Abstract
Barrett’s esophagus (BE)/Barrett’s metaplasia (BM) is a recognized precursor of esophageal adenocarcinoma (EA) with an intermediary stage of dysplasia. The low yield and high cost of endoscopic screening of patients with BE underscores the need for novel biomarkers, such as microRNA (miRNA), which have emerged as important players in neoplastic progression for risk assessment of developing dysplasia/adenocarcinoma. Recently, we reported highly elevated levels of miRNA-196a (miR-196a) in EA and demonstrated its growth-promoting and anti-apoptotic functions. Here, we evaluated miR-196a as a marker of BE progression to low-grade dysplasia, high-grade dysplasia, and EA using microdissected paraffin-embedded tissues from 11 patients. Higher levels of miR-196a were observed in EA, BE, and dysplastic lesions compared with normal squamous mucosa, and in high-grade dysplasia compared with BE and low-grade dysplasia. Using frozen tumor tissues from 10 additional patients who had advanced EA, we evaluated the correlation of miR-196a with its in silico-predicted targets, keratin 5 (KRT5), small proline-rich protein 2C (SPRR2C), and S100 calcium-binding protein A9 (S100A9), which are down-regulated during BE progression. MiR-196a levels inversely correlated with the predicted target mRNA levels in EA. We confirmed that miR-196a specifically targets KRT5, SPRR2C, and S100A9 3′ UTRs using miR-196a-mimic and luciferase reporter-based assays. In conclusion, this study identified miR-196a as a potential marker of progression of BE and KRT5, SPRR2C, and S100A9 as its targets.
The incidence of esophageal adenocarcinoma (EA) has increased by three-to fourfold in the past 3 decades exceeding all other type of cancers.1,2 Patients with EA generally present with advanced stage and despite aggressive treatment the overall 5-year survival is 25%.3 Barrett’s esophagus (BE)/Barrett metaplasia (BM) is a recognized precursor of EA with an intermediate step of dysplasia. At present, histological assessment is the gold standard for risk assessment of dysplasia or EA in BE patients. Esophagogastroduodenoscopy (EGD) with four quadrant biopsies to detect dysplasia and early invasive adenocarcinoma is the only regular screening method currently recommended for patients with BE. However, low yield and high cost of EGD screening necessitates a search for better methods and biomarkers to identify high-risk patients. Markers such as aneuploidy, tetraploidy, and p16 and p53 genetic abnormalities (loss of heterozygosity of chromosomes 9p and 17p, respectively) have been correlated with risk assessment.4,5 However, despite extensive studies these markers have been of limited clinical utility, warranting the exploration of novel biomarkers such as microRNA in BE and its progression to dysplasia and carcinoma.
MicroRNAs (miRNAs) are small (16 to 29 nucleotides) RNA molecules that are part of noncoding evolutionarily conserved class of endogenous riboregulators that alter the gene expression through target mRNA degradation or by reducing the translation of target mRNA by binding to their 3′ untranslated region.6 In cancer and metastasis, increasing numbers of miRNAs have been implicated in the deregulation of gene expression, thus establishing them as an important new class of oncogenes and tumor suppressors.7
Gene expression signature analysis, which involves the analysis of global gene expression at mRNA level is widely used for cancer detection and diagnosis and to study progression.8 The relatively recent identification of miRNAs as an additional level of posttranscriptional regulation of gene expression has shifted the focus to evaluating their value as diagnostic and prognostic markers in cancer. To date, every type of tumor analyzed has had a miRNA profile significantly different from that of the corresponding normal tissue.9 Further, a recent study showed that tissue-specific expression pattern of miRNAs also makes them valuable in the identification of tissue origin in unknown primary cancers.10 Recent studies have identified the tumor suppressor or oncogenic role of miRNAs, whose expression or functions are deregulated during the neoplastic transformation of tissues.11
Recently, we reported that miRNA-196a (miR-196a) levels were much higher in EA than in normal esophageal and gastric mucosal tissues and that ANXA1, a potential tumor suppressor gene is a direct target of miR-196a.12 miR-196a has been shown to have a defined biological function in hindlimb development by acting on HOXB and sonic hedgehog (Shh) signaling13 and in the pathogenesis of acute myeloid leukemia.14 Furthermore, recent studies have demonstrated that miR-196a levels allowed discrimination of normal pancreas from chronic pancreatitis and adenocarcinoma and that miR-196a levels inversely correlated with survival in pancreatic adenocarcinoma patients.15,16
We demonstrated growth-promoting and anti-apoptotic ability of miR-196a in esophageal cancer cell lines suggesting miR-196a may be an oncomir.12 Global gene expression profiling studies revealed that several genes, including ANXA1, show gradual decrease in expression during progression of BE to EA.17,18 Interestingly, many of these genes were potential in silico targets of miR-196a. These genes also included some that are down-regulated in tumors resistant to preoperative chemoradiation in our previous studies (Table 1).19,20
Table 1.
Potential Targets of miR-196a Down-Regulated during Progression of BE-EA
| Gene symbol | Gene name |
|---|---|
| ANXA1* | Annexin A1 |
| S100A9* | S100 calcium-binding protein A9 |
| SPRR2C* | Small proline-rich protein 2C |
| KRT5* | Keratin 5 |
| CLCA2* | CLCA family member 2, chloride channel regulator |
| CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 |
| KRT4* | Keratin 4 |
| LDOC1 | Leucine zipper, down-regulated in cancer 1 |
| LTA4H | Leukotriene A4 hydrolase |
| PTN* | Pleiotrophin |
| MAL | Mal, T-cell differentiation protein |
| TPD52L1 | Tumor protein D52-like 1 |
| VSNL1 | Visinin-like 1 |
Increased miR-196a levels in EA, the oncogenic potential of miR-196a,12 and the down-regulation of several in silico predicted miR-196a targets during progression of EA17,18 prompted us to investigate the potential of this miRNA as a marker of neoplastic progression from normal esophageal mucosa to EA. With this aim, we evaluated alterations in the levels of miR-196a in normal squamous mucosa (NSM), BE, dysplastic lesions, and EA.
We selected three miR-196a in silico targets, S100 calcium-binding protein A9 (S100A9), small proline-rich protein 2C (SPRR2C), and keratin 5 (KRT5), on the basis of their reported down-regulation during BE progression to EA and assessed correlation between their mRNA levels with miR-196a levels in cancers. S100A9 belongs to the S-100 family of calcium-binding proteins that are evolutionarily conserved and have a role in cell proliferation, differentiation, and migration.21 SPRR2C belongs to the family of small proline-rich (SPRR) proteins that function as crosslinkers of epidermal differentiation complex proteins.22 This family of genes is known to be up-regulated during inflammation, infection, and epithelial barrier remodeling23 and is functionally involved in protecting the epithelia from environmental insults. KRT5 is a structural protein that belongs to the family of cytokeratins and is down-regulated in EA.24 To evaluate whether KRT5, S100A9, and SPRR2C genes are true cellular targets of miR-196a, we transfected cells derived from EA with a miR-196a mimic and used luciferase reporter functional assays. Our results show gradual increase in miR-196a levels from NSM-BE-EA and suggest a role of miR-196a as a negative modulator of KRT5, S100A9, and SPRR2C genes in EA.
Materials and Methods
Clinical and Pathological Characteristics and Patient Samples in the Progression Study
We searched the institutional database of the Department of Pathology at The University of Texas M.D. Anderson Cancer Center and selected 11 patients who were diagnosed with early EA and underwent esophagogastrectomy without preoperative neoadjuvant therapy. A detailed retrospective chart review was performed to document stage, EGD findings, and treatment. The study was approved by the institutional review board with waiver of informed consent.
Endoscopic ultrasonography, EGD, computed tomography (CT), and positron emission tomography scan findings were reviewed for location and stage of tumor, and the presence and extent of BE. All patients were retrospectively clinically staged as per the sixth edition of the American Joint Commission on Cancer staging system.25
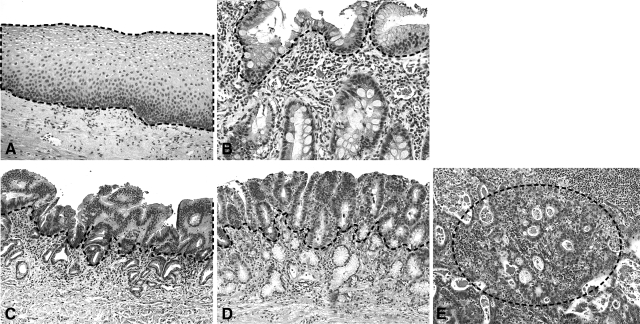
Hematoxylin and eosin (H&E)-stained slides from multiple preoperative biopsies and esophagogastrectomy specimens were reviewed for presence of BE, degree of dysplasia, and histological subtype of carcinoma. In addition, the depth of invasion and regional lymph node status were evaluated in 11 resection specimens for early EA with mucosal or submucosal invasion. The resection specimens were reviewed by two gastrointestinal pathologists to identify the foci of nonneoplastic squamous mucosa (NSM), BE, low-grade dysplasia (LGD), high-grade dysplasia (HGD), and invasive adenocarcinoma (a representative case is shown in Figure 1). In these 11 patients, adequate material was available from NSM, BE, LGD, and HGD. In addition, the adenocarcinoma component was available in six patients. Eight slides with 5-μm-thick sections were prepared from the formalin-fixed paraffin block without trimming the block for each lesion from the surgical resection specimens. These slides were H&E-stained and the lesion of interest was manually microdissected with minimal to absent contamination of stromal cells. Because the NSM, BE, and dysplastic lesions are surface lesions, pure epithelial cells are easily microdissected from a glass slide. From the cases of invasive adenocarcinoma, areas with more than 90% adenocarcinoma cells were selected. Figure 1 shows the plane of microdissection in NSM and in each lesion. In two patients (nos. 3 and 6), two to three areas from BE, LGD, HGD, and early EA were microdissected from sections prepared from paraffin blocks. In other patients, it was not possible to have multiple blocks of each lesion because extent of LGD, HGD, and EA was limited.
Figure 1.
Morphology of the different progression stages of EA. H&E-stained sections showing stages of progression from normal squamous (A), BE (B), LGD (C), HGD (D) to invasive adenocarcinoma (E) from a representative case. The dotted line indicates areas used in manual microdissection. Original magnifications, ×200.
Identification of miR-196a Targets and Clinical and Pathological Characteristics of Patient Samples for miR-196a and Its Target Correlative Study
From the list of genes down-regulated during BE progression to EA in the study of Kimchi and colleagues,17 we identified 13 genes (Table 1) as potential targets of miR-196a on the basis of the sequence complementarity between their 3′-untranslated regions (3′-UTR) and miR-196a using the Sanger microRNA database (http://microrna.sanger.ac.uk/sequences/). Interestingly, we noted that 7 of the 13 miR-196a in silico targets were also down-regulated in tumors resistant to preoperative chemoradiation (Table 1).19,20 To determine whether increased miR-196a levels correlate with decreased expression levels of computationally predicted targets, we measured S100A9, SPRR2C, and KRT5, mRNA levels in fresh frozen-tissue from 10 additional patients who had advanced EA. The selection of the target genes was based on their down-regulation during BE progression to EA17 and in EA.19,20,24
Because the specimens used in the current progression studies were from early cancers, it was not possible to harvest fresh tumor tissue for miR-196a and mRNA because of inability to identify invasive carcinoma by gross examination. Thus, specimens for target correlative study included fresh frozen and histologically confirmed adenocarcinomas from patients who had advanced loco-regional disease. Five of the ten patients for the target correlative study were included in our prior study.19
Real-Time Quantitative Polymerase Chain Reaction (Real-Time qPCR) for miRNA Expression Analysis
Total RNA from microdissected tissue was isolated by using the RecoverALL total nucleic acid isolation kit (Ambion/Applied Biosciences, Austin, TX) according to the manufacturer’s instructions. The RNA yield was measured using Nanodrop (Thermo Scientific, Wilmington, DE). The levels of miR-196a and miR-16 were determined by stem loop real-time qPCR using gene-specific TaqMan minor groove binding primers according to the TaqMan MicroRNA assay protocol (PE Applied Biosystems, Foster City, CA). miRNA levels in each sample were analyzed in duplicate reactions. The reverse transcription (RT) reaction was performed with 50 ng of total RNA from each specimen in a total volume of 7.5 μl using 50 nmol/L gene-specific stem-loop primer, 1× RT buffer, 0.25 mmol/L each of d NTPs, 3.33 U/μl MultiScribe reverse transcriptase, and 0.25 U/μl RNase inhibitor (PE Applied Biosystems). The reaction mix was incubated in an Applied Biosystems 9800 ThermoCycler in a 96-well plate for 30 minutes at 16°C, 30 minutes at 42°C, and 5 minutes at 85°C, and then held at 4°C. In two patients with multiple samples of each histology, miRNA levels were assessed from each separately microdissected sample.
Real-time qPCR was performed using an Applied Biosystems 7900 sequence detection system in a 10-μl volume that included 0.67 ml of RT product, 1× TaqMan Universal PCR master mix, and 1 μl of gene-specific primers and probe mix from the TaqMan microRNA assay. The PCR thermal cycling conditions were as follows: 10 minutes at 95°C for AmpliTaq Gold activation and 40 cycles for the melting (95°C, 15 seconds) and annealing/extension (60°C for 1 minute) steps. Each miRNA was analyzed in duplicate reactions. Default threshold settings were used to determine threshold cycle (CT).
Comparative CT Method (2−ΔCT) for Relative Quantification of miRNA Expression
We used miR-16 as normalizer because this miRNA showed minimal variation in esophageal, breast, and endometrial cell lines12 and also in our pilot miR profiling studies of esophageal cancers. The relative expression levels of each miRNA in comparison with the normalizer were then calculated using the formula 2−ΔCT where ΔCT represents the difference between each target gene and the normalizer (average CT for the target minus average CT for miR-16).
Quantitation of SPRR2C, S100A9, and KRT5 mRNA Levels in an EA Cell Line Transfected with miR-196a Mimic
The base complementarity of miR-196a with 3′-untranslated region (3′-UTR) of SPRR2C, S100A9, and KRT5 is shown in Figure 2. To evaluate if overexpression of miR-196a resulted in reduction of these potential target mRNA levels, the EA cell line BIC-1 was transfected with a RNA mimic of miR-196a purchased from Dharmacon, Inc. (Chicago, IL). The miRNA mimic is a synthetic double-stranded RNA oligonucleotide, which on delivery generates higher levels of respective miRNA in cells. The mimic was transfected into the cultured cells using DharmaFECT Duo transfection reagent (Dharmacon, Inc.) at a final concentration of 40 nmol/L. After 48 hours, total RNA was extracted from the cells and the levels of miR-196a and SPRR2C, S100A9, and KRT5 mRNA were measured by real-time qPCR analysis, as described above. A nonspecific miRNA mimic (Dharmacon, Inc.) was used as an appropriate negative control.
Figure 2.
Predicted base complementarity of miR-196a to S100A9, SPRR2C, and KRT5. The base complementarity of miR-196a to 3′-UTR binding sites of S100A9, SPRR2C, and KRT5 as predicted by Sanger miR-database (http://microrna.sanger.ac.uk/sequences/).
Generation of the Target 3′-Untranslated Region (3′-UTR) Luciferase Reporter Constructs
To further confirm that KRT5, SPR2C, and S100A9 are true in vitro targets of miR-196a, regions of 3′-UTR of these genes containing the putative miR-196a recognition site (Figure 2) were amplified using genomic DNA and then cloned into the pGL3 control vector (Promega Corp., Madison, WI) at the XbaI site and confirmed by sequencing. The size of the fragment amplified and the primers used for amplification are shown in Table 2. Two esophageal cancer cell lines, namely BIC1 (with relatively high endogenous miR-196a levels) and OE33 (with relatively low endogenous mir-196a levels) were used for luciferase assay. Co-transfection of the plasmid and the RNA mimics into the cells was accomplished using DharmaFECT Duo transfection reagent (Dharmacon, Inc.). A nonspecific miRNA mimic was used as an appropriate negative control. Luciferase activity was measured 48 hours after transfection using the Dual Luciferase Reporter Assay System kit (Promega Corp.). Luciferase assays were done in triplicate. For each target, the luciferase assay was performed twice in each cell line to confirm the effect of miR-196a overexpression.
Table 2.
Primers Used to Amplify the 3′-UTR of the miR-196a Target Genes
| Gene | Size | Primers |
|---|---|---|
| S100A9 | 365 bp | Forward: 5′-TAGTCTAGAGGTCATAGAACACATC-3′ |
| Reverse: 5′-TAGTCTAGAGACTTGGAGGAAGAGAC-3′ | ||
| SPRR2C | 365 bp | Forward: 5′-TAGTCTAGACAGCTTCGGAATTCATC-3′ |
| Reverse: 5′-TAGTCTAGAGCTACTTTATTCAGGGAG-3′ | ||
| KRT5 | 373 bp | Forward: 5′-TAGACTAGTGAACCTGCTGCAAGT-3′ |
| Reverse: 5′-TAGACTAGTATATTATAAAAGCAT-3′ |
Size of the 3′-UTR region of the three target genes with the potential binding region of miR-196a and the primers used to amplify these regions from the genomic DNA.
Quantitative Analysis of SPRR2C, S100A9, and KRT5 mRNA Levels in Tumors
For cDNA synthesis, 200 ng of total RNA from each adenocarcinoma sample was reverse-transcribed in a final volume of 20 μl using random primers and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). The TaqMan minor groove binder probe and the ABI Prism 7900 HT sequence detection system (PE Applied Biosystems) were used to perform real-time qPCR. The primers and probes for SPRR2C, S100A9, KRT5 and an internal control, glucuronidase-β (GUSB), were obtained from PE Applied Biosystems via their Assays-on-Demand gene expression products services. PCR assays included 10 μl of TaqMan Universal Master Mix No Amperase UNG (2×), 1 μl of 20× Assays-on-Demand Gene Expression Assay Mix, and 2 μl of cDNA diluted in RNase-free water in a final volume of 20 μl. The ABI Prism 7900 HT sequence detection system and cycling conditions identical to those described above for miRNA were used for mRNA expression analysis. Each target was amplified individually and in duplicate. The relative levels of each target were then calculated using average of the duplicates on the basis of the difference between amplification of the target and GUSB mRNA using the ΔCT method as described above. The qPCR data for both miR-196a as well as its targets were averages of duplicate reactions from a single experiment.
Statistical Methods
One-way within-subjects analysis of variance was used to test against the null hypothesis that there is no overall difference in relative expression levels among normal, precancerous, and cancerous tissue.26 For statistically significant analysis of variance results, Student’s t-tests were performed to further test the difference of miRNA expression between any two neoplastic progression stages. Holm’s method was applied to adjust P values of t-tests to correct multiple comparisons.27 Relative expression levels were log-transformed before analysis. P values ≤0.05 were considered statistically significant. Spearman’s rank correlation coefficient was used to measure the rank-based association between miR-196a levels and SPRR2C, S100A9, and KRT5 mRNA levels among the 10 cancer specimens. To assess the significance of coefficients, the P values were computed using algorithm AS 89.28
Results
The study population for miRNA analysis in progression specimens comprised men with an average age of 63 years (range, 57 to 74 years). All patients had long-segment BE on EGD and resection specimens with early EA. Seven cases had submucosal invasion and four cases had intramucosal invasion. Eight tumors were moderately differentiated, two were well-differentiated, and one tumor was poorly differentiated. Representative histology of different stages of progression of EA are shown in Figure 1.
The patient population for the correlative study of miR-196a and its targets included nine men and one woman with an average age of 62 years (range, 40 to 79 years). All patients had advanced loco-regional disease (stage II or III) on pretreatment staging. Nine tumors were moderately differentiated and one tumor was poorly differentiated. No tumor in either population had signet ring cell histology on pretreatment biopsies.
miR-196a Levels during Progression of BE-Dysplasia-EA
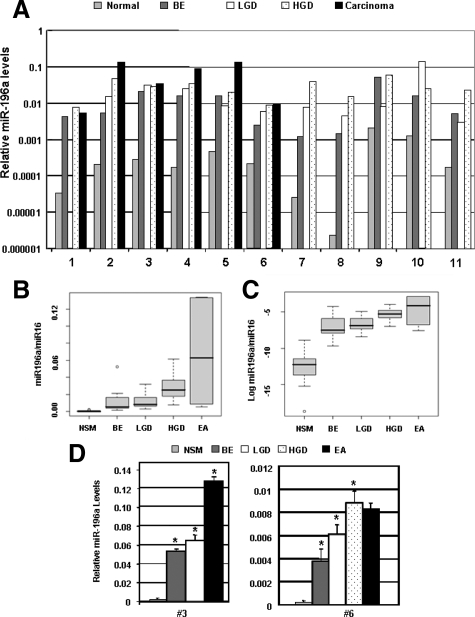
The relative expression levels of miR-196a in each histological type of lesion at different stages of progression in 11 patients are shown in Figure 3A and in Table 3. miR-196a levels increased incrementally with each stage of progression from normal mucosa to EA. The box plot of relative levels of miR-196a for the samples analyzed (Figure 3, B and C) reflects the progressive increase in miR-196a levels and illustrates that the progression of NSM-BE-LGD-HGD-EA was associated with a concomitant increase of miR-196a levels. The differences among different stages were statistically significant by a one-way within-subjects analysis of variance with P < 0.0001. In patients with EA available for microdissection, a 10- to 100-fold increase in miR-196a levels was present in EA when compared with the NSM as reflected by pair wise comparisons (P = 0.0005, Student’s t-test with P values adjusted by the Holm’s method). The pair wise comparisons also indicated that the miR-196a levels in precancerous lesion were significantly higher than the preceding precursor lesion or control squamous mucosa: NSM versus BE (P = 0.00001), versus LGD (P = 0.0005), versus HGD (P = 0.00002); BE versus HGD (P = 0.03); and LGD versus HGD (P = 0.01). To determine whether miR-196a levels varied among different areas with same histology in the same patient, we measured miR-196a in two to three separately microdissected sites for each histology in two additional patients (Figure 3D). As illustrated in the figure, there was minimal variation in the mean levels of miRNA among multiple sites of same histology. In case 3, the mean levels ± SD were 0.05 ± 0.002 in BE, 0.06 ± 0.006 in LGD, and 0.1 ± 0.005 in EA. The increases in miR-196a levels in BE compared with LGD and in EA compared with LGD were statistically significant with P = 0.03 and P = 0.0002, respectively. In case 6, the mean miRNA levels ± SD from different sites were 0.004 ± 0.001 in BE, 0.006 ± 0.0008 in LGD, 0.009 ± 0.001 in HGD, and 0.008 ± 0.0005 in EA. The miRNA levels were significantly higher in LGD compared with BE (P = 0.01) and in HGD compared with LGD (P = 0.02).
Figure 3.
Micro-RNA 196a levels are characteristically up-regulated with progression of EA. A: Real-time qRT PCR analysis of miR-196a levels in each lesion in each patient. Each bar represents average miRNA expression level of duplicate reactions of a single experiment. The figure shows an increase in the levels of miR-196a with neoplastic progression of normal esophageal mucosa (N) to adenocarcinoma (EA). The greatest increase in the levels of miR-196a was observed between N and BE with smaller increases in subsequent stages of neoplastic histological progression from BE to low-grade (LGD), LGD to high-grade dysplasia (HGD), and to EA. B: The box plots of the average levels of miR-196a at each stage of progression in the 11 patients. Each box shows the variation of relative values of miRs and the black horizontal bar shows the mean value in each box. C: Box plots of the log transformed relative values of miR-196a during progression. D: Analysis of miR-196a levels from multiple separately microdissected samples with same histology in two patients. RNA was isolated from two to three different areas of same histological stage of progression, ie, BE, LGD, HGD, and EA and miR-196a levels were measured by qRT-PCR separately from each site in duplicates. Each bar represents mean miR-196a level (±SD) from multiple sites within same histology from a single experiment. As illustrated in the figure, there was minimal variation in the levels of miRNA among multiple sites of same histology. Asterisks indicate miRNA levels were significantly higher in BE compared with NSM, LGD compared with BE, and in HGD/EA compared with LGD (P < 0.05).
Table 3.
MicroRNA-196a Levels in Progression from BE to Early Adenocarcinoma
| Mean ± SD | Range | |
|---|---|---|
| Normal squamous mucosa | 0.00045 ± 0.00065 | 2.4e-06 to 0.002 |
| Barrett’s esophagus (intestinal metaplasia) | 0.013 ± 0.015 | 0.001 to 0.052 |
| Low-grade dysplasia | 0.014 ± 0.010 | 0.003 to 0.032 |
| High-grade dysplasia | 0.028 ± 0.016 | 0.008 to 0.061 |
| Early adenocarcinoma | 0.068 ± 0.060 | 0.005 to 0.134 |
Summary of miR-196a levels in different progression stages leading to EA from the set of 11 patients. A trend of consistent increase in miR-196a levels during progression is evident.
Correlation of miR-196a Levels with mRNA Levels of Predicted Targets, SPRR2C, S100A9, and KRT5 in EA
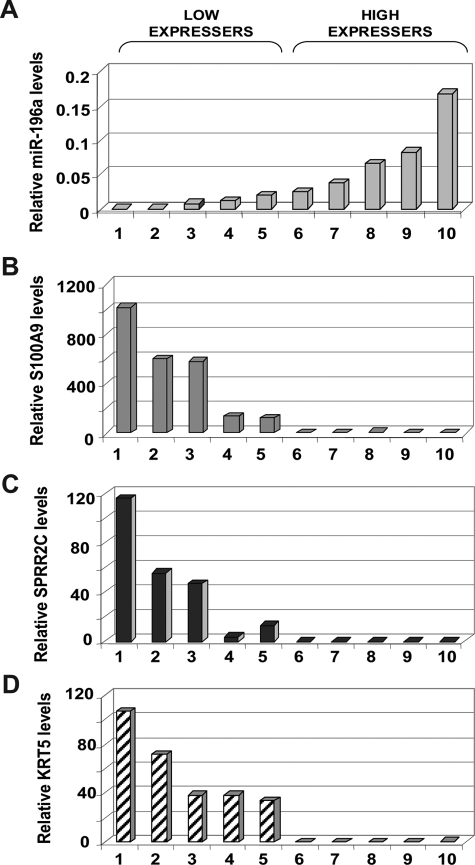
To test whether down-regulation of SPRR2C, S100A9, and KRT5 genes during the progression of BE-EA17 is attributable to increased miR-196a levels, we tested the correlation between their mRNA levels to mR-196a levels in 10 additional patients who had advanced EA. miR-196a levels varied considerably in these tumors specimens, ranging from 0.0009 to 1.7 and the specimens were arbitrarily separated into two categories expressing relatively low (samples 1 to 5) and high (samples 6 to 10) levels of miR-196a (Figure 4A). Although, the mean miR-196a level ± SD was comparatively low in the low-expressing category of tumors (0.008 ± 0.008), this level was substantially higher in comparison with the mean miR-196a levels in the normal mucosa (0.0005 ± 0.0007). miR-196a levels in early EA (paraffin-embedded tissue, 0.08 ± 0.06) and advanced EA (frozen tissue, 0.04 ± 0.05) were not significantly different (P = 0.26 by Student’s t-test).
Figure 4.
Inverse correlation of miR-196a levels with mRNA levels of S100A2, SPRR2C, and KRT5, three genes characteristically down-regulated in EA. A: Relative miR-196a levels in advanced EAs from 10 patients as measured by real-time qPCR. The relative levels showed considerable variation among EA, so the EA specimens were arbitrarily grouped into low- (samples 1 to 5) and high-expressing (6 to 10) tumors. The relative mRNA levels of S100A2 (B), SPRR2C (C), and KRT5 (D) in the same samples were inversely correlated with the relative levels of miR-196a. Each data point represents mean expression levels of duplicates from a single experiment.
The mRNA levels of S100A9, SPRR2C, and KRT5 also varied significantly among the tumors with levels ranging from 0.15 to 1021.87 (246.4 ± 359.3), from 0.0 to 115.3 (23.46 ± 38.36), and from 0.0 to 106.68 (29.0 ± 36.87), respectively (Figure 4, B–D). The tumor specimens with low miR-196a expression showed high mRNA levels of SPRR2C, S100A9, and KRT5. Conversely, specimens with high miR-196a levels showed low expression of its targets. For instance, the mean mRNA levels (±SD) of SPRR2C, S100A9, and KRT5 were 47.1 ± 44.2 (range, 3.78 to 115.3), 491.4 ± 374.8 (range, 123.6 to 1021.9), and 57.9 ± 31.2 (range, 33.63 to 106.7), respectively, in low expressers of miR-196a. On the other hand, in the miR-196a high-expressing category of tumors, the mean mRNA levels of SPRR2C, S100A9, and KRT5 were 0.003 ± 0.005, 1.5 ± 1.78, and 0.16 ± 0.35, respectively. The mRNA levels of the targets correlated inversely with miR-196a levels (Spearman’s rank correlation coefficients of −0.77, −0.81, and −0.78 for SPRR2C, S100A9, and KRT5, respectively, P < 0.01). The mRNA levels of these three genes were also analyzed in a set of three random NSM samples to compare their expression levels in NSM with that in tumors. Our analysis showed that the average mRNA levels of these genes in NSM were higher than the levels in the set of 10 tumors examined. The average levels (±SD) of S100A9 (1482.4 ± 344.0) and KRT5 (218.5 ± 120.7) in NSM were higher than the highest levels seen in the tumors, ie, 1021.9 for S100A9 and 106.7 for KRT5. In the case of SPRR2C, the average levels observed in NSM (58.7 ± 39.0) was higher than the levels in all tumor samples except for the highest expresser (115.3). This comparison indicated that the mRNA levels of S100A9, SPRR2, and KRT5 were suppressed during neoplastic transformation of esophageal tissue.
miR-196a Mimic Suppresses SPRR2C, S100A9, and KRT5 mRNA Levels in Esophageal Cancer Cell Line
The inverse correlation observed between the levels of miR-196a and mRNAs of the three genes suggests that these mRNAs are likely targets of mR-196a. The sequence complementarity of miR-196a and computationally identified 3′-UTR binding sites of these three mRNAs is shown in Figure 2. To further confirm that these mRNAs are indeed in vitro cellular targets, we tested the effect of increasing the levels of miR-196a on the mRNA levels of these genes in a esophageal cancer cell line. This was achieved by the transfection of a RNA mimic of miR-196a into BIC1 cell line derived from EA. The miR-196a miRNA mimic was a synthetic RNA oligonucleotide that on delivery generated higher levels of miR-196a in the cells. Increasing miR-196a levels in BIC-1 cells for 48 hours with the mimic resulted in 69%, 98%, and 20% decrease in S100A9, SPRR2C, and KRT5 mRNA levels, respectively, compared with the respective controls wherein the cells were transfected with a nonspecific negative control RNA mimic (Figure 5).
Figure 5.
Elevation of miR-196a levels in EA cells suppresses expression of S100A9, SPRR2C, and KRT5. Elevating the levels of miR-196a by transfection of RNA mimics in the EA cell line BIC1 resulted in suppression of the mRNA levels of all three genes, as measured by real-time qPCR assay. The relative mRNA levels of these genes in BIC1 cells transfected with a nonspecific miRNA mimic was considered as the respective control. Each data point represents average expression levels of duplicates from a single experiment.
RNA Mimics of miR-196a Directly Targets the 3′ Untranslated Regions (UTR) of SPRR2C, S100A9, and KRT5 mRNA
We used a luciferase-based assay to further confirm the direct targeting of SPRR2C, S100A9, and KRT5 mRNAs by miR-196a. We cloned the 3′-UTRs of these targets that included the miR-196a binding sites (shown in Figure 2), into a PGL3 luciferase reporter plasmid and tested the effect of increasing miR-196a levels on luciferase expression. The schematic representation of the principle behind the assay is depicted in Figure 6A. In esophageal cancer cell lines, BIC1 and OE33, increasing the level of miR-196a consistently resulted in a reproducible and considerable decrease (60 to 90%) in the luciferase activity compared with the negative mimic control in all three genes in two independent experiments (a representative experiment is shown in Figure 6B). The luciferase assay thus confirmed that these three genes are direct targets of miR-196a.
Figure 6.
SPRR2C, S100A9, and KRT5 mRNAs are direct targets of miR-196a. A: The schematic depiction of luciferase assay. To demonstrate the direct targeting of these mRNA by miR-196a, the 3′-UTRs of the genes with the predicted miR-196a binding sites were cloned upstream of the luciferase gene in PGL3 vector plasmid. When transfected into cells the mRNA transcript of the luciferase generated from the vector plasmid has the 3′-UTR of the gene of interest, and the additional miR-196a generated from the RNA mimics binds to the luciferase mRNA and suppresses its levels, resulting in a decrease in luciferase activity. B: The plasmid and RNA mimics of miR-196a were co-transfected into BIC1 and OE33 cells and the effect of miR-196a on luciferase gene transcription was measured by luciferase assay. The figure shows mean ± SD of triplicates from a single experiment. Repression of luciferase activity demonstrated the direct binding and repressive effect of miR-196a on the mRNA of these three genes. A nonspecific miRNA mimic co-transfected with the luciferase plasmid was used as the respective control. The decrease in the luciferase activity was statistically significant for all targets (P < 0.05). The luciferase assay for all three genes was repeated twice in both cell lines and a representative result is shown here.
Discussion
In the present study, we demonstrate that miR-196a has the strong potential of being a biomarker of BE progression. To our knowledge, this is the first study demonstrating a microRNA marker in the progression of EA. We have also shown that miR-196a potentially plays a role in the down-regulation of SPRR2C, S100A9, and KRT5 genes whose expression is characteristically decreased or lost during neoplastic transformation of esophageal tissue. We also demonstrated significance of miR-196a in EA using paraffin-embedded tissue from patients with BE-dysplasia-early EA sequence and in frozen tissue from patients who had advanced EA.
We quantitatively assessed levels of miR-196a in BE, different grades of dysplasia, and EA and found that miR-196a levels are 10- to 100-fold higher in precancerous lesions and EA than in NSM and that levels of miR-196a proportionally increase with higher histological grades of dysplasia. Higher levels of miR-196a in each lesion compared with control nonneoplastic squamous mucosa suggests that miR-196a alteration is an early event in carcinogenesis of EA. In addition, the higher miR-196a levels in HGD as compared with BE and LGD in patients who had invasive adenocarcinoma suggest that miR-196a levels can be helpful in differentiating LGD from HGD in a high-risk population with progression of BE to invasive adenocarcinoma. The marginal difference in miR-196a levels between HGD and invasive adenocarcinoma in the cases analyzed could be attributable to small number of early invasive carcinoma cases used in this study or to more stromal contamination in invasive component compared with that in dysplastic lesions, although these were carefully microdissected. However, it may also indicate that the increase in miR-196a is an early effect that is most marked at the NSM-BE transition and LGD-HGD/invasive EA transition and in advanced EA.
In this study all patients of progression cases had long segment BE comprising of intestinal metaplasia and extensive dysplasia. It would have been interesting to compare the miR-196a levels between intestinal metaplasia and cardia type mucosa (nondistinctive BE) in the tubular esophagus. However, the absence of cardia type mucosa in tubular esophagus separate from admixed intestinal metaplasia in the cases investigated here precluded such analysis in our study. The precancerous potential of cardia type mucosa in tubular esophagus (nondistinctive type BE) is not well established. However, miR-196a levels in cardia type mucosa may address the question if miR-196a is a marker of BE irrespective of presence or absence of intestinal metaplasia. This may be a valuable marker in short-segment BE or in differentiating nondistinctive type of BE in tubular esophagus from gastric cardia, a much debated and controversial subject but of great importance for selecting patients for screening and surveillance.
The functional role of miR-196a demonstrated in EA cell lines and decreased target mRNA levels in EA tissue specimens further support the role of miR-196a in pathogenesis of EA and in down-regulation of KRT5, SPRR2C, and S100A9 genes in EA. This group of genes has been shown to be down-regulated in EA in more than one study.17,20 S100A9 belongs to multigene family that codes for calcium-binding proteins and functions as a heterodimer with S100A8. Changes in the levels of both proteins are being increasingly implicated in epithelial cancers. Although in squamous cell esophageal carcinomas, the loss of S100A8 and S100A9 is very well documented.29 It has also been shown that the S100A9 levels correlate with the extent of tumor differentiation.30
In a recent study, we have demonstrated that annexin A1, a potential tumor suppressor is an in vivo target of miR-196a in EA.12 In that study, we also showed that miR-196a promotes cell proliferation and anchorage-independent growth and inhibits apoptosis in EA cell lines. These findings suggested that miR-196a plays an important role in carcinogenesis of EA.
It is interesting to note that some of the genes such as GATA-binding protein 6 (GATA6), and HOXB7 are also predicted targets of miR-196a (Table 1) but are up-regulated in normal versus BE, and BE versus EA in the study of Kimchi and colleagues.17 However, no negative correlation between miR-196a levels and HOXB7 or GATA6 mRNA levels was observed in tumors in our study (data not shown). This suggests that miR-196a may not be directly involved in the regulation of this set of genes during the progression of EA, and that these genes may have some other mode of regulation. Decreased levels of the target mRNA in miR-196a mimic transfected cell lines, despite partial complementarity between the two indicate that degradation of the target mRNA can occur even with partial complementarity between the miR and target mRNA, as recently reported in several studies.12,31,32,33
This study demonstrates successful utilization of paraffin tissue for quantitative miRNA assay. This is particularly important in esophageal cancer in which availability of fresh tissue is very limited because most of the patients, at least in our institution, who undergo esophagectomy have had prior chemoradiation. Minimal variation seen in the levels of miR-196a among multiple microdissected samples with the same histology from an individual patient further confirms that there is little heterogeneity among different areas within the same histology in a patient and the technique used in this study is suitable for miRNA studies in archival paraffin tissue.
We believe that the confirmation of miR-196a as a marker of progression of BE to EA would fill the lacuna in the identification of reliable molecular markers of progression in esophageal cancers. Also, in light of the potential growth-promoting effects of miR-196a, the observed incremental alterations in miR-196a levels during the progression from BE to EA suggest a causal rather than consequential effect of tumorigenesis in esophageal tissue. Thus, miR-196a could be a potential therapeutic target in the treatment of esophageal cancers as well as a valuable early detection marker. On the basis of these findings and the potential functions of miR-196a as an oncomir, further study of miR-196a in larger and clinically more diverse subsets of patients with BE is warranted.
Acknowledgments
We thank Dawn Chalaire for critical editing of the manuscript and James M. Gilbert for assistance with preparation of the manuscript and illustrations.
Footnotes
Address reprint requests to Rajyalakshmi Luthra, Ph.D., the University of Texas M.D. Anderson Cancer Center, 8515 Fannin St., NAO1.061a, Houston, TX 77054. E-mail: rluthra@mdanderson.org.
Supported by the University of Texas M.D. Anderson Cancer Center, Division of Pathology and Laboratory Medicine (fellow research funding to C.H.).
D.M.M., R.R.S., and C.H. contributed equally to the study.
References
- Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–2053. [PubMed] [Google Scholar]
- Shaheen NJ, Crosby MA, Bozymski EM, Sandler RS. Is there publication bias in the reporting of cancer risk in Barrett’s esophagus? Gastroenterology. 2000;119:333–338. doi: 10.1053/gast.2000.9302. [DOI] [PubMed] [Google Scholar]
- Swisher SG, Hunt KK, Holmes EC, Zinner MJ, McFadden DW. Changes in the surgical management of esophageal cancer from 1970 to 1993. Am J Surg. 1995;169:609–614. doi: 10.1016/s0002-9610(99)80231-1. [DOI] [PubMed] [Google Scholar]
- Reid BJ, Prevo LJ, Galipeau PC, Sanchez CA, Longton G, Levine DS, Blount PL, Rabinovitch PS. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BJ, Blount PL, Rabinovitch PS. Biomarkers in Barrett’s esophagus. Gastrointest Endosc Clin N Am. 2003;13:369–397. doi: 10.1016/s1052-5157(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Tricoli JV, Jacobson JW. MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru D, Broaddus RR, Rashid A, Albarracin CT. MicroRNA-196a targets annexin A1: a micro-RNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, Khodarev NN. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- Selaru FM, Zou T, Xu Y, Shustova V, Yin J, Mori Y, Sato F, Wang S, Olaru A, Shibata D, Greenwald BD, Krasna MJ, Abraham JM, Meltzer SJ. Global gene expression profiling in Barrett’s esophagus and esophageal cancer: a comparative analysis using cDNA microarrays. Oncogene. 2002;21:475–478. doi: 10.1038/sj.onc.1205111. [DOI] [PubMed] [Google Scholar]
- Luthra MG, Ajani JA, Izzo J, Ensor J, Wu TT, Rashid A, Zhang L, Phan A, Fukami N, Luthra R. Decreased expression of gene cluster at chromosome 1q21 defines molecular subgroups of chemoradiotherapy response in esophageal cancers. Clin Cancer Res. 2007;13:912–919. doi: 10.1158/1078-0432.CCR-06-1577. [DOI] [PubMed] [Google Scholar]
- Luthra R, Wu TT, Luthra MG, Izzo J, Lopez-Alvarez E, Zhang L, Bailey J, Lee JH, Bresalier R, Rashid A, Swisher SG, Ajani JA. Gene expression profiling of localized esophageal carcinomas: association with pathologic response to preoperative chemoradiation. J Clin Oncol. 2006;24:259–267. doi: 10.1200/JCO.2005.03.3688. [DOI] [PubMed] [Google Scholar]
- Schäfer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- Patel S, Kartasova T, Segre JA. Mouse Sprr locus: a tandem array of coordinately regulated genes. Mamm Genome. 2003;14:140–148. doi: 10.1007/s00335-002-2205-4. [DOI] [PubMed] [Google Scholar]
- Cabral A, Voskamp P, Cleton-Jansen AM, South A, Nizetic D, Backendorf C. Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J Biol Chem. 2001;276:19231–19237. doi: 10.1074/jbc.M100336200. [DOI] [PubMed] [Google Scholar]
- Sato F, Abraham JM, Yin J, Kan T, Ito T, Mori Y, Hamilton JP, Jin Z, Cheng Y, Paun B, Berki AT, Wang S, Shimada Y, Meltzer SJ. Polo-like kinase and survivin are esophageal tumor-specific promoters. Biochem Biophys Res Commun. 2006;342:465–471. doi: 10.1016/j.bbrc.2006.01.177. [DOI] [PubMed] [Google Scholar]
- New York: Springer; SpringerAmerican Joint Committee on Cancer Staging Atlas, (ed 6.) 2002:pp 91–95. [Google Scholar]
- Chambers JM, Hastie TJ. Statistical Models. Florence: Wadsworth and Brooks/Cole; 1992:pp 145–163. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Best DJ, Roberts DE. Algorithm AS 89: the upper tail probabilities for Spearman’s rho. Appl Stat. 1975;24:377–379. [Google Scholar]
- Wang J, Cai Y, Xu H, Zhao J, Xu X, Han YL, Xu ZX, Chen BS, Hu H, Wu M, Wang MR. Expression of MRP14 gene is frequently down-regulated in Chinese human esophageal cancer. Cell Res. 2004;14:46–53. doi: 10.1038/sj.cr.7290201. [DOI] [PubMed] [Google Scholar]
- Kong JP, Ding F, Zhou CN, Wang XQ, Miao XP, Wu M, Liu ZH. Loss of myeloid-related proteins 8 and myeloid-related proteins 14 expression in human esophageal squamous cell carcinoma correlates with poor differentiation. World J Gastroenterol. 2004;10:1093–1097. doi: 10.3748/wjg.v10.i8.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]