Abstract
There is growing support for the role of genetic factors in the development of retinopathy of prematurity (ROP), a serious visual morbidity resulting from preterm birth. We used both candidate gene and data-mining approaches to investigate the role of genetic polymorphisms in the development of ROP. Our study population consisted of 330 infants, less than 32 wk gestation, and their parents. We initially studied 24 single nucleotide polymorphisms (SNPs) in 11 candidate genes. Using a family-based analysis strategy, we found an association between SNPs in the EPAS1 gene and the development of ROP (p = 0.007). Logistic regression analysis showed three SNPs associated with development of ROP, two in the CFH gene (p = 0.01) and one in the EPAS1 gene (p = 0.001). Extending this analysis to include genotyping data from a larger genetic study of prematurity (455 SNPs in 153 genes), we found SNPs in five genes associated with the development of ROP: IHH (p = 0.003), AGTR1 (p = 0.005), TBX5 (p = 0.003), CETP (p = 0.004), and GP1BA (p = 0.005). Our data suggest that genetic risk factors contribute to the development of ROP.
Retinopathy of prematurity (ROP) is a proliferative vascular disorder of the retina that can lead to visual impairment or complete vision loss in premature infants. Low birth weight infants or those who are born at lower gestational age are particularly vulnerable, with an estimated incidence of 68% in infants who are less than 1250 g (1). In the United States, ROP accounts for 13% of childhood blindness and ranks as one of the top three causes of blindness in children (2,3).
Normal retinal vascularization begins at approximately 16 wk of gestation, beginning at the optic disc and proceeding peripherally, reaching the nasal ora serrata by 36 wk and the temporal ora serrata by 39–41 wk. Normally, retinal vascularization occurs in the fetal environment, characterized by a relatively low oxygen tension. After birth, premature neonates are exposed to a higher oxygen tension, which is required for their intact survival. In the premature extrauterine environment, ROP can be considered to occur in two phases (4). In the first phase, early oxygen exposure causes the developing retina to become hyperoxic, leading to arrest of normal retinal vascularization and regression of developing vessels. In the second phase, as the retina becomes more metabolically active, the avascular areas become relatively hypoxic, leading to the elaboration of angiogenic growth factors that induce abnormal retinal neo-vascularization (4). This vascularization can lead to the formation of a fibrous scar, which can cause retinal detachment and lead to vision impairment or vision loss (4).
In addition to well-established environmental factors, such as early oxygen exposure and gestational age, there is also a strong genetic susceptibility to ROP. A recent twin study evaluating the prevalence of ROP in monozygotic and dizygotic twin pairs demonstrated that when environmental factors were controlled for, there was a significant genetic contribution to the development of ROP (5).
Studies evaluating polymorphisms in specific candidate genes, such as VEGF, have also demonstrated an association between sequence variations and severity of ROP (6-8). Additionally, there have been multiple studies that have evaluated polymorphisms in other genes implicated in proliferative retinopathy-related diseases, such as X-linked familial exudative vitreoretinopathy, Norrie disease, and proliferative diabetic retinopathy, which have shown associations between specific polymorphisms and increased risk of severity of disease (9-12). Adult onset diseases of the retina, such as age-related macular degeneration (AMD), also exhibit a strong genetic component (13).
In this study, we used both hypothesis-generating and hypothesis-testing approaches to examine genetic polymorphisms that may be associated with the development of ROP and with the severity of disease. The infants in this analysis are a defined cohort from a larger study population evaluating possible genetic contributions to prematurity.
METHODS
Sample population
Since 2000, blood or buccal swabs from infants (and their parents) admitted to the neonatal intensive care unit at the University of Iowa Children's Hospital have been collected and banked after obtaining informed consent. This program has been approved by the Institutional Review Board (IRB) at the University of Iowa and is designed to generate samples for use in genetic studies evaluating the etiology and variability of diseases of the infant. A second IRB approval was obtained to access the patient samples and the clinical information required for this study.
DNA processing and genotyping
DNA was extracted from discarded umbilical cord blood for the infants in the analysis and from venous blood or buccal swabs for parental DNA. Allelic variation was determined using the TaqMan genotyping system (Applied Biosystems, Foster City, CA). The genotyping reactions were carried out in 384 well plates containing dried sample DNA, using a protocol slightly modified from the manufacturer's instructions, as previously described (14). Allele scoring was done using the Sequence Detection Systems (SDS, version 2.2) software preloaded on the ABI 7900HT. The genotype data were uploaded into a Progeny (Progeny Software, LLC, South Bend, IN) database containing demographic and clinical information for subsequent statistical analysis.
Genes analyzed
In the first phase of the study, we used a hypothesis-generating (data-mining) approach to examine possible genetic risk factors for ROP. Our study population was a subset of a larger group of preterm infants that underwent genotyping in an effort to find genetic contributions to preterm delivery. Using a subset of these samples (those less than 32 wk gestation), we were able to analyze genotype data on 455 single nucleotide polymorphisms (SNPS) in 153 genes to determine risk factors for ROP.
In the second phase of the study, candidate genes were chosen based on a review of the current literature and biologic plausibility. Candidate genes in the angiogenic, inflammation, and oxygen sensing pathways, as well as those previously associated with other proliferative retinopathic diseases, were specifically chosen for analysis of ROP in preterm infants. Once the gene was identified, SNPs were chosen within that gene for analysis based on haplotype blocks that gave the most thorough coverage of the gene with the fewest SNPs possible. SNPs within a haplotype block were chosen based on their minor allele frequency and their position within the gene. A minor allele frequency of 0.1 was chosen as a cutoff to ensure that an adequate number of individuals within the sample population would be carriers of the minor allele, thus, increasing informativity at that locus. In this phase, 24 SNPs in 11 genes were identified to be genotyped.
Statistical analysis
No one analysis paradigm is optimal for gene discovery in complex disorders like ROP. We chose to use both a family-based association strategy and regression analysis to fully characterize genetic contributions to ROP. A family-based association test (FBAT) was performed on all markers. Parental transmission of alleles was tested for association with the development of ROP and stage of disease. We also used a multiple logistic regression analysis to explore the association of particular alleles with ROP, while adjusting for other risk factors in the development of ROP, including gestational age, presence of a patent ductus arteriosus, and hyperglycemia.
RESULTS
Our study population consisted of 347 infants less than 32 wk gestational age, with 183 boys (57 with ROP) and 147 girls (45 with ROP) included. ROP data were unavailable on 17 neonates; generally, older infants (at the lowest risk for ROP) who were transferred back to the referring hospital before an initial ophthalmologic examination. Prethreshold ROP is pro-actively managed in our NICU and no cases of stage IV/V ROP were represented in the affected infants present in the study population. There was no statistical difference between the groups with respect to percentages of males, infants undergoing C-section, or percentage of infants who received antenatal steroids (Table 1). However, there was a significant difference in mean birthweight, gestational age, and mean days on oxygen between the two groups. This was expected as infants who have low birth weight and are born at a lower gestational age have a higher risk of developing ROP.
Table 1.
Demographic data of infants with and without ROP
| No ROP | ROP (All stages) |
p | ||
|---|---|---|---|---|
| Total number (n) | 228 | 102 | ||
| Mean birthweight (g) | 1192 ± 389 | 884 ± 337 | <0.0001 | |
| Mean gestational age (wks) | 28 ± 2 | 26 ±2 | <0.0001 | |
| % Males | 55.8 | 54.4 | NS | |
| % c-section | 66 | 71 | NS | |
| % Antenatal steroids | 88 | 88 | NS | |
| % SGA | 14 | 15.6 | NS | |
| Median 5 min. Apgar | 8 | 8 | ||
| Interquartile range | 6–8 | 7–8 | ||
| Days on O2 (mean) | 53.16 | 106.10 | <0.0001 | |
| Hyperglycemia days (mean) | 3.51 | 4.98 | <0.0001 | |
| Presence of PDA-number (%) |
64 (28) | 51 (50) | 0.002 | |
| Presence of IVH-number (%) | 37 (16.2) | 21 (20.6) | NS | |
NS, not significant.
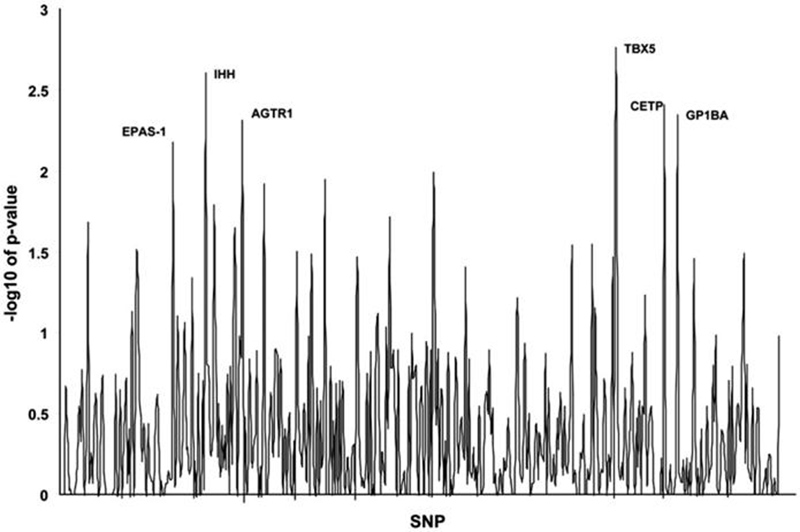
We first performed a data-mining investigation using genetic information available from a prior study focused on prematurity. We analyzed 455 SNPs in 153 genes to determine association with ROP (Fig. 1). As shown in Table 2, five SNPs had a p-value less than 0.01. Formally, a p-value of <0.0001 would be required for statistical significance using a stringent Bonferroni correction for the nearly 500 SNPs evaluated. There is currently debate as to whether this conservative correction is appropriate for genetic data-mining studies (15). A critical question is whether these types of studies represent multiple tests of a single hypothesis or the independent testing of multiple hypotheses. In either case, the ultimate goal of any type of statistical correction in genetic studies investigating multiple loci is to reduce the false positive rate, without missing true positives. We selected a p-value of 0.01 to minimize false positives without losing true positives in this initial hypothesis-generating investigation. Using this criteria, five genes were found to be associated with the development of ROP; Indian hedgehog, angiotensin II receptor (type1), T-box 5, cholesteryl ester transport protein, and glycoprotein Ib (platelet) alpha polypeptide.
Figure 1.
Graphical representation of study single nucleotide polymorphisms (SNPs). SNPs that were significantly associated with the development of retinopathy of prematurity (ROP) are labeled. A p-value of 2 corresponds to a −log10 of 0.01. Six SNPs were significant, one each in the following genes: EPAS-1, IHH, AGTR1, TBX5, CETP, and GP1BA.
Table 2.
Significant data mining genes and chromosome location
| Gene | SNP ID number | p | Location | Chromosome | Minimum allele frequency |
|---|---|---|---|---|---|
| IHH | rs3099 | 0.003 | Intergenic | 2 | 0.42 |
| AGTR1 | rs427832 | 0.005 | Intergenic | 3 | 0.43 |
| TBX5 | rs1895602 | 0.002 | Intron | 12 | 0.44 |
| CETP | rs289747 | 0.004 | Intergenic | 16 | — |
| GP1BA | rs2243093 | 0.005 | 5′ UTR | 17 | 0.11 |
We next used a true candidate gene approach, using both family-based and non–family-based statistical models to study genetic risk factors for ROP. The 11 candidate genes chosen, representing angiogenic, inflammation, and oxygen sensing pathways, are shown in Table 3. Using a family-based transmission disequilibrium test, we found an association between ROP and genetic variations in the EPAS1 gene (rs1867785) (p = 0.007). This association is not formally significant using the Bonferroni correction for the 24 SNPs analyzed, which would require a p-value of <0.002. When a reverse TDT was performed, none of the SNPs that were associated with the development of ROP were found to be significantly associated with those neonates who did not develop ROP.
Table 3.
Candidate genes chosen for analysis
| Gene | Gene product | No. SNPs |
|---|---|---|
| FZD4 | Frizzled 4 | 2 |
| VEGF | Vascular endothelial growth factor | 6 |
| EPO | Erythropoietin | 1 |
| SerpinF1 | Pigment epithelium derived factor | 2 |
| HIF1A | Hypoxia inducible factor 1 alpha | 2 |
| EPAS-1 | Endothelial PAD domain protein 1 | 4 |
| CFH | Complement factor H | 2 |
| ALDH2 | Aldehyde dehydrogenase | 2 |
| IL1A | Interleukin 1A (1) | 1 |
| IL1B | Interleukin 1B | 1 |
| TRAF2 | TNF-receptor-associated factor 2 | 1 |
We next investigated the genes listed in Tables 2 and 3 using a multiple logistic regression analysis. An initial univariate analysis was carried out, suggesting that clinical risk factors for ROP, including gestational age (p < 0.0001), presence of a PDA (p = 0.0003), days with a blood glucose over 150 mg/dL (p < 0.0001), and mean days on supplemental oxygen (p < 0.0001), should be included in the subsequent analysis. Gender (p = 0.97) and weight appropriate for gestational age (SGA p = 0.84, LGA p = 0.85) were not associated with ROP in the univariate analysis. Additionally, the development of intraventricular hemorrhage was also not associated with the development of ROP in the univariate analysis (p = 0.77). A multiple logistic regression model was then created, controlling the significant clinical risk factors described above to evaluate the effect candidate gene polymorphisms on the development of ROP. Analyses of genotypes between infants with ROP and those without ROP suggested that two genetic variations in the complement factor H (CFH) gene were associated with ROP. One T/C polymorphism (rs529825) showed protection against ROP as the number of T alleles increased (p = 0.01). A second T/C polymorphism in CFH (rs800292) also showed protection against ROP as the number of T alleles increased (p = 0.01). In an additional analysis comparing stages 0-I ROP (no disease and mild disease) with stages II-III ROP (more severe disease, requiring intervention), we found an association with a genetic variation in the EPAS1 gene. An A/G polymorphism (rs1868085), showed increased risk for ROP as the number of A alleles increased (p = 0.001). It is interesting that sequence variations in the EPAS1 gene were found to be associated with ROP when analyzed by either family-based (rs1867785) or regression (rs1868085) models. We recognize that analyses of subgroups of infants with ROP must be interpreted with caution, given that only 62 infants had stage II-III ROP, and none had stage IV-V ROP. None of the polymorphisms shown in Table 2 remained significantly associated with ROP after clinical factors were taken into consideration by multiple logistic regression analysis.
DISCUSSION
The approach taken in this study was to evaluate genetic polymorphisms within candidate genes and evaluating genes previously genotyped for association with preterm delivery. Using this approach, five genes (IHH, AGTR1, TBX5, CETP, and GP1BA) that had been previously genotyped for association with prematurity were found to have an association (p < 0.01) with the development of ROP. Additionally, using the candidate gene approach, the EPAS1 gene was found to be positively associated with the development of ROP via a family-based test. When a multiple logistic regression analysis was performed evaluating the genes and associated risk factors, EPAS1 and CFH were found to be associated with the development of ROP.
EPAS1 encodes a member of the basic helix-loop-helix/ PAS domain containing transcription factor family and is expressed mainly in vascular endothelial cells (16). Similar to hypoxia-inducible factor 1-alpha (HIF-1α), EPAS1 is stabilized during hypoxia and forms a heterodimer with aryl hydrocarbon nuclear receptor translocator (ARNT) and transactivates the VEGF promoter. Additionally, this heterodimer complex has also been shown to transactivate Flt-1, encoding VEGF receptor 1, by binding to the HIF-1 binding site upstream of the promoter region of Flt-1 (17). In a recent study evaluating the development of ROP in neonatal mice using a hyperoxia/normoxia model, HIF 2 alpha (the murine equivalent of EPAS1) knockdown mice demonstrated no evidence of retinal neovascularization when compared with control mice (18). It was also shown that erythropoietin (Epo) mRNA expression was significantly decreased when compared with wild-type mice (18). Epo has recently been shown to be a retinal angiogenic factor in proliferative diabetic retinopathy, which has retinal findings that are similar to ROP (19,20). Polymorphisms within this gene in the preterm infant could predispose to altered activity of EPAS1, increasing the expression of angiogenic factors such as VEGF and Epo.
AGTR1 encodes for angiotensin II type I receptor, also known as angiotensin type I receptor (AT1). This is part of the renin-angiotensin system and has been shown to be present in the retina (21). The role of angiotensin II on VEGF receptor expression and VEGF induced angiogenic activity in bovine retinal microcapillary endothelial cells (BRECs) was investigated and found that angiotensin II was a potent stimulant of VEGF-induced endothelial cell proliferation and tube formation and was mediated primarily through the VEGF receptor, KDR/Flk-1. The up-regulation of this receptor was transcriptionally regulated by the angiotensin type I receptor (22). In a hyperoxia/normoxia ROP rat model, administration of either ACE inhibitor lisinopril or the AT1 receptor blocker losartan to rat pups after exposure to hyperoxia demonstrated significantly decreased retinal neovascularization when compared with ROP pups who did not receive either medication. Additionally, VEGF mRNA levels were significantly reduced in rat pups that received lisinopril when compared with controls. However, blockade of AT1 via losartan did not alter VEGF mRNA levels when compared with untreated rat pups, suggesting that AT1 may not be the primary influence of VEGF expression (23).
Hedgehog proteins, such as Indian hedgehog (IHH) and Sonic hedgehog (SHH), act in a variety of tissues during embryonic development, including lung, gut, limb, and bone (24-26). Several recent studies have also demonstrated a role for hedgehog in angiogenesis. In aged mice that have had surgically induced hind limb ischemia, administration of Sonic hedgehog (SHH) led to increased limb salvage and increased capillary density when compared with control mice or those who received recombinant human VEGF (27). In cultured fibroblasts, administration of SHH stimulated up-regulation of patched-1, a receptor specific for hedgehog and increased expression of angiogenic factors such as VEGF, Ang-1, and Ang-2 (27). Later studies evaluating hind limb ischemia in aged mice also demonstrated increased expression of SHH and Ptc-1 in ischemic muscle (28). Additionally, after administration of a SHH neutralizing antibody into the ischemic hindlimb, there was no up-regulation of VEGF when compared with controls and decreased capillary density after prolonged SHH inhibition (28). In another study evaluating Indian hedgehog (IHH) in vascular development in mice yolk sac, IHH knockout mice were able to initiate vasculogenesis but had incomplete vascular remodeling when compared with wild-type or heterozygous mice (29). In our study, one SNP in IHH (rs3099) was found to be positively associated with development of ROP (p = 0.002) and was also found to be significantly associated with the development of severe ROP (p = 0.01).
Complement factor H (CFH) is an inhibitor of the complement pathway. CFH binds to complement component C3b, preventing production of C3 convertase and progression of the cascade. Multiple studies have demonstrated increased risk of developing AMD with specific polymorphisms within CFH (30-32). AMD can be divided into two stages. The early stage of AMD is characterized by hyperpigmentation in the outer retina or choroid and/or areas of depigmentation or hypopigmentation of the retinal pigment epithelium (RPE) associated with the deposition of drusen (33). Late stage AMD is characterized by geographic atrophy (“dry” AMD) and choroidal neovascularization (“wet” AMD) (33). In addition to choroidal neovascularization, there can also be associated formation of intraretinal or subretinal scars, subretinal hemorrhages, and detachment of the RPE and neurosensory retina (33).
Two recent studies have demonstrated that there were haplotypes that were protective against the development of AMD (34,35). Two SNPs in CFH that we found to be associated with the development of ROP (rs800292 and rs529825) are part of these haplotypes. In our multiple logistic regression analysis, we demonstrated that an increased number of T alleles were associated with protection against the development of ROP, similar to the protective haplotype as described for AMD (34,35).
T-Box 5 (TBX5) is a member of the T-Box transcription factor family that is involved in the transcriptional regulation of genes required for mesoderm differentiation. Mutations in TBX5 are associated with Holt-Oram syndrome, a developmental disorder characterized by thumb anomalies and atrial septal defects (36). A recent study demonstrated that TBX5 is expressed across the different layers of the developing retina in an asymmetric pattern at different stages of development (37). However, a role for TBX5 in the development of the retinal vasculature has yet to be identified.
Two additional genes, glycoprotein Ib (platelet) alpha polypeptide (GP1BA) and cholesterol ester transfer protein (CETP), were found to be associated with the development of ROP. GP1BA encodes for a surface membrane protein found on platelets that are involved in the formation of platelet plugs by binding to von Willebrand factor. Alterations in this protein function could potentially lead to easier formation of thrombi in the retinal vasculature leading to retinal ischemia, inducing hypoxia in the surrounding retinal tissue and serving as a signal for retinal neovascularization. Cholesteryl ester transfer protein (CETP) is involved in the transfer of lipids such as cholesterol esters and triglycerides between lipoproteins. Polymorphisms within CETP have recently been shown to be associated with different concentrations of HDL-C and activity of CETP (38). Additionally, certain polymorphisms within CETP were also shown to be associated with the development of coronary artery calcification, a measure of subclinical atherosclerosis (38). However, a role for either CETP or GP1BA in retinal vascular development has yet to be identified. In this study, these two genes may represent false positive but will need to be evaluated further.
An important limitation of this study is that although we have shown an association of sequence polymorphisms with the development of ROP, none of these polymorphisms have been demonstrated to have a direct effect on gene function that could impact the development of ROP. The identified variants are most likely genetically linked to other loci that are related to the development of this condition. With further analysis of these genes, it may be possible to gain a better understanding into the biologic pathways resulting in ROP.
In conclusion, through a candidate gene analysis and a data-mining analysis of previously genotyped genes, we have found six SNPs in six genes showing suggestive association with the development of ROP. Replication of these SNPs in a larger, independent population will need to be completed to confirm the associations between these common genetic variants and the development of ROP. Ultimately allelic associations may be used in modifying environmental components contributing to ROP such as the timing and amount of oxygen supplementation.
Acknowledgments
We thank the families who graciously agreed to participate in this study. Special thanks to our research nurses Gretchen Cress, R.N., B.S.N., Nancy Krutzfield, R.N., B.S.N., Ruthann Schrock, R.N., B.S.N., and Laura Knosp, R.N., B.S.N. Also, thanks to Jan Crewe, Susan Berends, and Jamie L'Heureux for their technical support.
Supported by grants from the Children's Miracle Network, University of Iowa GCRC the NIH (RO1 HD052953–01 and P30 ES05605), and the March of Dimes (FY08-260 and FY06-575).
Abbreviations
- AMD
age-related macular degeneration
- ROP
retinopathy of prematurity
REFERENCES
- 1.Good WV, Hardy RJ, Dobson V, Palmer EA, Phelps DL, Quintos M, Tung B. The incidence and course of retinopathy of prematurity: findings from the early treatment for retinopathy of prematurity study. Pediatrics. 2005;116:15–23. doi: 10.1542/peds.2004-1413. [DOI] [PubMed] [Google Scholar]
- 2.Steinkuller PG, Du L, Gilbert C, Foster A, Collins ML, Coats DK. Childhood blindness. J AAPOS. 1999;3:26–32. doi: 10.1016/s1091-8531(99)70091-1. [DOI] [PubMed] [Google Scholar]
- 3.Hatton DD, Schwietz E, Boyer B, Rychwalski P. Babies Count: the national registry for children with visual impairments, birth to 3 years. J AAPOS. 2007;11:351–355. doi: 10.1016/j.jaapos.2007.01.107. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Smith LE. Retinopathy of prematurity. Angiogenesis. 2007;10:133–140. doi: 10.1007/s10456-007-9066-0. [DOI] [PubMed] [Google Scholar]
- 5.Bizzarro MJ, Hussain N, Jonsson B, Feng R, Ment LR, Gruen JR, Zhang H, Bhandari V. Genetic susceptibility to retinopathy of prematurity. Pediatrics. 2006;118:1858–1863. doi: 10.1542/peds.2006-1088. [DOI] [PubMed] [Google Scholar]
- 6.Cooke RW, Drury JA, Mountford R, Clark D. Genetic polymorphisms and retinopathy of prematurity. Invest Ophthalmol Vis Sci. 2004;45:1712–1715. doi: 10.1167/iovs.03-1303. [DOI] [PubMed] [Google Scholar]
- 7.Vannay A, Dunai G, Banyasz I, Szabo M, Vamos R, Treszl A, Hajdu J, Tulassay T, Vasarhelyi B. Association of genetic polymorphisms of vascular endothelial growth factor and risk for proliferative retinopathy of prematurity. Pediatr Res. 2005;57:396–398. doi: 10.1203/01.PDR.0000153867.80238.E0. [DOI] [PubMed] [Google Scholar]
- 8.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–1235. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 9.Hanis CL, Hallman D. Genetics of diabetic retinopathy. Curr Diab Rep. 2006;6:155–161. doi: 10.1007/s11892-006-0027-6. [DOI] [PubMed] [Google Scholar]
- 10.Hutcheson KA, Paluru PC, Bernstein SL, Koh J, Rappaport EF, Leach RA, Young TL. Norrie disease gene sequence variants in an ethnically diverse population with retinopathy of prematurity. Mol Vis. 2005;11:501–508. [PubMed] [Google Scholar]
- 11.MacDonald ML, Goldberg YP, Macfarlane J, Samuels ME, Trese MT, Shastry BS. Genetic variants of frizzled-4 gene in familial exudative vitreoretinopathy and advanced retinopathy of prematurity. Clin Genet. 2005;67:363–366. doi: 10.1111/j.1399-0004.2005.00408.x. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson JL, Sale MM, Passmore A, FitzGerald LM, Wheatley CM, Burdon KP, Craig JE, Tengtrisorn S, Carden SM, Maclean H, Mackey DA. Mutations in the NDP gene: contribution to Norrie disease, familial exudative vitreoretinopathy and retinopathy of prematurity. Clin Experiment Ophthalmol. 2006;34:682–688. doi: 10.1111/j.1442-9071.2006.01314.x. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehn NL, Cooper ME, Orr K, Shi M, Johnson MK, Caprau D, Dagle J, Steffen K, Johnson K, Marazita ML, Merrill D, Murray JC. Evaluation of fetal and maternal genetic variation in the progesterone receptor gene for contributions to preterm birth. Pediatr Res. 2007;62:630–635. doi: 10.1203/PDR.0b013e3181567bfc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 17.Takeda N, Maemura K, Imai Y, Harada T, Kawanami D, Nojiri T, Manabe I, Nagai R. Endothelial PAS domain protein 1 gene promotes angiogenesis through the transactivation of both vascular endothelial growth factor and its receptor, Flt-1. Circ Res. 2004;95:146–153. doi: 10.1161/01.RES.0000134920.10128.b4. [DOI] [PubMed] [Google Scholar]
- 18.Morita M, Ohneda O, Yamashita T, Takahashi S, Suzuki N, Nakajima O, Kawauchi S, Ema M, Shibahara S, Udono T, Tomita K, Tamai M, Sogawa K, Yamamoto M, Fujii-Kuriyama Y. HLF/HIF-2alpha is a key factor in retinopathy of prematurity in association with erythropoietin. EMBO J. 2003;22:1134–1146. doi: 10.1093/emboj/cdg117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe D, Suzuma K, Matsui S, Kurimoto M, Kiryu J, Kita M, Suzuma I, Ohashi H, Ojima T, Murakami T, Kobayashi T, Masuda S, Nagao M, Yoshimura N, Takagi H. Erythropoietin as a retinal angiogenic factor in proliferative diabetic retinopathy. N Engl J Med. 2005;353:782–792. doi: 10.1056/NEJMoa041773. [DOI] [PubMed] [Google Scholar]
- 20.Takagi H, Watanabe D, Suzuma K, Kurimoto M, Suzuma I, Ohashi H, Ojima T, Murakami T. Novel role of erythropoietin in proliferative diabetic retinopathy. Diabetes Res Clin Pract. 2007;77:S62–S64. doi: 10.1016/j.diabres.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 21.Sarlos S, Wilkinson-Berka JL. The renin-angiotensin system and the developing retinal vasculature. Invest Ophthalmol Vis Sci. 2005;46:1069–1077. doi: 10.1167/iovs.04-0885. [DOI] [PubMed] [Google Scholar]
- 22.Otani A, Takagi H, Suzuma K, Honda Y. Angiotensin II potentiates vascular endothelial growth factor-induced angiogenic activity in retinal microcapillary endothelial cells. Circ Res. 1998;82:619–628. doi: 10.1161/01.res.82.5.619. [DOI] [PubMed] [Google Scholar]
- 23.Moravski CJ, Kelly DJ, Cooper ME, Gilbert RE, Bertram JF, Shahinfar S, Skinner SL, Wilkinson-Berka JL. Retinal neovascularization is prevented by blockade of the renin-angiotensin system. Hypertension. 2000;36:1099–1104. doi: 10.1161/01.hyp.36.6.1099. [DOI] [PubMed] [Google Scholar]
- 24.Johnson RL, Tabin CJ. Molecular models for vertebrate limb development. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- 25.Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 26.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pola R, Ling LE, Silver M, Corbley MJ, Kearney M, Blake Pepinsky R, Shapiro R, Taylor FR, Baker DP, Asahara T, Isner JM. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med. 2001;7:706–711. doi: 10.1038/89083. [DOI] [PubMed] [Google Scholar]
- 28.Pola R, Ling LE, Aprahamian TR, Barban E, Bosch-Marce M, Curry C, Corbley M, Kearney M, Isner JM, Losordo DW. Postnatal recapitulation of embryonic hedgehog pathway in response to skeletal muscle ischemia. Circulation. 2003;108:479–485. doi: 10.1161/01.CIR.0000080338.60981.FA. [DOI] [PubMed] [Google Scholar]
- 29.Byrd N, Becker S, Maye P, Narasimhaiah R, St-Jacques B, Zhang X, McMahon J, McMahon A, Grabel L. Hedgehog is required for murine yolk sac angiogenesis. Development. 2002;129:361–372. doi: 10.1242/dev.129.2.361. [DOI] [PubMed] [Google Scholar]
- 30.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 31.Despriet DD, Klaver CC, Witteman JC, Bergen AA, Kardys I, de Maat MP, Boekhoorn SS, Vingerling JR, Hofman A, Oostra BA, Uitterlinden AG, Stijnen T, van Duijn CM, de Jong PT. Complement factor H polymorphism, complement activators, and risk of age-related macular degeneration. JAMA. 2006;296:301–309. doi: 10.1001/jama.296.3.301. [DOI] [PubMed] [Google Scholar]
- 32.Wegscheider BJ, Weger M, Renner W, Steinbrugger I, Marz W, Mossbock G, Temmel W, El-Shabrawi Y, Schmut O, Jahrbacher R, Haas A. Association of complement factor H Y402H gene polymorphism with different subtypes of exudative age-related macular degeneration. Ophthalmology. 2007;114:738–742. doi: 10.1016/j.ophtha.2006.07.048. [DOI] [PubMed] [Google Scholar]
- 33.Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, de Jong PT, Klaver CC, Klein BE, Klein R, Mitchell P, Sarks JP, Sarks SH, Soubrane G, Taylor HR, Vingerling JR. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39:367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 34.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer KL, Hauser MA, Olson LM, Schnetz-Boutaud N, Scott WK, Schmidt S, Gallins P, Agarwal A, Postel EA, Pericak-Vance MA, Haines JL. Haplotypes spanning the complement factor H gene are protective against age-related macular degeneration. Invest Ophthalmol Vis Sci. 2007;48:4277–4283. doi: 10.1167/iovs.06-1427. [DOI] [PubMed] [Google Scholar]
- 36.Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet. 2003;12:R37–R44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- 37.Sowden JC, Holt JK, Meins M, Smith HK, Bhattacharya SS. Expression of Drosophila omb-related T-box genes in the developing human and mouse neural retina. Invest Ophthalmol Vis Sci. 2001;42:3095–3102. [PubMed] [Google Scholar]
- 38.Tsai MY, Johnson C, Kao WH, Sharrett AR, Arends VL, Kronmal R, Jenny NS, Jacobs DR, Jr, Arnett D, O'Leary D, Post W. Cholesteryl ester transfer protein genetic polymorphisms, HDL cholesterol, and subclinical cardiovascular disease in the Multi-Ethnic Study of Atherosclerosis. Atherosclerosis. 2008;200:359–367. doi: 10.1016/j.atherosclerosis.2007.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]