Abstract
Although the biological functions of tumor-originated cell-free DNA have not been previously clarified, such molecular characteristics as mutations, hypermethylation, and microsatellite instability have confirmed its tumor origin. Therefore, to investigate the use of plasma DNA level as a biomarker of lung cancer, we compared plasma DNA concentrations in 102 patients with lung cancer and 105 healthy individuals using quantitative PCR analyses. The median plasma DNA concentrations for the healthy and cancer groups were 10.4 and 22.6 ng/ml, respectively (P < 0.0001), and elevated plasma DNA levels were also detected in patients with either stage I or II disease. Neither smoking status nor the number of packs per year had an effect on the level of circulating cell-free DNA. Increased concentrations of circulating cell-free DNA showed the potential power to discriminate lung cancer (area under the receiver operating characteristic curve = 0.86, 95% CI = 0.81 to 0.91). When subjects were classified into three groups based on their plasma DNA concentrations, subjects in the upper tertile (ie, those with the highest concentration) had a significantly increased risk of lung cancer as compared with those in the lowest tertile (adjusted odds ratio = 50.6, P < 0.001). These results suggest that elevated circulating plasma DNA levels may serve as a potential diagnostic indicator and be an important risk factor for lung cancer.
Lung cancer is the leading cause of cancer-related death in Korea and across the globe. Despite the development of several advanced technologies with enhanced sensitivity for detecting cancer, current screening methods for the diagnosis of lung cancer at an early stage are insufficient. Recently, circulating cell-free DNA was detected in blood fractions from patients with cancer. Since then, efforts have been made to use circulating cell-free DNA as a diagnostic or prognostic factor for lung cancer.1,2,3,4 Although the biological mechanisms leading to its presence in the blood have not been elucidated, several genetic or epigenetic studies have shown that circulating DNA has the features of tumor DNA.5,6 The same somatic mutations in oncogenes or tumor suppressor genes such as K-ras or p53 that are present in primary tumors were also detected in the circulating cell-free DNA samples of patients with cancer.7,8,9 Additionally, the cancer-specific hypermethylation of the p16INK4A, CDH1, and DAPK1 promoter regions in plasma DNA isolated from patients with cancer suggested that circulating methylated DNA could be used as a diagnostic and prognostic marker for the disease.10,11,12
While the quantification of plasma DNA has been proposed as a diagnostic tool for cancer, current results suggest that additional controlled studies are required to establish this possibility.13,14,15 Although some studies reported increased levels of DNA in patients with lung cancer, the experimental methods used in those studies and environmental factors such as smoking status should also be carefully considered. Furthermore, the development of reproducible, standardized methods for the detection and quantification of cell-free DNA is important for improving the sensitivity, specificity, and relevance of this potential biomarker.
If the quantitative analysis of circulating cell-free DNA can be used to distinguish those with lung cancer, the simplicity and non-invasiveness of sample collection would make it a very appealing method of cancer diagnosis. In this study, we investigated the use of plasma DNA as a potential diagnostic factor for identifying patients with lung cancer by quantifying its presence using quantitative PCR (qPCR) in 102 patients with lung cancer and 105 healthy individuals.
Materials and Methods
Study Population
In this case-control study, blood plasma samples were collected from 102 patients with histologically confirmed lung cancer (LC group) and 105 healthy control individuals (NC group) from May 2002 to July 2003. None of the patients had received chemotherapy or radiotherapy before their recruitment. For comparison with the patients with cancer, healthy individuals with no prior history of cancer were recruited from the visitors of the cancer-screening program at our institution. Information on smoking habits was collected by means of self-reporting; the staging of lung cancer was confirmed by computed tomography at the time of sample collection. Blood samples were collected from the LC and NC groups for genetic testing after obtaining written informed consent. This study was approved by the institutional review board. Those patients with lung cancer were appropriately matched with the control subjects with respect to age and sex.
Quantification of Circulating Cell-Free DNA
Blood samples were collected in EDTA-coated tubes and centrifuged immediately at 1000 × g for 15 minutes. Plasma was carefully separated and stored at −80°C. A total 50 μl of DNA was extracted from 200 μl of plasma using QIAamp DNA Blood Mini Kits (Qiagen, Valencia, CA) according to the manufacturer's instructions. To measure the plasma DNA concentration, the genomic sequence of β-actin was amplified by quantitative real-time PCR. The primers and fluorescent probe used for qPCR were as described in Herrera et al.14 Two microliters of purified plasma DNA was amplified using 0.3 μM/L of each primer (5′-CCACACTGTGCCCATCTACG-3′ and 5′-AGGATCTTCATGAGGTAGTCAGTCAG-3′) and a 0.25 μM/L fluorescent probe (5′FAM-ATGCCCTCCCCCATGCCATCCTGCGT-TAMRA-3′). PCR was performed using the following program: 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Quantitative standard curves were prepared using serial diluted control genomic human DNA (Applied Biosystems, Foster City, CA). The fluorescence of the amplified PCR products was detected using an ABI 7900HT Sequence Detection System (Applied Biosystems). The results of qPCR assay represent the mean of three independent experiments, each consisting of duplicate samples.
Statistical Analysis
To test for demographic differences between the patients with lung cancer and controls, we used Pearson's χ2-test for categorical variables and the Wilcoxon rank-sum test for continuous variables. The association between the plasma DNA concentration and lung cancer risk was analyzed with unconditional multiple logistic regression models using smoking status as a covariate. The receiver operating characteristic curve and area under the curve were estimated to test the discrimination power of the plasma DNA concentration. A P value of 5% (α = 0.05) was applied for all statistic analyses; all P values were two-sided. STATA version 9.1 was used for all statistical analyses (StataCorp LP, College Station, TX).
Results
The demographics of the 102 patients with lung cancer and 105 healthy controls enrolled in this study are presented in Table 1. Adenocarcinoma (65.7%) was the predominant histological type of cancer among the patients with lung cancer. At the time of recruitment, the majority of patients had stage IV disease (62.7%). No significant difference existed between the cases and controls in terms of age and sex, but the control group had a higher prevalence of smoking history than the patient group (P = 0.06).
Table 1.
Demographic Features of the Patients with Lung Cancer and Healthy Controls
| Lung cancer (n = 102) | Healthy controls (n = 105) | P value | |
|---|---|---|---|
| Age, median (range) | 49 (28–70) | 46 (27–69) | 0.27 |
| Sex (n, %) | 0.64 | ||
| Male | 54 (52.9%) | 59 (56.2%) | |
| Female | 48 (47.1%) | 46 (43.8%) | |
| Smoking status (n, %) | 0.06 | ||
| Never smoker | 49 (48.0%) | 37 (35.2%) | |
| Ever smoker | 53 (52.0%) | 68 (64.8%) | |
| Pack years, median (range) | 25.5 (0.08–102.5) | 25 (0.1–102.5) | 0.7 |
| Histological type | |||
| Adenocarcinoma | 67 (65.7%) | ||
| Squamous cell caricnoma | 16 (15.7%) | ||
| Small cell lung cancer | 10 (9.8%) | ||
| Others | 9 (8.8%) | ||
| Stage | |||
| NSCLC | |||
| Stage I | 8 (7.8%) | ||
| Stage II | 2 (2.0%) | ||
| Stage III | 19 (18.6%) | ||
| Stage IV | 64 (62.7%) | ||
| SCLC | |||
| LD | 5 (4.9%) | ||
| ED | 4 (3.9%) |
The median plasma DNA concentration of healthy controls was 10.4 ng/ml (range, 1.6 to 89.8 ng/ml), which was substantially lower than that of the lung cancer patients. The plasma DNA concentrations among the 102 patients with lung cancer varied from 3.1 to 730.5 ng/ml, with a median concentration of 22.6 ng/ml. The median DNA concentration difference between the two groups was statistically significant (P < 0.0001; Figure 1A). The elevated plasma DNA level was already apparent at early disease stages (Figure 1B). Although the number of patients with early-stage disease was limited, the plasma DNA level was significantly increased in patients with non-small cell lung cancer with stage I/II disease compared with the healthy controls (P = 0.0003). The median plasma DNA concentration for the ever- and never-smokers among lung cancer patients was 23.1 and 21.2 ng/ml, respectively (P = 0.62). Among the healthy controls, the median plasma DNA concentration of ever-smokers (10.5 ng/ml) was also not notably different from that of never-smokers (9.1 ng/ml), demonstrating that smoking status was not significantly correlated with the plasma DNA concentration (Figure 1C).
Figure 1.
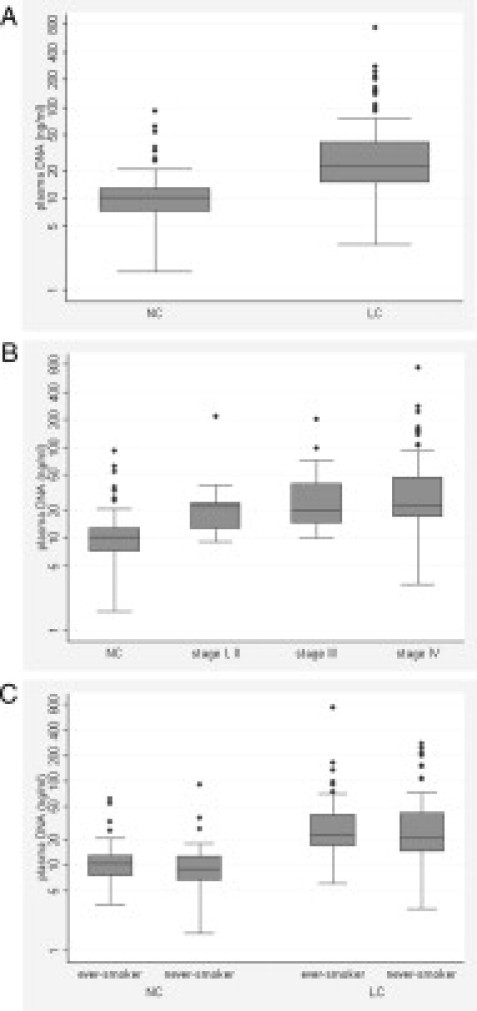
A: Distribution of the plasma DNA concentrations in patients with lung cancer (LC) and healthy controls (NC) as shown by box plots. The y axis has a log scale. B: The plasma DNA concentration was compared between healthy controls (NC) and patients with non-small cell lung cancer (NSCLC; n = 93) classified by disease stage. Tumor stages are indicated with roman numerals. C: The plasma DNA concentration was compared according to smoking status.
The discrimination power of plasma DNA concentration was tested by estimating the receiver operating characteristic and area under the curve. Increased levels of plasma DNA showed the power to discriminate patients with lung cancer from healthy controls (receiver operating characteristic area under the curve = 0.86, 95% CI = 0.81 to 0.91; Figure 2). To analyze the association between the plasma DNA concentration and lung cancer risk, the subjects were classified into three groups according to their plasma DNA concentrations. As shown in Table 2, those individuals with the highest DNA concentrations exhibited a significantly increased lung cancer risk compared with those individuals with the lowest DNA concentrations (adjusted odds ratio = 50.6, P < 0.001).
Figure 2.
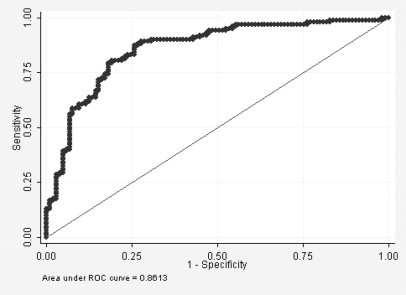
The efficacy of using plasma DNA levels to distinguish lung cancer patients from healthy controls was tested using a receiver operating characteristic curve.
Table 2.
Association of the Circulating DNA Level with Lung Cancer Risk
| Group | CFDNA concentration (ng/ml) | Lung Cancer N (%) | Healthy Controls N (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| 1 | ≤11 | 9 (8.8) | 60 (57.1) | 1 | |
| 2 | >11, ≤20 | 33 (32.4) | 35 (33.3) | 6.8 (2.8–16.5) | <0.001 |
| 3 | >20 | 60 (58.8) | 10 (9.5) | 50.6 (18.1–141.9) | <0.001 |
Discussion
The utility of blood samples for cancer diagnosis or surveillance is extremely appealing due to the simplicity and noninvasiveness of sample collecting. Previous studies have demonstrated the use of DNA extracted from blood plasma or serum in analyzing mutations and methylation in multiple genes. Several recent studies have reported increased levels of cell-free DNA among patients with various types of cancer such as prostate, breast, and colorectal, and they have suggested that DNA levels could have prognostic value.16,17,18 However, since the quantification of cell-free DNA as a diagnostic factor for lung cancer was suggested by Sozzi et al, few case-control studies have been conducted to demonstrate the feasibility of this method for Asian populations.13
In this study, we focused on the presence of cell-free DNA in blood plasma using blood samples collected from patients with lung cancer and healthy controls. Despite the use of frozen plasma samples for DNA extraction, the samples were stored under the same conditions and for the same time period; thus, no variation existed among the samples in terms of storage. To test the effect of specimen storage on plasma DNA, we analyzed the association between DNA concentration and storage time and found no correlation. Previous studies on the quantification of circulating DNA suggested that a standardized experimental method for quantitative analysis is important to increase the sensitivity and specificity of cancer detection. The analysis of β-actin gene by real-time PCR was used for the quantification of plasma DNA due to its high PCR efficiency and reproducibility as already reported by Herrera et al.14
As our results revealed significantly increased levels of plasma DNA among patients with lung cancer, we also examined the DNA level with respect to histological type or tumor stage. The plasma DNA concentration did not differ among histological groups, including adenocarcinoma, squamous cell carcinoma, and small cell lung cancer (data not shown). In addition, we found considerably elevated plasma DNA levels in the patients with early-stage lung cancer as compared with the healthy controls. Although the median plasma DNA level was highest in those patients with stage IV disease, our results did not show a critical association between an increased DNA concentration and advanced tumor stages. These results support the quantification of plasma DNA as a diagnostic approach for lung cancer, even at early disease stages. The efficiency of the quantification of plasma DNA for the early detection of cancer should be examined further using a larger number of patients with stage I or II disease.
In conclusion, the quantitative analysis of plasma DNA may be useful in distinguishing patients with lung cancer from healthy individuals. Furthermore, the availability of noninvasive materials and the ease of sample collection make the use of plasma DNA as an attractive diagnostic tool. This case-control study with 102 patients and 105 healthy controls demonstrates the application of measuring plasma DNA level as a diagnostic factor for lung cancer. Additional studies are being conducted to characterize the tumor-associated features of increased circulating DNA in patients with lung cancer.
Footnotes
Supported by National Cancer Center Research Grant (0710222).
References
- 1.Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev. 2002;28:255–271. doi: 10.1016/s0305-7372(02)00077-4. [DOI] [PubMed] [Google Scholar]
- 2.Bremnes RM, Sirera R, Camps C. Circulating tumour-derived DNA and RNA markers in blood: a tool for early detection, diagnostics, and follow-up? Lung Cancer. 2005;49:1–12. doi: 10.1016/j.lungcan.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Xue X, Zhu YM, Woll PJ. Circulating DNA and lung cancer. Ann NY Acad Sci. 2006;1075:154–164. doi: 10.1196/annals.1368.021. [DOI] [PubMed] [Google Scholar]
- 4.Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, Rossier A, Chen XQ, Anker P. The origin and mechanism of circulating DNA. Ann NY Acad Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x. [DOI] [PubMed] [Google Scholar]
- 6.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez R, Silva JM, Sanchez A, Dominguez G, Garcia JM, Chen XQ, Stroun M, Provencio M, Espana P, Anker P, Bonilla F. Microsatellite alterations and TP53 mutations in plasma DNA of small-cell lung cancer patients: follow-up study and prognostic significance. Ann Oncol. 2000;11:1097–1104. doi: 10.1023/a:1008305412635. [DOI] [PubMed] [Google Scholar]
- 8.Kopreski MS, Benko FA, Borys DJ, Khan A, McGarrity TJ, Gocke CD. Somatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNA. J Natl Cancer Inst. 2000;92:918–923. doi: 10.1093/jnci/92.11.918. [DOI] [PubMed] [Google Scholar]
- 9.Shao ZM, Wu J, Shen ZZ, Nguyen M. p53 mutation in plasma DNA and its prognostic value in breast cancer patients. Clin Cancer Res. 2001;7:2222–2227. [PubMed] [Google Scholar]
- 10.An Q, Liu Y, Gao Y, Huang J, Fong X, Li L, Zhang D, Cheng S. Detection of p16 hypermethylation in circulating plasma DNA of non-small cell lung cancer patients. Cancer Lett. 2002;188:109–114. doi: 10.1016/s0304-3835(02)00496-2. [DOI] [PubMed] [Google Scholar]
- 11.Wong TS, Kwong DL, Sham JS, Wei WI, Kwong YL, Yuen AP. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin Cancer Res. 2004;10:2401–2406. doi: 10.1158/1078-0432.ccr-03-0139. [DOI] [PubMed] [Google Scholar]
- 12.Bearzatto A, Conte D, Frattini M, Zaffaroni N, Andriani F, Balestra D, Tavecchio L, Daidone MG, Sozzi G. p16(INK4A) Hypermethylation detected by fluorescent methylation-specific PCR in plasmas from non-small cell lung cancer. Clin Cancer Res. 2002;8:3782–3787. [PubMed] [Google Scholar]
- 13.Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Herrera LJ, Raja S, Gooding WE, El-Hefnawy T, Kelly L, Luketich JD, Godfrey TE. Quantitative analysis of circulating plasma DNA as a tumor marker in thoracic malignancies. Clin Chem. 2005;51:113–118. doi: 10.1373/clinchem.2004.039263. [DOI] [PubMed] [Google Scholar]
- 15.Camps C, Sirera R, Bremnes RM, Rodenas V, Blasco A, Safont MJ, Garde J, Juarez A, Caballero C, Sanchez JJ, Taron M, Rosell R. Quantification in the serum of the catalytic fraction of reverse telomerase: a useful prognostic factor in advanced non-small cell lung cancer. Anticancer Res. 2006;26:4905–4909. [PubMed] [Google Scholar]
- 16.Papadopoulou E, Davilas E, Sotiriou V, Georgakopoulos E, Georgakopoulou S, Koliopanos A, Aggelakis F, Dardoufas K, Agnanti NJ, Karydas I, Nasioulas G. Cell-free DNA and RNA in plasma as a new molecular marker for prostate and breast cancer. Ann NY Acad Sci. 2006;1075:235–243. doi: 10.1196/annals.1368.032. [DOI] [PubMed] [Google Scholar]
- 17.Frattini M, Gallino G, Signoroni S, Balestra D, Battaglia L, Sozzi G, Leo E, Pilotti S, Pierotti MA. Quantitative analysis of plasma DNA in colorectal cancer patients: a novel prognostic tool. Ann NY Acad Sci. 2006;1075:185–190. doi: 10.1196/annals.1368.025. [DOI] [PubMed] [Google Scholar]
- 18.Kamat AA, Sood AK, Dang D, Gershenson DM, Simpson JL, Bischoff FZ. Quantification of total plasma cell-free DNA in ovarian cancer using real-time PCR. Ann NY Acad Sci. 2006;1075:230–234. doi: 10.1196/annals.1368.031. [DOI] [PubMed] [Google Scholar]


