for a placental origin of infantile hemangioma. Proc Natl Acad Sci USA. 2005;102:19097–19102. doi: 10.1073/pnas.0509579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick MB, Sabo E, Meitner PA, Kim SS, Cho Y, Kim HK, Tavares R, Moss SF. Global analysis of the human gastric epithelial transcriptome altered by Helicobacter pylori eradication in vivo. Gut. 2006;55:1717–1724. doi: 10.1136/gut.2006.095646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagzag D, Hooper A, Friedlander DR, Chan W, Holash J, Weigland SJ, Yancopoulos GD, Grumet M. In situ expression of angiogenesis in astrocytomas identifies angiopoietin-2 as an early marker of tumor angiogenesis. Exp Neurol. 1999;159:391–400. doi: 10.1006/exnr.1999.7162. [DOI] [PubMed] [Google Scholar]
- García-Fernández MO, Bodega G, Ruiz-Villaespesa A, Cortes J, Prieto JC, Carmena MJ. PACAP expression and distribution in human breast cancer and healthy tissue. Cancer Lett. 2004;205:189–195. doi: 10.1016/j.canlet.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Moretti C, Mammi C, Frajese GV, Mariani S, Gnessi L, Arizzi M, Wannenes F, Frajese G. PACAP and type I PACAP receptors in human prostate cancer tissue. Ann NY Acad Sci. 2006;1070:440–449. doi: 10.1196/annals.1317.059. [DOI] [PubMed] [Google Scholar]
- Tomii K, Tsukuda K, Toyooka S, Dote H, Hanafusa T, Asano H, Naitou M, Doihara H, Kisimoto K, Katayama H, Pass HI, Date H, Shimizu N. Aberrant promoter methylation of insulin-like growth factor protein-3 gene in human cancers. Int J Cancer. 2007;120:566–573. doi: 10.1002/ijc.22341. [DOI] [PubMed] [Google Scholar]
- Yatomi Y. Sphingosine 1-phosphate in vascular biology: possible therapeutic strategies to control vascular disease. Curr Pharm Des. 2006;12:575–587. doi: 10.2174/138161206775474404. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- Hasan Q, Ruger BM, Tan ST, Gush J, Davis PF. Clusterin.apoJ expression during the development of hemangioma. Hum Pathol. 2000;31:691–697. doi: 10.1053/hupa.2000.7638. [DOI] [PubMed] [Google Scholar]
- Shannan B, Seifert M, Leskov K, Willis J, Boothman D, Tilgen W, Reichrath J. Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006;13:12–19. doi: 10.1038/sj.cdd.4401779. [DOI] [PubMed] [Google Scholar]
- Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, Ratajczak J, Ratajczak MZ. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23:879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- Umehara H, Bloom ET, Okazaki T, Nagano Y, Yoshie O, Imai T. Fractalkine in vascular biology: from basic research to clinical disease. Arterioscler Thromb Vasc Biol. 2004;24:34–40. doi: 10.1161/01.ATV.0000095360.62479.1F. [DOI] [PubMed] [Google Scholar]
- McClure BJ, Hercus TR, Cambareri BA, Woodcock JM, Bagley CJ, Howlett GL, Lopez AF. Molecular assembly of the ternary granulocyte-macrophage colony-stimulating factor receptor complex. Blood. 2003;101:1308–1315. doi: 10.1182/blood-2002-06-1903. [DOI] [PubMed] [Google Scholar]
- Fuentes JJ, Genesca L, Kingsbury TJ, Cunningham KW, Perez-Riba M, Estivill X, de la Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet. 2000;9:1681–1690. doi: 10.1093/hmg/9.11.1681. [DOI] [PubMed] [Google Scholar]
- Minami T, Horiuchi K, Miura M, Abid MR, Takabe W, Boguchi N, Kohro T, Ge X, Aburatani H, Hamakubo T, Kodama T, Aird WC. Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1 attenuates endothelial cell proliferation and angiogenesis. J Biol Chem. 2004;279:50537–50564. doi: 10.1074/jbc.M406454200. [DOI] [PubMed] [Google Scholar]
- Minami T, Miura M, Aird WC, Kodama T. Thrombin-induced autoinhibitory factor, Down syndrome critical region-1, attenuates NFAT-dependent vascular cell adhesion molecule-1 expression and inflammation in the endothelium. J Biol Chem. 2006;281:20503–20520. doi: 10.1074/jbc.M513112200. [DOI] [PubMed] [Google Scholar]
- Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase polymerase-chain reaction assay. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughna S, Sato TN. Angiopoietin and Tie signaling pathways in vascular development. Matrix Biol. 2001;20:319–325. doi: 10.1016/s0945-053x(01)00149-4. [DOI] [PubMed] [Google Scholar]
- Perry BN, Govindarajan B, Bhandarkar SS, Knaus UG, Valo M, Sturk C, Carrillo CO, Sohn A, Cerimele F, Dumont D, Losken A, Williams J, Brown LF, Tan X, Ioffe E, Yancopoulos GD, Arbiser JL. Pharmacologic blockade of angiopoietin-2 is efficacious against model hemangiomas in mice. J Invest Dermatol. 2006;126:2316–2322. doi: 10.1038/sj.jid.5700413. [DOI] [PubMed] [Google Scholar]
- Yu Y, Varughese J, Brown LF, Mulliken JB, Bischoff J. Increased Tie2 expression, enhanced response to angiopoietin-1 and dysregulated angiopoietin-2 expression in hemangioma-derived endothelial cells. Am J Pathol. 2001;159:2271–2280. doi: 10.1016/S0002-9440(10)63077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerimele F, Brown LF, Bravo F, Ihler GM, Kouadio P, Arbiser JL. Infectious angiogenesis: Bartonella bacilliformis infection results in endothelial production of angiopoetin-2 and epidermal production of vascular endothelial growth factor. Am J Pathol. 2003;163:1321–1327. doi: 10.1016/S0002-9440(10)63491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait CR, Jones PF. Angiopoietins in tumours: the angiogenic switch. J Pathol. 2004;204:1–10. doi: 10.1002/path.1618. [DOI] [PubMed] [Google Scholar]
- Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Cazaubon S, Deshayes F, Couraud PO, Nahmias C. [Endothelin-1, angiotensin II and cancer]. Med Sci (Paris) 2006;22:416–422. doi: 10.1051/medsci/2006224416. [DOI] [PubMed] [Google Scholar]
- Nelson JB, Nabulsi AA, Vogelzang NHJ, Breul J, Zonnenberg BA, Daliani DD, Schulman CC, Carducci MA. Suppression of prostate cancer induced bone remodeling by the endothelin receptor A antagonist atrasentan. J Urol. 2003;169:1143–1149. doi: 10.1097/01.ju.0000042162.08938.27. [DOI] [PubMed] [Google Scholar]
- Zonnenberg BA, Groenewegen G, Janus TJ, Leahy TW, Humerickhouse RA, Isaacson JD, Carr RA, Voest E. Phase I dose-escalation study of the safety and pharmacokinetics of atrasentan: an endothelin receptor antagonist for refractory prostate cancer. Clin Cancer Res. 2003;9:2965–2972. [PubMed] [Google Scholar]
- Ali O, Cohen P, Lee KW. Epidemiology and biology of insulin-like growth factor binding protein-3 (IGFBP-3) as an anti-cancer molecule. Horm Metab Res. 2003;35:726–733. doi: 10.1055/s-2004-814146. [DOI] [PubMed] [Google Scholar]
- Rajah R, Valentinis B, Cohen P. Insulin-like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta1 on programmed cell death through a p53- and IGF-independent mechanism. J Biol Chem. 1997;272:12181–12188. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]
- Grimberg A. P53 and IGFBP-3: apoptosis and cancer protection. Mol Genet Metab. 2000;70:85–98. doi: 10.1006/mgme.2000.3008. [DOI] [PubMed] [Google Scholar]
- Yuan L, Moyon D, Pardanaud L, Bréant C, Karkkainen MJ, Alitalo K, Eichmann A. Abnormal lymphatic vessel development in neuropilin 2 mutant mice. Development. 2002;129:4797–4806. doi: 10.1242/dev.129.20.4797. [DOI] [PubMed] [Google Scholar]
- Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, Risau K, Klein R. Roles of ephrin B ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains, vascular morphogenesis and sprouting angiogenesis. Genes Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchting S, Heal P, Tahtis K, Stewart LM, Bicknell R. Soluble Robo4 receptor inhibits in vivo angiogenesis and endothelial cell migration. FASEB J. 2005;19:121–123. doi: 10.1096/fj.04-1991fje. [DOI] [PubMed] [Google Scholar]
- Park KW, Morrison CM, Sorensen LK, Jones CA, Rao Y, Chien CB, Wu JY, Urness LD, Li DY. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol. 2003;261:251–267. doi: 10.1016/s0012-1606(03)00258-6. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Yeo SY, Park KW, Chung J, Seth P, Shivalingappa V, Zhao J, Obara T, Sukhatme VP, Drummond IA, Li DY, Ramchandran R. Roundabout4 is essential for angiogenesis in vivo. Proc Natl Acad Sci USA. 2005;102:6373–6378. doi: 10.1073/pnas.0408318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Connolly JO, Soga N, Guo XL, Alvarez U, Hruska KA. Rac is essential in the transformation of endothelial cells by polyoma middle T. Cell Adhes Commun. 2000;7:409–422. doi: 10.3109/15419060009109022. [DOI] [PubMed] [Google Scholar]
- Hernández GL, Volpert OV, Iniquez MA, Lorenzo E, Martinez-Martinez S, Grau R, Fresno M, Redondo JM. Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: roles of the nuclear factor of activated T cells and cyclooxygenase 2. J Exp Med. 2001;193:607–620. doi: 10.1084/jem.193.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollogly LK, Ryeom SW, Yoon SS. Down syndrome candidate region 1-like 1 (DSCR1-L1) mimics the inhibitory effects of DSCR1 on calcineurin signaling in endothelial cells and inhibits angiogenesis. J Surg Res. 2007;142:129–136. doi: 10.1016/j.jss.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Duh EJ. VEGF selectively induces Down syndrome critical region 1 gene expression in endothelial cells: a mechanism for feedback regulation of angiogenesis? Biochem Biophys Res Commun. 2004;321:648–656. doi: 10.1016/j.bbrc.2004.06.176. [DOI] [PubMed] [Google Scholar]
- Hesser BA, Liang XH, Camenisch G, Lewin DA, Scheller R, Ferrara N, Gerber HP. Down syndrome critical region protein 1 (DCSR1), a novel VEGF target that regulates expression of inflammatory markers on activated endothelial cells. Blood. 2004;104:149–159. doi: 10.1182/blood-2004-01-0273. [DOI] [PubMed] [Google Scholar]