Abstract
In the current standard procedure for preparation of mammalian rhodopsin mutants, transfected COS-1 cells expressing the mutant opsin genes are treated with 5 μM 11-cis-retinal before detergent solubilization for purification. We found that binding of 11-cis-retinal to opsin mutants with single amino acid changes at Trp-265 (W265F,Y,A) and a retinitis pigmentosa mutant (A164V) was far from complete and required much higher concentrations of 11-cis-retinal. By isolation of the expressed opsins in a stable form, kinetic studies of retinal binding to the opsins in vitro have been carried out by using defined phospholipid–detergent mixtures. The results show wide variation in the rates of 11-cis-retinal binding. Thus, the in vitro reconstitution procedure serves as a probe of the retinal-binding pocket in the opsins. Further, a method is described for purification and characterization of the rhodopsin mutants after retinal binding to the opsins in vitro.
Keywords: G-protein-coupled receptors, signal transduction, 11-cis-retinal, binding pocket, retinitis pigmentosa
Structure and function studies of bacteriorhodopsin were aided greatly by kinetic studies of retinal binding to purified bacterio-opsin mutants in defined phospholipid–detergent mixtures (1, 2). However, the standard procedure used in essentially all the work reported so far on the mammalian rhodopsin mutants has involved treatment of the expressed proteins in COS-1 cells with 11-cis-retinal before solubilization in detergents (3–6). The rationale for this has been that binding of retinal to the opsin mutants to form the corresponding rhodopsins increases stability and offers an advantage during subsequent purification procedures. Observations made recently with certain amino acid substitution mutants, in particular W265F,Y,A (7), indicated that at the retinal concentration usually used, the binding of retinal does not reach completion. This presumably is because of reduced affinity of 11-cis-retinal for the altered binding pockets in the opsins. The alternative approach to ensure completion of retinal binding to the opsin mutants would be to isolate the opsins in stable form and subsequently treat them with retinal in vitro under defined conditions. The isolation of certain opsins from COS-1 cells in a stable form has been reported recently by Oprian and coworkers by including phospholipids during solubilization of the COS-1 cell-expressed proteins in detergents (8). In the present work, by using a slight modification of the detergent–phospholipid isolation procedure, we have prepared pure opsin mutants and have developed a general procedure for their in vitro constitution to the corresponding rhodopsins by treatment with 11-cis-retinal in defined phospholipid–detergent mixtures. We show by kinetic studies of retinal binding that the mutants show large variation in rates of retinal binding. We have chosen as examples the tryptophan 265 mutants, W265F,Y,A (7), as well as the retinitis pigmentosa (RP) mutant A164V (9, 10) in the transmembrane domain (Fig. 1). Further, we describe a method for the purification and characterization of the fully constituted rhodopsin mutants after in vitro retinal binding.†
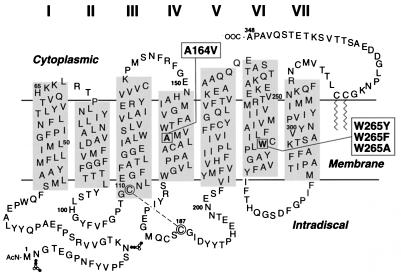
Figure 1.
A secondary-structure model of bovine rhodopsin with single-letter abbreviations for amino acids showing the positions of mutants studied (boxed). Tryptophan-265 was mutated to phenylalananine, tyrosine, and alanine (7). Alanine 164 to valine is a naturally occurring RP mutation (9). The disulfide bond between C110 and C187 (circled) is shown by a dashed line. The oligosaccharides attached at asparagines N-2 and N-15 are shown by the chain of symbols. ●, N-acetylglucosamine; ○, mannose.
MATERIALS AND METHODS
Materials.
11-cis-retinal was a gift from Rosalie Crouch (University of South Carolina and the National Eye Institute, National Institutes of Health). The detergents dodecyl maltoside (DM) and 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS) were from Anatrace (Maumee, OH). The phospholipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was from Avanti Polar Lipids. Antirhodopsin monoclonal antibody rho-1D4 was purified from myeloma cell lines provided by R. S. Molday (University of British Columbia) and was coupled to cyanogen bromide-activated Sepharose 4B (Amersham Pharmacia), as described (3). The buffers were: buffer A, 137 mM NaCl/2.7 mM KCl/1.8 mM KH2PO4/10 mM Na2HPO4, pH 7.2; buffer B, buffer A containing 1% (wt/vol) DM; buffer C, buffer A containing 0.1% (wt/vol) DM; buffer D, 10 mM 1,3-bis[tris(hydroxymethyl)-methylamino]propane, pH 6.0/0.1% (wt/vol) DM; buffer E, buffer D containing 100 μM nonapeptide, corresponding to the C-terminal rhodopsin sequence; buffer F, buffer E containing 140 mM NaCl; buffer G, 10 mM 1,3-bis[tris(hydroxymethyl)-methylamino]propane, pH 6.0/140 mM NaCl/1% (wt/vol) CHAPS/1% (wt/vol) DMPC: buffer H, buffer G containing 100 μM nonapeptide, corresponding to the C-terminal rhodopsin sequence.
Expression of the Mutant Opsin Genes and Purification of the Mutant Rhodopsin and Opsins.
The construction of the mutant genes corresponding to W265 (F,Y,A) and A164V has been described previously (7, 10).
The procedure for the transient transfection of COS-1 cells by using pMT4/5 vectors expressing the synthetic opsin gene has been described previously (3, 4).
(i) In Vivo Constitution Using Different Concentrations of 11-cis-Retinal.
The cells from 15-cm cell culture dishes were harvested 52–56 hr after transfection and resuspended in buffer A (2 ml per dish) containing phenylmethyl sulfonyl fluoride (0.1 mM). Cell suspensions (from three dishes) were each treated with 5, 50, or 100 μM retinal for 6 hr at 4°C in the dark. Mutant rhodopsins were then purified in the dark as described previously (3, 11). Briefly, cells were solubilized in 6 ml of buffer B containing phenylmethyl sulfonyl fluoride (0.1 mM) and cleared supernatants were applied to a column containing 100 μl rho-1D4-Sepharose (capacity 1 mg rhodopsin per ml settled beads). The column was washed with 500 column volumes of buffer C and 100 column volumes of buffer D before elution of rhodopsin by using buffer E (7 × 300 μl) followed by buffer F (7 × 300 μl).
(ii) Isolation of Stable Opsins.
Transfected cells from five 15-cm culture dishes were solubilized in 10 ml buffer G (containing 0.1 mM phenylmethyl sulfonyl fluoride) by end-over-end mixing at 4°C for 1 hr. The cleared supernatant was incubated with rho-1D4-Sepharose (200 μl settled beads) with mixing for 1 hr at 4°C. The beads were washed by using 50 bed volumes of buffer G and opsin was eluted by using buffer H (2 × 500 μl). Each elution was performed at 4°C for 1 hr and samples were stored on ice before use.
(iii) Purification and Characterization of in Vitro-Constituted Rhodopsin.
For repurification of in vitro-generated rhodopsins, the elution peptide was first removed from the samples (150 μl) by using a G-50 Sephadex column (1.5 ml bed volume settled beads) and centrifugation for 10 min at 3,000 rpm by using an IEC clinical centrifuge. The rhodopsins were then repurified by using 1D4-Sepharose immunoaffinity chromatography essentially as described above.
UV–Visible (UV–Vis) Absorption Spectroscopy.
UV–vis absorption spectra were recorded by using a Perkin–Elmer λ-6 UV–vis spectrophotometer. All spectra were recorded at 20°C by using a bandwidth of 2 nm, a response time of 1 sec, and a scan speed of 480 nm/min.
Measurement of in Vitro Rhodopsin Formation Kinetics.
Opsin was diluted to an A280 of about 0.04 (0.7 μM) in 300 μl of buffer G. 11-cis-Retinal (1 μl) was added to the cuvette from ethanol-dissolved stocks (10 mM, 1 mM, and 100 μM) to a final concentration of 33, 3.3, or 0.3 μM, respectively. UV–vis absorption spectra were recorded by using a Perkin–Elmer λ6 spectrophotometer by using the above parameters, programmed for repeated scans. Increases in absorbance at 500 nm were plotted as a function of time. Data manipulations and curve fittings were performed by using microcal origin software.
Measurement of Rate of Metarhodopsin II Decay Rates by Retinal Release.
Purified rhodopsin samples were in buffer D. They were bleached (for 30 sec) by using light (>495 nm) from a fiber optic source (Dolan Jenner, Lawrence, MA) and metarhodopsin II decay was measured by following the retinal release by the fluorescence assay (12).
RESULTS
11-cis-Retinal Binding to Mutant Opsins W265Y and W265A in Vivo Is Dependent on Retinal Concentration.
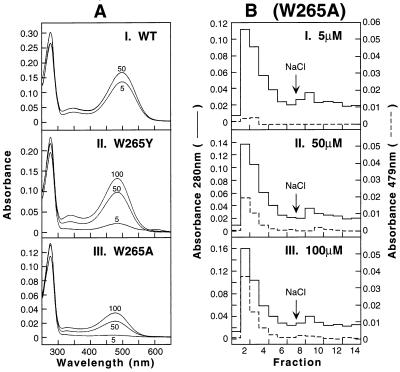
Wild-type (WT) opsin and the mutant opsins, W265Y and W265A, were expressed in COS-1 cells by transient transfection and the harvested cells were treated with 5-, 50-, and 100-μM concentrations of 11-cis-retinal at 4°C for 6 hr. After solubilization in buffer B, the proteins were purified by immunoaffinity chromatography by using the antibody 1D4-Sepharose, as in Methods. The UV–vis spectra of the main fractions eluted in the presence of the nonapeptide epitope are shown in Fig. 2A. As seen in A–I for the WT opsin, 11-cis-retinal binding to form the 500-nm chromophore was not complete at 5 μM retinal concentration, and an increase in binding at 50 μM was found. For W265Y, A–II, chromophore formation was inefficient at 5 μM 11-cis-retinal concentration; however, progressively more chromophore formation occurred at 50 and 100 μM 11-cis-retinal. The results of in vivo retinal binding to the opsin mutant W265A were even more striking. As seen in A–III, even at 100 μM 11-cis-retinal concentration, the binding was not complete, whereas at 5 μM 11-cis-retinal concentration, very little of 479-nm-absorbing chromophore was formed, and the bulk of the 280 nm-absorbing protein corresponded to free opsin. The conclusion that the free opsin and the constituted rhodopsin are eluted together at pH 6 (low salt) and that no misfolded opsin is present was confirmed by the results in Fig. 2 B. B I–III show complete column elution profiles for W265A treated with retinal concentrations at 5, 50, and 100 μM. As is seen, at 5 μM retinal (B–I) mainly opsin absorbing at 280 nm was eluted at pH 6 in low salt. The amount of 479-nm-absorbing pigment eluted in low salt progressively increased at 50 μM (B–II) and 100 μM (B–III) retinal concentration. The procedure used for elution from the column did not discriminate between the constituted rhodopsin and free opsin. In all three experiments of Fig. 2B (B I–III), the amount of opsin eluting at 140 mM NaCl was very small. Although not shown, the mutant W265F also showed concentration dependence for 11-cis-retinal binding very similar to W265Y.
Figure 2.
UV–vis spectra of rhodopsin and rhodopsin mutants formed on retinal treatment of COS-1 cells expressing opsins. Whole-cell suspensions of transfected COS-1 cells were divided and treated with the 11-cis-retinal at concentrations indicated. Pigments were solubilized and purified by immunoaffinity chromatography as described in Methods. (A) The UV–vis absorption spectra are shown, I, WT; II, W265Y, and III, W265A. (B) (W265A), elution profiles from 1D-4 Sepharose column of pigments formed after treatment with 11-cis-retinal at 5 μM (I), 50 μM (II), and 100 μM (III). Shown are the absorbances at 280 nm (solid line) and 479 nm (dotted line) for each fraction. Fractions 1–7 in every case were as eluted by using buffer E (no salt), whereas fractions 8–14 were obtained by using buffer F (140 mM NaCl).
11-cis-Retinal Binding to Retinitis Pigmentosa Mutant A164V and Characterization of the Products.
In the work previously described with this mutant, COS-1 cells harvested after expression of the gene were treated with 5 μM 11-cis-retinal for 3 hr and the products obtained by subsequent immunoaffinity chromatography, including separation by differential elution, gave patterns (Fig. 2D, ref. 10) that were concluded to be correctly folded and misfolded nonretinal binding opsins. Work with this mutant was now extended as shown in Fig. 3. Equal amounts of COS-1 cells expressing the opsin were treated in parallel with 5 and 50 μM 11-cis-retinal. The products, after solubilization and purification, were analyzed by UV–vis spectroscopy. The results are in Fig. 3A. The proteins from both retinal treatments were further analyzed by differential elution from 1D4-Sepharose columns as shown in Fig. 3 B and C. The pattern shown in Fig. 3C shows that 280 nm absorbing but nonretinal binding material is eluted with salt. This material has been confirmed to be misfolded opsin by being immunopositive to the antibody 1D4 (not shown).
Figure 3.
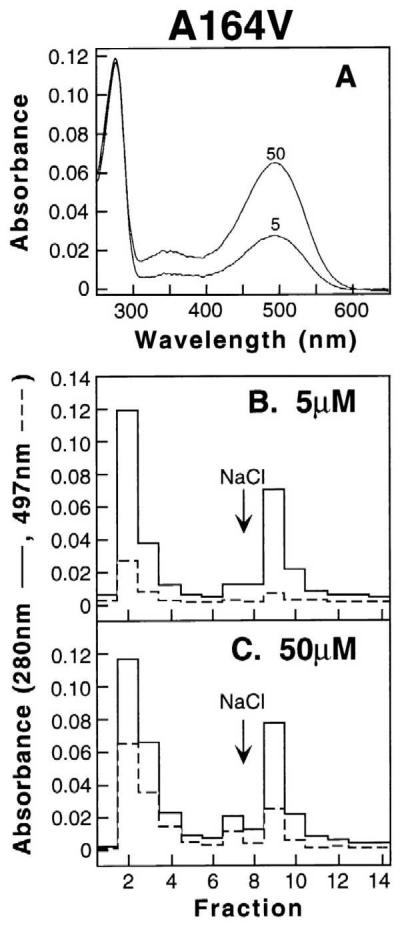
(A) Concentration-dependent retinal binding in vivo to expressed A164V opsin in COS-1 cells. Transfected COS-1 cells expressing A164V opsin were treated with 11-cis-retinal (5 and 50 μM) and purified as described in Methods. UV–vis absorption spectra of the pigments obtained are shown. (B and C) Elution profile from 1D-4 Sepharose column of A164V proteins obtained after retinal treatments (5 or 50 μM). Shown are absorbances at 280 nm (solid line) and 497 nm (dotted line) for each fraction. Fractions 1–7 were as eluted by using buffer E (no salt) and fractions 8–14 were eluted by using buffer F (140 mM NaCl).
Kinetics of 11-cis-Retinal Binding in Vitro to the Mutant Opsins.
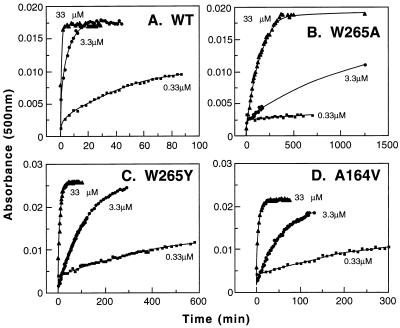
WT opsin and the opsin mutants W265Y, W265A, and A164V were purified by using CHAPS/DMPC mixture by 1D4-Sepharose chromatography (Methods). They were incubated with different concentrations of 11-cis-retinal (0.33–33 μM) and monitored by time-resolved UV–vis absorption spectroscopy. The rates of chromophore formation were measured by increase in A500, and the results are shown in Fig. 4. The data fit well by using a single exponential equation, and from this the rates of rhodopsin formation were calculated (Table 1). Thus, for WT (Table 1 and Fig. 4A), the t1/2 of rhodopsin formation decreases with increasing retinal concentration. The kinetics observed for retinal binding by the mutants W265A, W265Y, and A164V are shown in Fig. 4 B, C, and D, respectively. The t1/2 values for the formation of rhodopsin from these mutant opsins and W265F are shown in Table 1. The rates for W265F and W265Y are about 20 times slower than that for WT at 3.3 μM 11-cis-retinal. Increasing the retinal concentration to 33 μM increases the rates to a t1/2 of about 5 min for each W265F and W265Y. For the W265A mutant, rhodopsin formation was inefficient even at 33 μM 11-cis-retinal with a t1/2 value of 92.5 min. Mutant A164V displays a t1/2 of rhodopsin formation that is between WT and W265(F,Y). This explains the in vivo rhodopsin formation results presented in the previous section.
Figure 4.
Kinetics of rhodopsin formation by using purified WT and mutant opsins. WT opsin and mutant opsins at positions W265 and A164V were prepared in buffer containing CHAPS [1%(wt/vol)] and DMPC [1%(wt/vol)], as described in Methods. Opsins at about 0.7 μM concentrations in the mixtures were treated with 11-cis-retinal and increases in absorbance at 500 nm were measured as a function of time. Plots obtained with different opsins are shown at the indicated 11-cis-retinal concentrations (μm). (A) WT opsin. (B) W265A. (C) W265Y. (D) A164V.
Table 1.
Rates of rhodopsin chromophore formation from opsin mutants at different 11-cis-retinal concentrations
| Mutant |
t1/2 of Rhodopsin formation, min∗
|
||
|---|---|---|---|
| 0.33 μM | 3.3 μM | 33 μM | |
| WT | 28.7 | 2.7 | 0.5 |
| W265F | 648.4 | 79.8 | 6.1 |
| W265Y | 346.0 | 69.9 | 5.0 |
| W265A | ND† | 453.1 | 92.5 |
| A164V | 293.5 | 38.8 | 2.8 |
t1/2 values for A500 formation were calculated from the curve-fitted data presented in Fig. 3.
Could not be determined because of undetectable absorption increase.
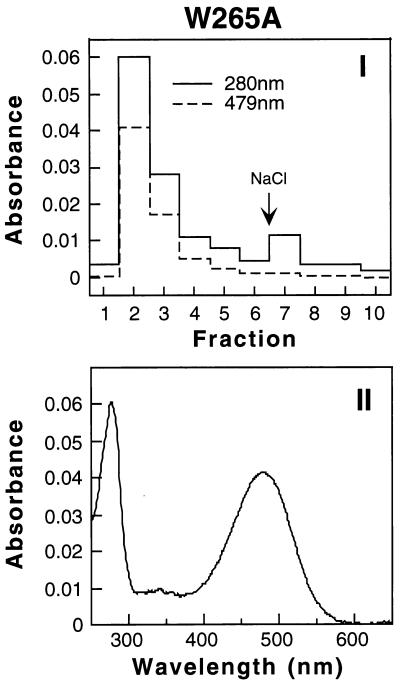
Isolation and Characterization of the Mutant Rhodopsin W265A After Retinal Binding in Vitro.
A procedure for the purification of the rhodopsin mutants after retinal treatment of the corresponding opsins in vitro was developed by using W265A as an example. This involved principally the removal of the nonapeptide by using Sephadex G-50 filtration as described in Methods. The constituted rhodopsin was then repurified by rho 1D4-Sepharose immunoaffinity chromatography. The main fraction eluted with buffer E (Fig. 5I) showed an A280/A479 ratio of 1.5, the UV–vis spectrum ratio of the peak being shown in Fig. 5II.
Figure 5.
Purification of W265A rhodopsin after constitution from opsin and 11-cis-retinal in vitro. W265A opsin was prepared from transfected COS-1 cells, as described in Methods. Pigment was generated by using 33 μM 11-cis-retinal, and after purification, the epitope peptide was removed as in Methods. Pigment was repurified by 1D4-Sepharose chromatography. (I) Absorbance at 280 nm (solid line) and 479 nm (dotted line) for each fraction eluted. (II) UV–vis absorption spectrum of W265A from fraction 2 of I.
Rates of Metarhodopsin II Decay.
These were measured as the rates of all-trans-retinal release (12) and the results are shown in Table 2. The rates of retinal release for mutations at W265 and for A164V are four to eight times higher than WT rhodopsin. Thus, the altered retinal binding pocket in these mutants also affects the stability of metarhodopsin II.
Table 2.
Metarhodopsin II decay rates as measured by retinal release*
| Rhodopsin mutants | t1/2 Meta II decay, min |
|---|---|
| WT | 16 |
| W265F | 3.9 |
| W265Y | 4.4 |
| W265A | 1.9 |
| A164V† | 2.1 |
As in Materials and Methods.
This sample was in 0.05% DM.
DISCUSSION
The 11-cis-retinal-binding pocket in rhodopsin and, indeed, in all the visual pigments represents the ultimate evolution of a sensory molecule. The interactions between all parts of the retinal and the amino acids that contact it have been optimized for the stabilization of the molecule in the dark state and for optimal signal transduction after light activation. Retinal serves as a reverse agonist, keeping the sensory molecule absolutely noise-free in the dark yet showing sensitivity to single photons such that, after retinal isomerization, the highest catalysis in activating transducin molecules is achieved. Mutations in amino acids interacting with retinal also provide the basis for spectral tuning (6). Previously, efforts have been made to map the amino acids that line the retinal binding pocket. These involved covalent crosslinking by using a photoactivatable analog of retinal (13) and by mutagenesis of amino acids in the binding pocket (7). More recently, similar studies have been reported by Han et al. (14, 15) and Lin et al. (16) and molecular models for retinal–amino acid interactions to understand spectral tuning in cone pigments have been proposed (6, 16).
Striking effects on the affinity for binding of retinal to opsin mutants expressed in COS-1 cells have been highlighted by the work now described. A large variation in rates of retinal binding has been found for different amino acids substituting a single native amino acid, Trp 265, a highly conserved amino acid in the visual pigments. Quantitation of the rates of binding of 11-cis-retinal has been achieved by in vitro constitution of the corresponding rhodopsins under standardized conditions. Previously, a similar procedure for studies of binding of all-trans-retinal to bacterio-opsin mutants proved extremely useful in structure–function studies of bacteriorhodopsin, the light-driven biological proton pump (1, 2).
In previous studies of folding and misfolding caused by RP mutations, the extent of misfolding was determined after treatment of COS-1 cell expressed opsin mutants at 5 μM 11-cis-retinal (10). Although the basic conclusions regarding total misfolding and partial misfolding in RP mutants (10, 17–19) are not in doubt, the extent of misfolding was likely overestimated in certain cases. Detailed study of the A164V mutation in the present work confirmed the presence of misfolded opsin in the COS-1 cell-expressed protein, although its proportion was less than that estimated in the earlier work.
The results now reported allow the following recommendations for processing of rhodopsin mutants after expression of the corresponding opsins in mammalian cells. Treatment with 11-cis-retinal in vivo should be at two levels, 5 and 25 μM concentrations. This should bring out the difference in extent of retinal binding if retinal binding affinity has been affected in the mutant. Assurance of completeness of binding can be achieved by retinal binding in vitro as now described.
Acknowledgments
We thank Professor Daniel Oprian (Brandeis University) for reading this manuscript. We have benefited greatly from discussions with Professor U. L. RajBhandary (Biology Department, Massachusetts Institute of Technology) and our laboratory colleagues. We thank Ms. Judy Carlin for her expert assistance in the preparation of the manuscript. This work was supported by both a National Institute of General Medical Sciences (NIGMS) grant R01-GM28289 and by a National Eye Institute grant R01-EY11716 from the National Institutes of Health. J.H. is the recipient of a Howard Hughes Medical Institute Physician Postdoctoral Fellowship.
ABBREVIATIONS
- CHAPS
3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate
- DM
dodecyl maltoside
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- WT
wild type
- RP
retinitis pigmentosa
- UV–vis
UV–visible. Mutant rhodopsins with amino acid substitutions are designated by the one letter abbreviations for amino acids. The amino acid to the left of the residue number is the original, whereas that to the right is the substituted amino acid
Footnotes
This is paper no. 34 in the series “Structure and Function in Rhodopsin.” Paper no. 33 is ref. 20.
References
- 1.Khorana H G. J Biol Chem. 1988;263:7439–7442. [PubMed] [Google Scholar]
- 2.Khorana H G. Proc Natl Acad Sci USA. 1993;90:1166–1171. doi: 10.1073/pnas.90.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franke R R, Sakmar T P, Oprain D D, Khorana H G. J Biol Chem. 1988;26:2119–2122. [PubMed] [Google Scholar]
- 5.Oprian D D. In: Methods in Neurosciences, Photoreceptor Cells. Hargrave P A, editor. Vol. 15. New York: Academic; 1993. pp. 301–306. [Google Scholar]
- 6.Sakmar T P. Prog Nucleic Acid Res Mol Biol. 1998;59:1–34. doi: 10.1016/s0079-6603(08)61027-2. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama T, Khorana H G. J Biol Chem. 1991;266:4269–4275. [PubMed] [Google Scholar]
- 8.Rim J, Oprian D D. Biochemistry. 1995;34:11938–11945. doi: 10.1021/bi00037a035. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs S, Kranich H, Denton M J, Zrenner E, Bhattacharya S S, Humphries P, Gal A. Hum Mol Genet. 1994;3:1203. doi: 10.1093/hmg/3.7.1203. [DOI] [PubMed] [Google Scholar]
- 10.Hwa J, Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridge K D, Lu Z, Liu X, Khorana H G. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 12.Farrens D L, Khorana H G. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama T A, Khorana H G. J Biol Chem. 1990;265:15762–15769. [PubMed] [Google Scholar]
- 14.Han M, Lin S W, Minkova M, Smith S O, Sakmar T P. J Biol Chem. 1996;271:32337–32342. doi: 10.1074/jbc.271.50.32337. [DOI] [PubMed] [Google Scholar]
- 15.Han M, Lin S W, Smith S O, Sakmar T P. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 16.Lin S W, Kochendoerfer G G, Carroll K S, Wang D, Mathies R A, Sakmar T P. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 17.Kaushal S, Ridge K D, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4560–4564. doi: 10.1073/pnas.93.10.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Garriga P, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altenbach, C., Klein-Seetheraman, J, Khorana, H. G. & Hubbell, W. L. (1999) Biochemistry, in press.