Sulfated tyrosines are present in a wide array of proteins, such as G-protein-coupled receptors (GPCRs),[1] anticoagulation/coagulation factors,[2,3] antibodies,[4] and bioactive peptides, such as phyllokinin, phytosulfokine, and cholecystokinin.[1] The post-translational addition of a sulfate group to tyrosine residues on peptides and proteins is catalyzed by membrane-bound tyrosylprotein sulfotransferases (TPSTs)[5] in the trans-Golgi network.[5] Sulfation occurs following protein translocation to the endoplasmic reticulum, and thus, there is spatial separation between the two common forms of post-translationally modified tyrosine residues, namely, phosphorylation in the cytosol, and sulfation in the extracellular space.[3]
Although tyrosine-O-sulfation plays a critical role in protein–protein interactions, in cell function, and in certain disease states,[3] the elucidation of sulfation sites on proteins and peptides, and consequently the understanding of their function, is challenging.[1] There are no consensus sequences for tyrosine sulfation other than the presence of neighboring acidic residues. Furthermore, the sulfate group is hydrolyzed at low-pH conditions typically used for chemical analysis[6] and during analysis under positive/negative mode MS/MS.[7,8]
We have been pursuing an enzyme-engineering-based[9,10] approach to detect post-translationally modified tyrosines by altering the substrate specificity of the Escherichia coli outer membrane protease OmpT to cleave only those proteins with modified tyrosine residues. To realize this goal, an engineered protease must cleave at a modified tyrosine residue while being able to discriminate between the chemically similar sulfotyrosine (sTyr) and phosphotyrosine (pTyr) modifications. Herein is reported a highly active engineered OmpT variant (kcat/KM > 1 × 105M−1s−1, where kcat is the catalytic rate constant and KM is the Michaelis constant) exhibiting specific recognition of sulfotyrosine in the P1 position. Significantly, this OmpT variant showed greater than 200-fold and tenfold preferences in favor of sulfotyrosine over phosphotyrosine and unmodified tyrosine, respectively. Recently, a rational engineering effort to expand the substrate selectivity of the bacterial protease subtilisin BPN′ resulted in an enzyme capable of cleaving substrates containing either sulfo- or phosphotyrosine, with the latter being preferred by roughly a factor of two.[11]
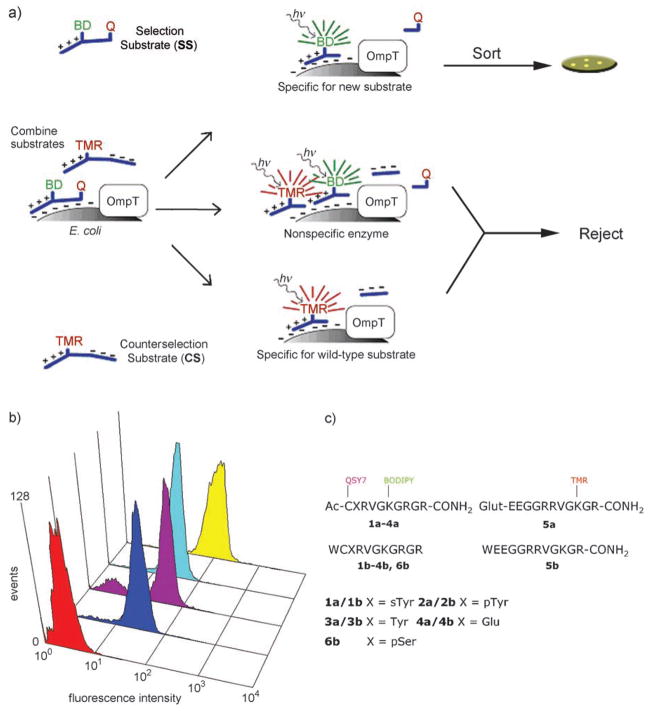
To engineer OmpT to recognize selectively sulfotyrosine-containing substrates, a sulfotyrosine selection peptide was synthesized, in which the sulfotyrosine residue was flanked by a fluorophore (BODIPY) and a positively-charged tail on one side, and a quencher (QSY 7) on the other.[12] Cleavage at the sulfotyrosine residue by enzyme variants displayed on the surface of E. coli resulted in capture of the positively-charged fluorescent moiety. Simultaneous counterselection using a zwitterionic, fluorescently labeled (tetramethyl rhodamine) peptide containing the wild-type (WT) OmpT-preferred dibasic sequence (ArgArg) was used to eliminate nonspecific protease variants (Figure 1a).
Figure 1.
a) Two-color flow-cytometric scheme for the isolation of sulfotyrosine-specific OmpT variants. BD =BODIPY, TMR =tetramethylrhodamine, Q =QSY 7. b) Fluorescence profiles (emission at λ =530 nm with a 30 nm filter) of cells expressing no enzyme (BL21(DE3), red trace), sT1 (blue), sT2 (purple), sT3 (cyan), and sT4 (yellow) labeled with the sTyr↓Arg screening substrate 1 a. c) Sequences of the substrates used for screening and characterization of the OmpT variants.
A partial saturation library (the targeted amino acid is randomly encoded for by either the wild-type or NNS codon (N =guanine, adenine, thymine, or cytosine; S =guanine or cytosine) targeting the 21 amino acids (Figure S1 in the Supporting Information) lining the entire OmpT active site[13] (excluding the putative catalytic residues Asp83, Asp85, Asp210, and His212) was constructed by oligonucleotide-based gene assembly in which degenerate NNS oligonucleotides (90 mol%) were mixed with WT oligonucleotides (10 mol%).[12] The library was cloned into pDUCE19, a plasmid that expresses OmpT under the control of its native promoter, and transformed into electrocompetent E. coli MC1061 to generate 3 × 108 transformants. Plasmid was isolated from pooled cells and retransformed into E. coli BL21(DE3), an ompT ompP deficient strain. The cells were grown for 6–8 h at 37°C to an optical density at λ =600 nm OD600 ≈ 2. A 1 mL aliquot of the culture (ca. 109 cells) was washed and resuspended in 1% sucrose with 20 nM selection substrate 1a and 100 nM 5 a for ten minutes and sorted by flow cytometry (MoFlo, Dako, Fort Collins, CO). Gates were set based on forward/side scatter and FL-1 (BODIPY fluorescence)/FL-2 (TMR fluorescence) to collect E. coli cells expressing OmpT variants with high BODIPY and low TMR fluorescence. After five rounds of regrowth and sorting, the isolated cells were plated onto lysogeny broth (Difco) agar plates containing ampicillin. Three unique clones (designated sT1, sT2, and sT3) were isolated after the final round of sorting. Fluorescence analysis of cells labeled with 1a–4a demonstrated that the enzyme variants were selective, but the overall fluorescence with 1a was not indicative of a highly active enzyme variant (Figure 1b). To isolate an OmpT variant exhibiting more efficient hydrolysis of 1a, the three clones above were backcrossed with WT OmpT using DNA shuffling[14] to yield a library of 2 × 106 independent transformants. After three rounds of flow cytometric sorting as above, six clones were isolated, and DNA sequencing revealed three unique variants. One clone (designated sT4) exhibited a selective fluorescence profile when labeled individually with 1a–5a. Clone sT4 contains a total of nine amino acid changes, and notably, both aromatic and basic amino acids had been introduced in the putative S1 binding pocket (Table 1).
Table 1.
Amino acid changes and kinetic parameters for WT OmpT and the sulfotyrosine variant sT4, measured at room temperature (25°C).
| Enzyme | Mutations | Substrate, cleavage site | kcat/KM [M−1s−1] |
|---|---|---|---|
| WT OmpT | – | 5b, Arg↓Arg | 1.7 ± 0.4 × 105 |
| sT4 | E27F, V29A, M87R, Y126C, E153D, I170V, D208R, Y221A, I282H | 1b, sTyr↓Arg | 1.1 ± 0.2 × 105 |
| 2b, pTyr↓Arg | 5 ± 3 × 102 | ||
| 3b, Tyr↓Arg | Non-MMK[a] | ||
| 4b, Glu↓Arg | 3 ± 1 × 102 |
The enzyme exhibited non-Michaelis–Menten kinetics with the Tyr↓Arg substrate 3b. However, the selectivity based on competition experiments was found to be tenfold for substrate 1b over 3b at 90 μM.
The sT4 variant was expressed and purified (to greater than 90% purity) with approximately the same yield as the WT protein (Figure S2 in the Supporting Information) using extraction with n-octylglucoside,[15] and the kinetics of hydrolysis of unlabeled peptide substrates 1 b–6 b were determined (Figure 1c). In accordance with its fluorescence profile, sT4 demonstrated efficient hydrolysis of the sulfotyrosine peptide 1b, with kcat/KM = 1.1 ± 0.2 × 105M−1s−1 (Figure S3 in the Supporting Information). The engineered enzyme did not exhibit Michaelis–Menten behavior with the unmodified tyrosine peptide, and therefore a direct comparison of catalytic parameters for the respective substrates is not possible. For a qualitative estimate of substrate discrimination, a competition experiment was performed by incubating 2.5 nM sT4 with equimolar mixtures of the sTyr↓Arg and Tyr↓Arg substrates (1b and 3b) at five different substrate concentrations (50, 60, 70, 80, 90 μM each substrate; Figure S4 in the Supporting Information). The results indicated that sT4 exhibits about tenfold selectivity for the cleavage of the sulfated peptide.
Consistent with its fluorescence profile, sT4 displayed only modest hydrolysis of the 2b pTyr↓Arg substrate, with kcat/KM = 5 ± 3 × 102M−1s−1, indicating a remarkable 200-fold selectivity in favor of sulfotyrosine over phosphotyrosine. Also, the enzyme showed no cleavage between pSer↓Arg even after overnight incubation (14 h) of the substrate 6b with a high concentration of the enzyme variant (0.5 μM). Finally, kcat/KM of sT4 for the hydrolysis of Glu↓Arg substrate (4b) was measured to be 3 ± 1 × 102M−1s−1, thus confirming only minor cross-reactivity with acidic residues. It is noteworthy that the selectivity of sT4 was not engineered at the expense of catalytic efficiency, since its kinetic parameters with its preferred sulfotyrosine substrate, 1b, are almost the same as WT OmpT with its preferred dibasic substrate 5b (Table 1). Although the reasons for sT4 discriminating between sulfotyrosine and phosphotyrosine are not known, a plausible explanation is that overall charge is important in substrate discrimination, as sulfotyrosine possesses an overall charge of −1 at neutral pH, while the overall charge on phosphotyrosine is −2. We are currently trying to crystallize the purified sT4 variant in complex with substrate analogue to identify the atomic interactions responsible for its selectivity, but that is beyond the scope of the current work.
The ability to remodel OmpT activity to recognize selectively sulfotyrosine in P1 raises an intriguing issue regarding natural protease specificity. Could it be that certain natural proteases recognize post-translationally modified amino acids in biologically significant ways? There are at least two examples of proteases cleaving at post-translationally modified amino acids: Subtilisin BPN′ possesses appreciable activity towards phosphotyrosine-containing substrates,[11] and aminopeptidase Ey from chicken egg yolks can processively digest sulfotyrosine containing chemotactic peptides.[16]
The successful engineering of a protease that selectively cleaves at sulfotyrosine residues in peptides marks the critical first step towards creating a practical enzyme useful for the detection of this interesting post-translational modification. Additional mutagenesis and screening of both targeted and random-error-prone mutant libraries of sT4 will be employed to achieve two important goals: 1) relax specificity for P1′-P3′ to accommodate all residues, especially acidic amino acids often found adjacent to sulfotyrosine and 2) increase the preference for sTyr/Tyr from tenfold to at least a 100-fold. These goals appear to be readily tractable given that OmpT exhibits relaxed selectivity for P1′ and P2′ and mutants[17] and our success in selectively altering the amino acid preference at P1′.[12]
Experimental Section
Flow cytometric OmpT activity assays: Single colonies were used to inoculate 2 mL 2xYT cultures supplemented with ampicillin (200 μgmL−1). The cells were harvested and resuspended in 1% sucrose as previously described.[12] For labeling, the cells (50 μL) were diluted into sucrose (949 μL) and the substrate (1 μL; final concentration 20 nM for 1a and 100 nM for 5a). An aliquot of the labeling reaction (20 μL) was diluted into 1% sucrose (0.5 mL) and analyzed on the MoFlo flow cytometer (Dako, Fort Collins, CO).
Detailed protocols for substrate conjugation, library screening, enzyme purification, and kinetics are described in the Supporting Information.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200800736.
We thank J. Cantor, M. Pogson, and M. Rani for reading the manuscript, M. Pogson for assistance in preparing the graphical abstract. This work was supported by US National Institutes of Health grants R01 GM065551 and RO1 GM073089.
References
- 1.Seibert C, Sakmar TP. Biopolymers. 2008;90:459. doi: 10.1002/bip.20821. [DOI] [PubMed] [Google Scholar]
- 2.Liu CC, Brustad E, Liu W, Schultz PG. J Am Chem Soc. 2007;129:10648. doi: 10.1021/ja0735002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh G, Jefferis R. Nat Biotechnol. 2006;24:1241. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 4.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. Science. 2007;317:1930. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishiro E, Sakakibara Y, Liu MC, Suiko M. J Biochem. 2006;140:731. doi: 10.1093/jb/mvj206. [DOI] [PubMed] [Google Scholar]
- 6.Liu MC, Lipmann F. Proc Natl Acad Sci USA. 1984;81:3695. doi: 10.1073/pnas.81.12.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salek M, Costagliola S, Lehmann WD. Anal Chem. 2004;76:5136. doi: 10.1021/ac0400414. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, Hoffhines AJ, Moore KL, Leary JA. Nat Methods. 2007;4:583. doi: 10.1038/nmeth1056. [DOI] [PubMed] [Google Scholar]
- 9.Taglieber A, Hobenreich H, Carballeira JD, Mondiere RJ, Reetz MT. Angew Chem. 2007;119:8751. [Google Scholar]; Angew Chem Int Ed. 2007;46:8597. [Google Scholar]
- 10.Toscano MD, Woycechowsky KJ, Hilvert D. Angew Chem. 2007;119:3274. [Google Scholar]; Angew Chem Int Ed. 2007;46:3212. [Google Scholar]
- 11.Knight ZA, Garrison JL, Chan K, King DS, Shokat KM. J Am Chem Soc. 2007;129:11672. doi: 10.1021/ja073875n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varadarajan N, Rodriguez S, Hwang BY, Georgiou G, Iverson BL. Nat Chem Biol. 2008;4:290. doi: 10.1038/nchembio.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmond MR, Gros P. EMBO J. 2001;20:5033. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stemmer WP. Proc Natl Acad Sci USA. 1994;91:10747. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varadarajan N, Gam J, Olsen MJ, Georgiou G, Iverson BL. Proc Natl Acad Sci USA. 2005;102:6855. doi: 10.1073/pnas.0500063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T, Ichishima E. Comp Biochem Physiol Part B. 1994;107:533. doi: 10.1016/0305-0491(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 17.McCarter JD, Stephens D, Shoemaker K, Rosenberg S, Kirsch JF, Georgiou G. J Bacteriol. 2004;186:5919. doi: 10.1128/JB.186.17.5919-5925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]